ABSTRACT
Using atomic force microscopy imaging and nanoindentation measurements, we investigated the effect of the minor capsid proteins pUL17 and pUL25 on the structural stability of icosahedral herpes simplex virus capsids. pUL17 and pUL25, which form the capsid vertex-specific component (CVSC), particularly contributed to capsid resilience along the 5-fold and 2-fold but not along the 3-fold icosahedral axes. Our detailed analyses, including quantitative mass spectrometry of the protein composition of the capsids, revealed that both pUL17 and pUL25 are required to stabilize the capsid shells at the vertices. This indicates that herpesviruses withstand the internal pressure that is generated during DNA genome packaging by locally reinforcing the mechanical sturdiness of the vertices, the most stressed part of the capsids.
IMPORTANCE In this study, the structural, material properties of herpes simplex virus 1 were investigated. The capsid of herpes simplex virus is built up of a variety of proteins, and we scrutinized the influence of two of these proteins on the stability of the capsid. For this, we used a scanning force microscope that makes detailed, topographic images of the particles and that is able to perform mechanical deformation measurements. Using this approach, we revealed that both studied proteins play an essential role in viral stability. These new insights support us in forming a complete view on viral structure and furthermore could possibly help not only to develop specific antivirals but also to build protein shells with improved stability for drug delivery purposes.
KEYWORDS: AFM, capsid, herpes simplex virus, stability
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is an important human pathogen that causes a variety of diseases ranging from common cold sores to life-threatening encephalitis (1–3). Herpesvirus particles are enveloped virions with T=16 icosahedral capsids harboring double-stranded DNA (dsDNA) genomes. After the synthesis and nuclear import of the capsid proteins, they initially assemble into rather spherical immature procapsids (4, 5). Upon proteolytic cleavage of the internal scaffold, consisting mostly of the protein VP22a, these procapsids mature into three icosahedral capsid types (6–9). B-type capsids have failed to expel the protein scaffold; A-type capsids are considered to have aborted DNA packaging and lack both DNA and the internal scaffold; and C-type capsids, also called nucleocapsids, result from the successful replacement of the internal protein scaffold with the 152-kb dsDNA genome of HSV-1. C capsids then leave the nucleus and undergo secondary envelopment in the cytoplasm to generate mature, infectious, enveloped virions (10, 11). Recent nanoindentation experiments using atomic force microscopy (AFM) have revealed remarkable insights into the mechanical basis of HSV-1 genome packaging and capsid maturation (12–14).
In AFM-nanoindentation experiments, viral capsids are deposited on a glass surface, imaged by AFM, and subsequently indented to probe the mechanical resilience of the particle (15, 16). Such AFM studies have revealed how the structural stability of capsids depends on environmental conditions, the length of the packaged genome, and the protein composition of the particle (17–23). Moreover, it has been shown that the mechanical resilience of viral capsids is directly related to (i) local conformational dynamics (minute virus of mice) (24), (ii) the virus's infectivity (HIV-1) (25), and (iii) the particle's propensity for efficient uncoating (adenovirus) (26, 27).
In the case of HSV-1 capsids, we have shown that scaffold expulsion and genome packaging result in molecular changes that strengthen the particles (12). This is reflected by an increase in the threshold for the breaking force, Fbreak, required for structural collapse. By treating HSV-1 capsids with a moderate, partially denaturing concentration of guanidine hydrochloride (GuHCl), the penton fraction of the major capsid protein VP5, the small capsid protein VP26 located on the tips of the VP5 hexons, the scaffold protein VP22a, the minor capsid proteins pUL17 and pUL25, as well as the DNA genomes are extracted (12, 28, 29). Using such pentonless B, A, and C capsids, we showed that their stiffness is reduced, indicating that the vertex proteins of HSV-1 capsids are especially important for the mechanical resilience of the capsids (12, 13). In addition, it was recently reported that the protein pUL25 reinforces the capsid (30). The two minor capsid proteins pUL25 and pUL17 form heterodimers that are attached to the capsid vertices (Fig. 1a) and hence have been called capsid vertex-specific components (CVSC) (31–44).
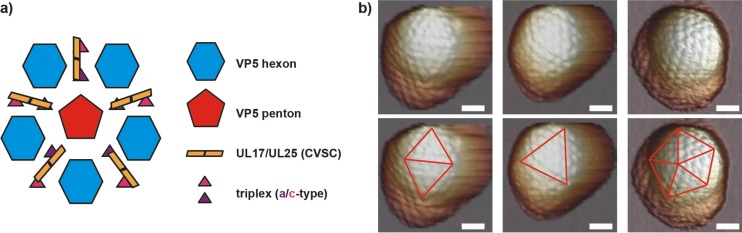
FIG 1.
Atomic force microscopy imaging of HSV-1 capsids. (a) Schematic of the HSV-1 capsid vertex region (41, 44). The UL25 part of the CVSC is proposed to be closest to the vertex, likely touching it (45). (b) AFM images of HSV-1 capsids. Based on the facet orientation and capsomer morphology, particles deposited on the 2-, 3-, and 5-fold icosahedral symmetry axes can be distinguished. Bar, 50 nm.
Next to HSV-1, similar CVSC complexes are present on purified capsids of the swine alphaherpesvirus pseudorabies virus with even higher occupancy levels (45–47). Furthermore, homologs of these minor capsid components exist in other alphaherpesviruses, the betaherpesviruses (e.g., pUL77 and pUL93 in human cytomegalovirus) (48), and the gammaherpesviruses (e.g., open reading frame 32 [ORF32] and ORF19 in Kaposi sarcoma-associated virus) (44), suggesting that functional, stabilizing CVSCs are a feature of all herpesviruses (49).
In HSV-1, the CVSCs also mediate interactions with the inner tegument protein pUL36 and the outer tegument protein VP13/14 that link the capsids to envelope components during assembly (50–52). Previous studies have shown that pUL17 and pUL25 depend on each other for optimal capsid binding, since capsids derived from either UL17 or UL25 deletion mutants lack most of the CVSC altogether (53). Furthermore, a recent study using cryo-electron microscopy reconstructions clearly shows that the CVSCs directly link the pentons to the adjacent triplexes (45). In the present study, we used AFM to determine at the single-particle level how the CVSC contributes to the mechanical properties of HSV-1 capsids.
RESULTS
From AFM images taken immediately prior to the nanoindentation experiments, we determined the orientation of each capsid based on its capsomer morphology and the orientation of the triangular facets on the capsid surface. Figure 1b shows a projection of the facets on the AFM images. UL17- and UL25-null capsids that adhered to the surface in different orientations were compared to similarly oriented B-, A-, and C-type capsids of the wild-type (WT) strain. There was a marked decrease in the spring constants, k, of the capsids of both deletion strains (Fig. 2a).
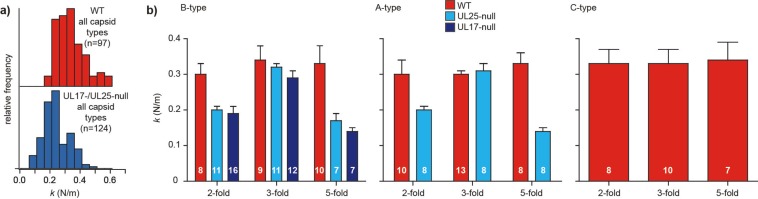
FIG 2.
Both the CVSCs pUL17 and pUL25 contribute to the mechanical vertex stabilization of HSV-1 capsids. (a) Frequency distributions of particle spring constants (k) for particles with or without the CVSC, showing the shift to lower k values for the latter particles. (b) The average spring constants (k) for each orientation are shown for all three capsid types, comparing capsids in UL17- or UL25-null backgrounds to WT capsids. Error bars represent standard errors of the means, and the numbers of particles per type/orientation are indicated in white on each bar.
We then stratified these data into B, A, and C capsids, and based on our AFM images, we further stratified them into measurements along the 2-fold, the 3-fold, or the 5-fold axes. Deposition onto the 2-, 3-, and 5-fold axes occurred at a ratio of 61:63:47 (Fig. 2b). In an icosahedral particle, there are 30 2-fold axes, 20 3-fold axes, and 12 5-fold axes. This ratio of deposition was determined previously by using hepatitis B virus (HBV) capsids (54). In the present study, there were roughly the same numbers of particles deposited on the 2- and 3-fold axes. Thus, compared to the T=3 and T=4 HBV capsids with a diameter of ∼30 nm, the larger T=16 HSV-1 capsids of 125 nm likely elicit additional surface interaction effects that slightly favor a stable deposition on a 3-fold axis over a 2-fold axis. Analysis of spring constants revealed that the reduction in stiffness was particularly prominent for certain icosahedral orientations (Fig. 2b). Capsids that had been deposited on a triangular facet of the icosahedral shell, and, thus, probed along the 3-fold icosahedral symmetry axis of the capsid, exhibited no significant loss of stiffness for the UL25-null or UL17-null mutant compared to WT capsids. However, there was a significant decrease in the stiffness of UL25- or UL17-null capsids compared to WT capsids when the particle had been deposited on the edge between two facets (i.e., 2-fold icosahedral symmetry axis) or on a vertex (5-fold icosahedral symmetry axis).
We then determined the protein compositions of the different capsid types of the wild type and the two deletion mutants by quantitative mass spectrometry (MS) using a label-free approach in which the number of peptide spectrum matches (PSMs) serves as a proxy for the relative protein amounts (see Table S1 in the supplemental material). We used the major capsid protein VP5 (pUL19) that forms the pentons and hexons in each capsid for normalization since it is considered to be present in constant amounts among different capsid types (55) (Fig. 3). Based on this normalization, we then determined the amounts of the other capsid proteins in the different samples. As expected, the abundances of two triplex proteins, VP19c (pUL38) and VP23 (pUL18), were also similar in the different samples, indicating that the different preparations from wild-type HSV-1 and the mutants indeed contained capsids with identical backbone architectures. In contrast, neither of the CVSC proteins pUL17 and pUL25 could be detected in either of the deletion mutants. This indicates that none of the CVSCs were recruited or maintained on the capsids if one of them had been missing. Data from this analysis of the protein compositions of all capsid types fit our measurements of capsid stability, since both deletion mutants displayed identical mechanics of their HSV capsids.
FIG 3.
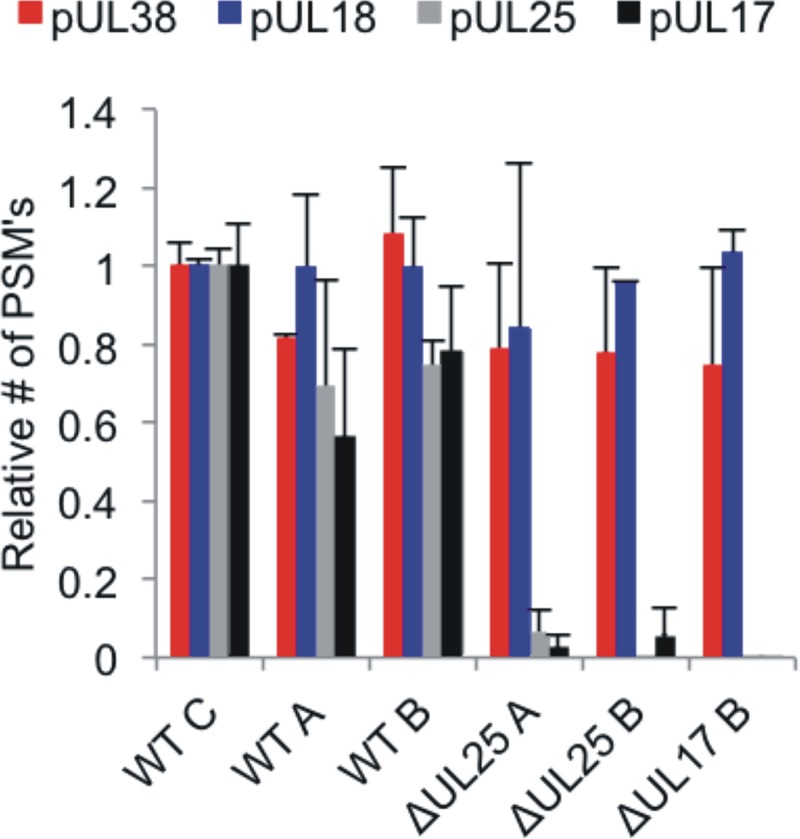
Protein copy numbers on capsids. Shown are quantitative mass spectrometry results for the abundance of pUL38, pUL18, pUL25, and pUL17 on the different capsids. The relative numbers of PSMs (67) are indicated on the y axis.
DISCUSSION
Our results on the B and A capsids of the UL25-null mutant corroborate and extend recent findings by Sae-Ueng et al. (30), who also reported a reduced stability of HSV-1 capsids upon the deletion of pUL25. However, those researchers did not detect any changes in the mechanical resilience of the B capsids upon the deletion of UL17. In contrast, we measured a significant decrease in stiffness for the B capsids of the UL17-null mutant (Fig. 2b, dark blue columns). Furthermore, we have been able, for the first time for herpesviruses, to separately analyze the spring constants along the different icosahedral axes. As our data show that the spring constants, k, along the 3-fold axis remain largely unaffected by the deletion of either UL17 or UL25, it is possible that Sae-Ueng et al. (30) predominantly measured the spring constants of the UL17-null mutant upon probing the triangular sides but not capsids with their 2-fold or 5-fold axes oriented toward the AFM tip. Moreover, using quantitative mass spectrometry analysis, we have corroborated previous findings that the capsid levels of pUL17 and pUL25 largely depend on each other for stable capsid association (42, 53). In contrast, the immunoblot analyses reported by Sae-Ueng et al. (30) and Huet et al. (45) revealed residual amounts of pUL17 on the capsids of the UL25-null mutant. The reasons for this difference are unclear; it may be due to the presence of dithiothreitol (DTT) in our purification buffers to generate a reducing environment similar to that in the nucleoplasm or the cytoplasm.
Our new data and those reported previously Sae-Ueng et al. (30) support the notion that the CVSCs provide substantial mechanical resilience to HSV-1 capsids, and here we also show that both pUL17 and pUL25 are required to increase vertex resilience. Our finding that the deletion of either pUL17 or UL25 results in a reduced strength of capsids corroborates data from a recent report of the structure of the CVSC that clearly shows how both proteins are intimately linked to each other in the CVSC (45). As the CVSC is located at the 5-fold vertices and oriented along the 2-fold symmetry axis, it is very likely to impact capsid resilience along these symmetry axes, which is exactly what we find. The 3-fold axis, on the other hand, does not appear to be affected by the presence or absence of the CVSC (45). This also correlates with our findings, explaining the differences in the observed impact of the removal of the CVSC on the different icosahedral orientations. The vertices are first removed from the capsid when the particles are stressed, e.g., nanoindentation or partial denaturation with urea or GuHCl (12, 28). Moreover, in the absence of the capsid-stabilizing CVSCs, e.g., in mutants lacking UL25, the capsids cannot maintain the viral genomes in their lumena, presumably because the capsids are not stably sealed (32). Actually, herpesviruses depend on the DNA terminase complex consisting of pUL15, pUL28, and pUL33 and ATP hydrolysis to package their genomes into capsids and to work against the repulsive force of the highly confined, negatively charged DNA (56–58). Thus, one major function of the CVSCs could be to reinforce the vertices of the nucleocapsids to ensure the retention of the genome inside the particle. Recent experimental and theoretical studies of virus capsid nanoindentation have demonstrated that the mechanical response of a capsid is basically a local property of the capsid structure (24, 59). The local reinforcement of the capsid vertices by the CVSC is therefore an example of a virus specifically adapting to mechanical limitations imposed by packaging large genomes to near-liquid crystalline density.
MATERIALS AND METHODS
Capsid purification.
Nuclear capsids were isolated from cells infected with WT HSV-1 (strain F; ATCC VR-733), the HSV1-ΔUL17 mutants (derived from HSV-1 strain F [see reference 38]), or the HSV1-ΔUL25 mutants (HSV-1 strain KUL25NS, derived from strain KOS [see reference 32]) after cell homogenization and purification on a linear 20 to 50% (wt/wt) sucrose gradient in a solution containing 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 1 mM EDTA supplemented with 10 mM dithiothreitol, as described previously (12, 55, 60). While during WT infection B, A, and C capsids are assembled, B- and A-type capsids are formed in the absence of pUL25 (32), and only B-type capsids are formed in the absence of pUL17 (35, 38, 61).
AFM imaging and nanoindentation.
The capsids were deposited onto silanized glass substrates and analyzed at room temperature in a solution containing 50 mM Tris buffer (pH 7.5) and 150 mM sodium chloride by AFM imaging and nanoindentation, as described in detail previously (12, 62, 63). The experiments were performed with a Nanotec AFM instrument (Nanotec, Tres Cantos, Spain), using cantilevers with an approximate tip radius of 15 nm and a spring constant of 0.05 N/m (OMCL-RC800PSA; Olympus). Imaging was performed by jumping-mode AFM, which is a very gentle imaging mode where lateral forces are almost absent and which is therefore ideally suited for imaging proteinaceous assemblies such as viral capsids (64). The probe velocity during nanoindentation was 60 nm/s. The data were analyzed with WSxM software (version 4; Nanotec) and a home-written Labview program (63). Capsid absorption to the surface was expected to be random with respect to the icosahedral orientation; in addition to absorption to the 2-, 3-, or 5-fold symmetry axes, we also detected intermediate positions. As the intermediate positions were difficult to classify, we focused on particles that adhered to the 2-, 3-, or 5-fold symmetry axes.
Protein extraction, liquid chromatography-tandem mass spectrometry, and data analysis.
HSV-1 capsids were resuspended in a solution containing 50 mM ammonium bicarbonate and 5% (wt/vol) sodium deoxycholate and heated at 90°C for 5 min. For each reaction, 100 μg of protein was reduced by using DTT for 30 min at 56°C and then alkylated with iodoacetamide for 30 min in the dark. After dilution to a final concentration of 0.5% sodium deoxycholate, each sample was digested overnight at 37°C with trypsin at an enzyme-to-protein ratio of 1:50. Sodium deoxycholate was precipitated, and the reaction/digestion mixture was quenched by adding formic acid to a final concentration of 2% (vol/vol). The samples were centrifuged for 20 min at 20,000 × g, and the supernatants were analyzed on a mass spectrometer (Q-Exactive Plus instrument coupled to an Agilent 1290 Infinity Ultra-high-pressure liquid chromatography [UHPLC] system). Briefly, the peptides were loaded onto a trapping column (Reprosil C18 [3 μm, 2 cm by 100 μm]; Dr. Maisch) with a flow rate of 5 μl/min for 10 min with reversed-phase solvent A, whereas peptide separation was performed with a column flow rate of ∼300 nl/min (Poroshell 120 EC-C18 [2.7 μm, 50 cm by 75 μm]; Agilent). Nanospray was achieved with an in-house-pulled and gold-coated fused silica capillary (360-μm outer diameter, 20-μm inner diameter, and 10-μm tip inner diameter) and an applied voltage of 1.9 kV. Full-scan MS spectra (from m/z 350 to 1,500) were acquired with the Orbitrap instrument at a resolution of 35,000. High-energy collision dissociation (HCD) fragmentation was performed in a data-dependent mode, as previously described (65).
Peak lists were generated (Proteome Discoverer, version 1.4; Thermo Scientific, Bremen, Germany) and searched against a database containing the human herpesvirus 1 strain 17 sequences (77 protein entries) by using Mascot (version 2.4; Matrix Science, London, UK) and mass tolerances of 50 ppm for precursor masses and ±0.05 Da for fragment ions. Enzyme specificity was set to trypsin with 2 missed cleavages allowed. Carbamidomethylation of cysteines was set as a fixed modification, while oxidation of methionine was used as a variable modification. The false discovery rate was set to <1%. To further filter for high-quality data, we used the following parameters: high-confidence peptide spectrum matches, a minimal Mascot score of 20, a minimal peptide length of 6, and only unique rank 1 peptides.
Accession number(s).
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (66) with data set accession number PXD005104 (http://www.ebi.ac.uk/pride/archive/projects/PXD005104).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Fred Homa (University of Pittsburgh School of Medicine, USA) and to Valerie Preston (MRC-University of Glasgow Centre for Virus Research, United Kingdom) for providing the UL25 deletion mutant KUL25NS and the complementing Vero 8-1 cell line.
This study was supported by FOM Projectruimte grants to G.J.L.W. and W.H.R.; an NWO-VICI grant to G.J.L.W.; a grant from the Deutsche Forschungsgemeinschaft (DFG) (SFB 900, project C2) and the Deutsches Zentrum für Infektionsforschung (DZIF) (TTU IICH) to B.S.; a VIDI grant from the NWO to W.H.R.; National Institutes of Health award GM 507401 to J.B.; and a grant from the EraNet NanoSci-E+ Initiative to G.J.L.W., W.H.R., and B.S. (DFG, So303/4).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00123-17.
REFERENCES
- 1.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 2.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. 2000. Seeing the herpesvirus capsid at 8.5 angstrom. Science 288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 3.Brown JC, Newcomb WW. 2011. Herpesvirus capsid assembly: insights from structural analysis. Curr Opin Virol 1:142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng L, Ryazantsev S, Sun R, Zhou ZH. 2010. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure 18:47–58. doi: 10.1016/j.str.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heymann JB, Cheng NQ, Newcomb WW, Trus BL, Brown JC, Steven AC. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol 10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 6.Perdue ML, Cohen JC, Kemp MC, Randall CC, Ocallaghan DJ. 1975. Characterization of 3 species of nucleocapsids of equine herpesvirus type-1 (EHV-1). Virology 64:187–204. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 7.Perdue ML, Cohen JC, Randall CC, O'Callaghan DJ. 1976. Biochemical studies of maturation of herpesvirus nucleocapsid species. Virology 74:194–208. doi: 10.1016/0042-6822(76)90141-0. [DOI] [PubMed] [Google Scholar]
- 8.Rixon FJ. 1993. Structure and assembly of herpesviruses. Semin Virol 4:135–144. doi: 10.1006/smvy.1993.1009. [DOI] [Google Scholar]
- 9.Homa FL, Brown JC. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol 7:107–122. doi:. [DOI] [PubMed] [Google Scholar]
- 10.Henaff D, Radtke K, Lippe R. 2012. Herpesviruses exploit several host compartments for envelopment. Traffic 13:1443–1449. doi: 10.1111/j.1600-0854.2012.01399.x. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 12.Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJ. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc Natl Acad Sci U S A 106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klug WS, Roos WH, Wuite GJL. 2012. Unlocking internal prestress from protein nanoshells. Phys Rev Lett 109:168104. doi: 10.1103/PhysRevLett.109.168104. [DOI] [PubMed] [Google Scholar]
- 14.Liashkovich I, Hafezi W, Kuhn JE, Oberleithner H, Kramer A, Shahin V. 2008. Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J Cell Sci 121:2287–2292. doi: 10.1242/jcs.032284. [DOI] [PubMed] [Google Scholar]
- 15.Roos WH, Bruinsma R, Wuite GJL. 2010. Physical virology. Nat Phys 6:733–743. doi: 10.1038/nphys1797. [DOI] [Google Scholar]
- 16.Marchetti M, Wuite GJL, Roos WH. 2016. Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation. Curr Opin Virol 18:82–88. doi: 10.1016/j.coviro.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Hernando-Perez M, Miranda R, Aznar M, Carrascosa JL, Schaap IAT, Reguera D, de Pablo PJ. 2012. Direct measurement of phage phi29 stiffness provides evidence of internal pressure. Small 8:2366–2370. doi: 10.1002/smll.201200664. [DOI] [PubMed] [Google Scholar]
- 18.Carrasco C, Carreira A, Schaap IAT, Serena PA, Gomez-Herrero J, Mateu MG, Pablo PJ. 2006. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci U S A 103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanovska IL, de Pablo PJ, Ibarra B, Sgalari G, MacKintosh FC, Carrascosa JL, Schmidt CF, Wuite GJL. 2004. Bacteriophage capsids: tough nanoshells with complex elastic properties. Proc Natl Acad Sci U S A 101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snijder J, Uetrecht C, Rose R, Sanchez R, Marti G, Agirre J, Guérin DM, Wuite GJ, Heck AJ, Roos WH. 2013. Probing the biophysical interplay between a viral genome and its capsid. Nat Chem 5:502–509. doi: 10.1038/nchem.1627. [DOI] [PubMed] [Google Scholar]
- 21.Evilevitch A, Roos WH, Ivanovska IL, Jeembaeva M, Jonsson B, Wuite GJ. 2011. Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J Mol Biol 405:18–23. doi: 10.1016/j.jmb.2010.10.039. [DOI] [PubMed] [Google Scholar]
- 22.Ivanovska I, Wuite G, Jonsson B, Evilevitch A. 2007. Internal DNA pressure modifies stability of WT phage. Proc Natl Acad Sci U S A 104:9603–9608. doi: 10.1073/pnas.0703166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. 2006. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci U S A 103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellanos M, Perez R, Carrasco C, Hernando-Perez M, Gomez-Herrero J, de Pablo PJ, Mateu MG. 2012. Mechanical elasticity as a physical signature of conformational dynamics in a virus particle. Proc Natl Acad Sci U S A 109:12028–12033. doi: 10.1073/pnas.1207437109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I. 2007. A stiffness switch in human immunodeficiency virus. Biophys J 92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Berna AJ, Ortega-Esteban A, Menendez-Conejero R, Winkler DC, Menendez M, Steven AC, Flint SJ, de Pablo PJ, San Martin C. 2012. The role of capsid maturation on adenovirus priming for sequential uncoating. J Biol Chem 287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder J, Reddy VS, May ER, Roos WH, Nemerow GR, Wuite GJL. 2013. Integrin and defensin modulate the mechanical properties of adenovirus. J Virol 87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb WW, Brown JC. 1991. Structure of the herpes simplex virus capsid—effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol 65:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol 232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 30.Sae-Ueng U, Liu T, Catalano CE, Huffman JB, Homa FL, Evilevitch A. 2014. Major capsid reinforcement by a minor protein in herpesviruses and phage. Nucleic Acids Res 42:9096–9107. doi: 10.1093/nar/gku634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb WW, Homa FL, Brown JC. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol 80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol 72:1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogasawara M, Suzutani T, Yoshida I, Azuma M. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol 75:1427–1436. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klupp BG, Granzow H, Karger A, Mettenleiter TC. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J Virol 79:13442–13453. doi: 10.1128/JVI.79.21.13442-13453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cockrell SK, Sanchez ME, Erazo A, Homa FL. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol 83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J Virol 82:6654–6666. doi: 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol 72:3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholtes L, Baines JD. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pU(L)17/pU(L)25 complex of herpes simplex virus 1. J Virol 83:12725–12737. doi: 10.1128/JVI.01658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J Virol 75:10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trus BL, Newcomb WW, Cheng NQ, Cardone G, Marekov L, Horna FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wills E, Scholtes L, Baines JD. 2006. Herpes simplex virus 1 DNA packaging proteins encoded by U(L)6, U(L)15, U(L)17, U(L)28, and U(L)33 are located on the external surface of the viral capsid. J Virol 80:10894–10899. doi: 10.1128/JVI.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai XH, Gong DY, Wu TT, Sun R, Zhou ZH. 2014. Organization of capsid-associated tegument components in Kaposi's sarcoma-associated herpesvirus. J Virol 88:12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huet A, Makhov AM, Huffman JB, Vos M, Homa FL, Conway JF. 2016. Extensive subunit contacts underpin herpesvirus capsid stability and interior-to-exterior allostery. Nat Struct Mol Biol 23:531–539. doi: 10.1038/nsmb.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn J, Leege T, Granzow H, Fuchs W, Mettenleiter TC, Klupp BG. 2010. Analysis of pseudorabies and herpes simplex virus recombinants simultaneously lacking the pUL17 and pUL25 components of the C-capsid specific component. Virus Res 153:20–28. doi: 10.1016/j.virusres.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 47.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol 425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borst EM, Bauerfeind R, Binz A, Stephan TM, Neuber S, Wagner K, Steinbruck L, Sodeik B, Rovis TL, Jonjic S, Messerle M. 2016. The essential human cytomegalovirus proteins pUL77 and pUL93 are structural components necessary for viral genome encapsidation. J Virol 90:5860–5875. doi: 10.1128/JVI.00384-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeRussy BM, Boland MT, Tandon R. 2016. Human cytomegalovirus pUL93 links nucleocapsid maturation and nuclear egress. J Virol 90:7109–7117. doi: 10.1128/JVI.00728-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholtes LD, Yang K, Li LX, Baines JD. 2010. The capsid protein encoded by U(L)17 of herpes simplex virus 1 interacts with tegument protein VP13/14. J Virol 84:7642–7650. doi: 10.1128/JVI.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coller KE, Lee JIH, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol 81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan WH, Roberts APE, McElwee M, Bhella D, Rixon FJ, Lauder R. 2015. The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J Virol 89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thurlow JK, Murphy M, Stow ND, Preston VG. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol 80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roos WH, Gibbons MM, Arkhipov A, Uetrecht C, Watts NR, Wingfield PT, Steven AC, Heck AJR, Schulten K, Klug WS, Wuite GJL. 2010. Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys J 99:1175–1181. doi: 10.1016/j.bpj.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJL. 2007. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci 64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sigamani SS, Zhao HY, Kamau YN, Baines JD, Tang L. 2013. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J Virol 87:7140–7148. doi: 10.1128/JVI.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang K, Homa F, Baines JD. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J Virol 81:6419–6433. doi: 10.1128/JVI.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kononova O, Snijder J, Brasch M, Cornelissen J, Dima RI, Marx KA, Wuite GJL, Roos WH, Barsegov V. 2013. Structural transitions and energy landscape for cowpea chlorotic mottle virus capsid mechanics from nanomanipulation in vitro and in silico. Biophys J 105:1893–1903. doi: 10.1016/j.bpj.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radtke K, Anderson F, Sodeik B. 2014. A precipitation-based assay to analyze interactions of viral particles with cytosolic host factors. Methods Mol Biol 1144:191–208. doi: 10.1007/978-1-4939-0428-0_13. [DOI] [PubMed] [Google Scholar]
- 61.Taus NS, Salmon B, Baines JD. 1998. The herpes simplex virus 1 U(L)17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115–125. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- 62.Roos WH. 2011. How to perform a nanoindentation experiment on a virus. Methods Mol Biol 783:251–264. doi: 10.1007/978-1-61779-282-3_14. [DOI] [PubMed] [Google Scholar]
- 63.Snijder J, Ivanovska IL, Baclayon M, Roos WH, Wuite GJL. 2012. Probing the impact of loading rate on the mechanical properties of viral nanoparticles. Micron 43:1343–1350. doi: 10.1016/j.micron.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 64.de Pablo PJ, Colchero J, Gomez-Herrero J, Baro AM. 1998. Jumping mode scanning force microscopy. Appl Phys Lett 73:3300–3302. doi: 10.1063/1.122751. [DOI] [Google Scholar]
- 65.Zhou HJ, Ye ML, Dong J, Corradini E, Cristobal A, Heck AJR, Zou HF, Mohammed S. 2013. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat Protoc 8:461–480. doi: 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- 66.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bantscheff M, Lemeer S, Savitski MM, Kuster B. 2012. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem 404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.