ABSTRACT
H5N6 avian influenza virus (AIV) has posed a potential threat to public health since its emergence in China in 2013. To understand the evolution and emergence of H5N6 AIV in the avian population, we performed molecular surveillance of live poultry markets (LPMs) in Wugang Prefecture, Hunan Province, in central China, during 2014 and 2015. Wugang Prefecture is located on the Eastern Asian-Australian migratory bird flyway, and a human death due to an H5N6 virus was reported in the prefecture on 21 November 2016. In total, we sampled and sequenced the complete genomes of 175 H5N6 AIVs. Notably, our analysis revealed that H5N6 AIVs contain at least six genotypes arising from segment reassortment, including a rare variant that possesses an HA gene derived from H5N1 clade 2.3.2 and a novel NP gene that has its origins with H7N3 viruses. In addition, phylogenetic analysis revealed that genetically similar H5N6 AIVs tend to cluster according to their geographic regions of origin. These results help to reveal the evolutionary behavior of influenza viruses prior to their emergence in humans.
IMPORTANCE The newly emerged H5N6 influenza A virus has caused more than 10 human deaths in China since 2013. In November 2016, a human death due to an H5N6 virus, in Wugang Prefecture, Hunan Province, was confirmed by the WHO. To better understand the evolution and emergence of H5N6 viruses, we surveyed live poultry markets (LPMs) in Wugang Prefecture before the reported human death, with a focus on revealing the diversity and genomic origins of H5N6 in birds during 2014 and 2015. In general, H5N6 viruses in this region were most closely related to H5N1 clade 2.3.4.4, with the exception of one virus with an HA gene derived from clade 2.3.2 such that it represents a novel reassortant. Clearly, the ongoing surveillance of LPMs is central to monitoring the emergence of pathogenic influenza viruses.
KEYWORDS: H5N6, avian influenza virus, evolution, molecular epidemiology, reassortment
INTRODUCTION
Since its emergence in Guangdong Province in 1996, the highly pathogenic H5N1 avian influenza A virus (AIV) has become endemic in poultry in multiple regions of China, developing into both genetically and antigenically distinct lineages (1) and causing a number of human deaths (2). This H5 subtype virus has also undergone frequent reassortment with cocirculating AIV subtypes, generating a number of novel viruses (3, 4).
Recently, a novel AIV type—H5N6—was isolated in Asia and was initially identified as a reassortant between H5N1 and H6N6 viruses (5–7). This virus has caused more than 10 human deaths since 2014 (8; http://www.who.int/csr/don/archive/country/chn/en/). Recently, with more H5N6 genomes sequenced, it has become clear that the gene segments of these viruses derived from a variety of AIV subtypes. Specifically, the PB2 gene of some H5N6 AIVs derived from that of H6N6 viruses, while the PB1 gene originated in H3 subtype viruses (9); a novel H5N6 virus isolated from migratory waterfowl possessed a hemagglutinin (HA) gene that was related to that of H5N2 viruses (10); the internal genes of viruses isolated from two human cases in 2015 derived from H7N9/H9N2 viruses in chickens (11); human infections in Guangdong Province, China, in 2015 were due to a newly emerged H5N6 virus that was a reassortant between H6N6 and H9N2 viruses (12); and a human-associated H5N6 (clade 2.3.3.4) virus sampled in China during 2014 to 2015 was a triple reassortant derived from H6N6, H5N1 (clade 2.3.1.1), and H5N6 (2.3.4.4) viruses (13). In addition, a recent study of multiple live poultry markets (LPMs) in southern China revealed that the HA gene of H5N6 viruses derived from clade 2.3.4.4, the NA gene derived from H6N6 viruses, and all six internal gene segments had their origins in clades 2.3.4.4 and 2.3.2.1 of H9N2/H7N9 viruses (14).
To better understand the complex molecular evolution and emergence of H5N6 viruses at the local level, we sampled AIVs in LPMs in Wugang Prefecture, Hunan Province, in central China. Notably, these LPMs are located on the Eastern Asian-Australian migratory bird flyway, and a human death in this prefecture that was caused by an H5N6 virus was reported and confirmed by the WHO on 21 November 2016 (http://www.who.int/csr/don/07-december-2016-ah5n6-china/en/). In addition, the LPMs in this area are located in close proximity to each other and hence provide an ideal sampling framework to understand the local evolution of H5N6 in central China since its emergence in 2013.
RESULTS
H5N6 AIV in Wugang Prefecture.
Of the 175 H5N6 AIVs reported in this study (see Table S1 in the supplemental material), 169 were isolated from LPMs in Wugang Prefecture, Hunan Province. Ducks (n = 79) and Muscovy ducks (n = 37) were the major hosts, representing 68.6% of the H5N6 AIVs. Other hosts were chickens (n = 26; 15.4%) and geese (n = 26; 15.4%). Unsurprisingly, all isolated H5N6 viruses fell into clade 2.3.4.4 in the HA phylogeny. However, it was striking that our phylogenetic analysis revealed that one of the newly sequenced H5N6 strains—A/duck/Hunan/HN232/2015 (H5N6)—had an HA gene seemingly derived from clade 2.3.2 (Fig. S1) (see below).
Geographic clustering of Wugang H5N6 AIVs.
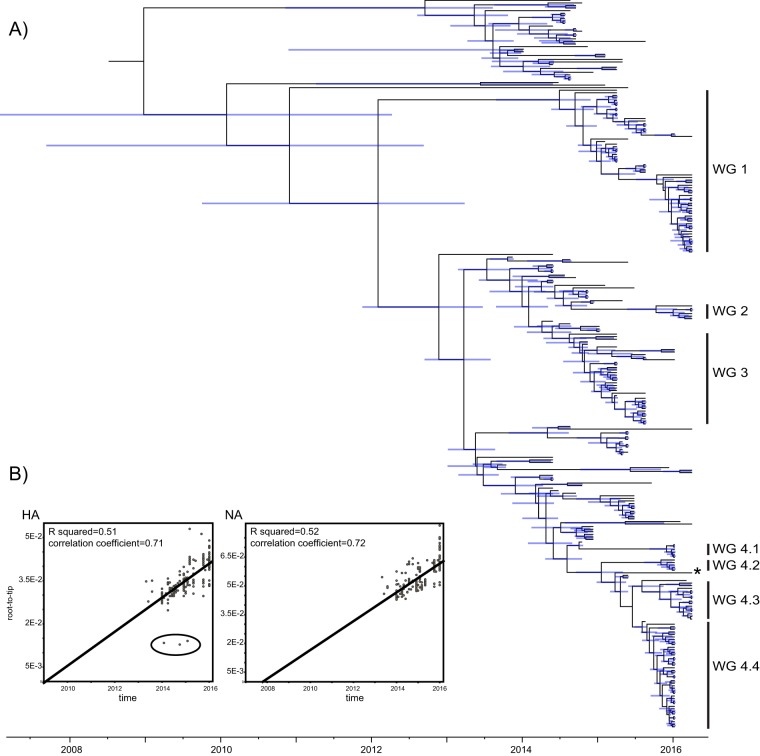
Our phylogenetic analysis of HA suggested that H5N6 AIVs tend to cluster geographically (Fig. 1A). In particular, several major geographic clusters with high Bayesian posterior probability values (>95%) were identified in Wugang Prefecture. These were denoted WG1 (with “WG” denoting Wugang Prefecture; n = 60), WG2 (n = 6), WG3 (n = 34), and WG4, which contained four subclusters: WG4.1 (n = 6), WG4.2 (n = 5), WG4.3 (n = 15), and WG4.4 (n = 42). With the exception of WG2, all these clusters also contained viruses sampled outside Hunan Province (shown by the mix of colors in Fig. 1). In this context, it is noteworthy that a virus isolated from a human case of H5N6 (A/Guangdong/ZQ874/2015) infection from a different (Guangdong) province was embedded within cluster WG4.
FIG 1.
Time-scaled evolutionary history of H5N6 AIVs. (A) An MCC tree of the HA sequences of viruses sampled in China (n = 281) is shown, with the viruses collected in Wugang Prefecture (WG) highlighted in blue. A virus from a human case of H5N6 (A/Guangdong/ZQ874/2015) infection distributed within WG4 is marked with an asterisk. Shaded bars represent the 95% highest probability distribution for the age of each node. (B) Analysis of root-to-tip divergence against sampling date for the HA and NA gene segments. A group of clear outliers in the HA graph is marked (and was removed from the BEAST analysis).
Phylogenetic history of the H5N6 NA segment.
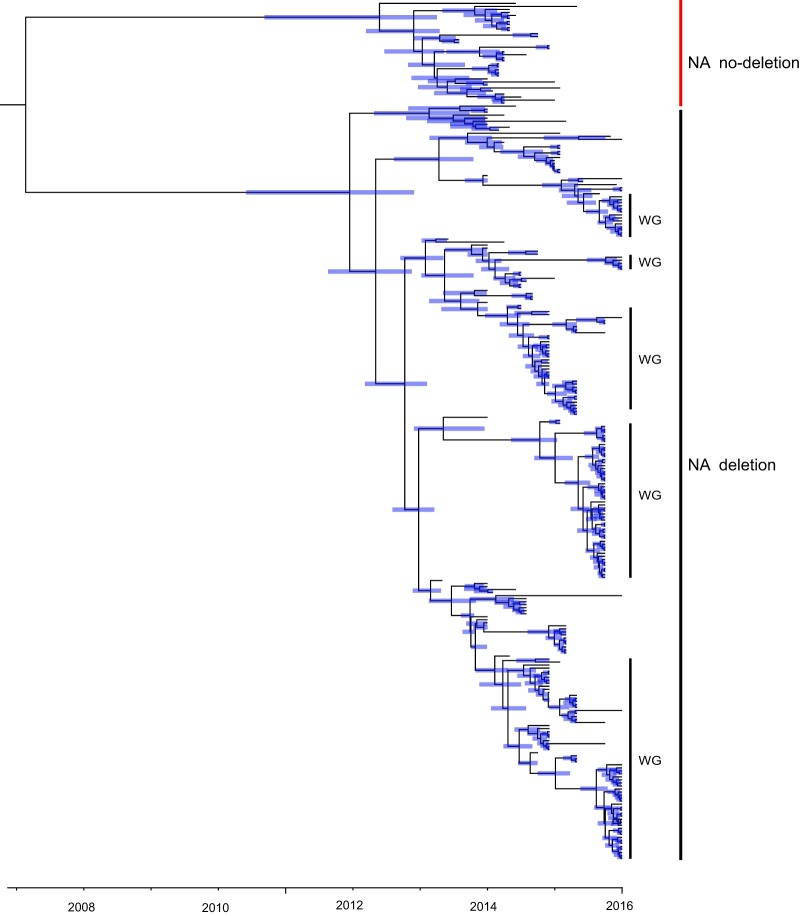
Two major clusters of H5N6 viruses were observed in the NA phylogeny (Fig. 2); one was characterized by an 11-amino-acid deletion at residues 58 to 68, while the other, less frequent cluster had no such deletion. All the viruses sampled from Wugang Prefecture belonged to the former cluster, suggesting that the deletion mutant is dominant in central China.
FIG 2.
Phylogenetic history of the H5N6 NA gene segment. This time-scaled phylogeny reveals two major groups: those with and without an 11-amino-acid deletion. The Wugang strains determined here are marked “WG.” Shaded bars represent the 95% highest probability distribution for the age of each node.
Reassortment and genesis of H5N6 genomic diversity.
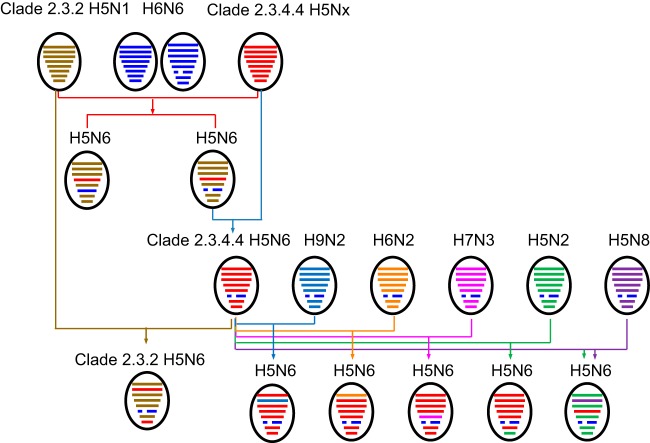
We next used phylogenetic analysis to determine the extent of reassortment in the H5N6 viruses sampled for the present study. This revealed segments with complex evolutionary origins (Table 1; Fig. 3; Fig. S1). Specifically, PB2 gene segments were derived from H5N1, H5N2, and H6N2 viruses; PB1 from H5N8 and H9N2 viruses; PA from H5N1 and H5N2 viruses; NP from H5N1, H5N2, and H7N3 viruses; M from H5N1 and H5N2 viruses; and NS from H5N2 viruses. To our knowledge, the H7N3 origin of the H5N6 NP gene segment has not been reported previously (Fig. 3). Strikingly, as noted above, we also found one virus—A/duck/Hunan/HN232/2015 (H5N6) (Fig. 3)—that possessed an HA gene derived from H5N1 clade 2.3.2 (exhibiting 98.9% nucleotide identity to A/chicken/Wuhan/HAQL07/2014 [H5N1]).
TABLE 1.
H5N6 avian influenza viruses isolated for this study and published viruses to which they show the highest nucleotide identities
| Segment | Virus | Closest GenBank virus by genetic distance | Identity (%) |
|---|---|---|---|
| HA | A/duck/Hunan/HN232/2015 | A/chicken/Wuhan/HAQL07/2014 (H5N1) | 98.8 |
| PB2 | A/duck/Hunan/HN232/2015 | A/chicken/Wuhan/HAQL07/2014 (H5N1) | 97.4 |
| A/muscovy duck/Hunan/HN359/2015 | A/chicken/Guangdong/C273/2011 (H6N2) | 93.2 | |
| A/goose/Hunan/HN325/2015 | A/duck/Hubei/SZY250/2016 (H5N2) | 99.4 | |
| PB1 | A/goose/Hunan/HN325/2015 | A/duck/Eastern China/L0423/2011 (H5N8) | 93.1 |
| A/muscovy duck/Hunan/102/2014 | A/chicken/Jiangsu/245/2004 (H9N2) | 95.8 | |
| PA | A/duck/Hunan/HN232/2015 | A/chicken/Wuhan/HAQL07/2014 (H5N1) | 99.3 |
| A/goose/Hunan/HN325/2015 | A/duck/Hubei/SZY250/2016 (H5N2) | 99.3 | |
| NP | A/duck/Hunan/HN232/2015 | A/duck/Wenzhou/HAYXLG07/2015 (H5N1) | 99.4 |
| A/goose/Hunan/HN325/2015 | A/duck/Hubei/SZY250/2016 (H5N2) | 99.3 | |
| A/duck/Hunan/HN214/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| A/duck/Hunan/HN234/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| A/duck/Hunan/HN236/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| A/duck/Hunan/HN238/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| A/duck/Hunan/HN240/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| A/duck/Hunan/HN242/2015 | A/duck/Jiangxi/31028/2013 (H7N3) | 99.3 | |
| M | A/duck/Hunan/HN232/2015 | A/chicken/Wuhan/HAQL07/2014 (H5N1) | 99.6 |
| A/goose/Hunan/HN325/2015 | A/duck/Hubei/SZY250/2016 (H5N2) | 99.7 | |
| NS | A/goose/Hunan/HN325/2015 | A/duck/Hubei/SZY250/2016 (H5N2) | 99.3 |
| A/chicken/Anhui/AH325/2015 | A/mallard/Italy/4223-2/2006 (H5N2) | 97.3 |
FIG 3.
Genesis of H5N6 AIV. Virus particles are shown as colored ovals containing horizontal bars that represent the eight gene segments (from top to bottom: PB2, PB1, PA, HA, NP, NA, M, and NS). To illustrate the history of reassortant events, segments in descendant viruses are colored according to their corresponding source viruses.
To reveal the phylogenetic positions of the newly isolated AIVs, we performed a phylogenetic analysis using all H5N6 AIVs isolated in different Asian countries, including China (n = 487). Such analysis revealed that the viruses from Wugang (i.e., Hunan Province) generally formed distinct clusters indicative of largely in situ evolution (Fig. 4). A notable exception was several H5N6 AIVs sampled in Vietnam at the end of 2014 and in early 2015 that fell within a Hunan cluster. All reassortant strains detected in this study (Table 1) fell within the Hunan or Anhui clusters and were phylogenetically distinct from those sampled outside China (Fig. 4), suggesting that they arose within China.
FIG 4.
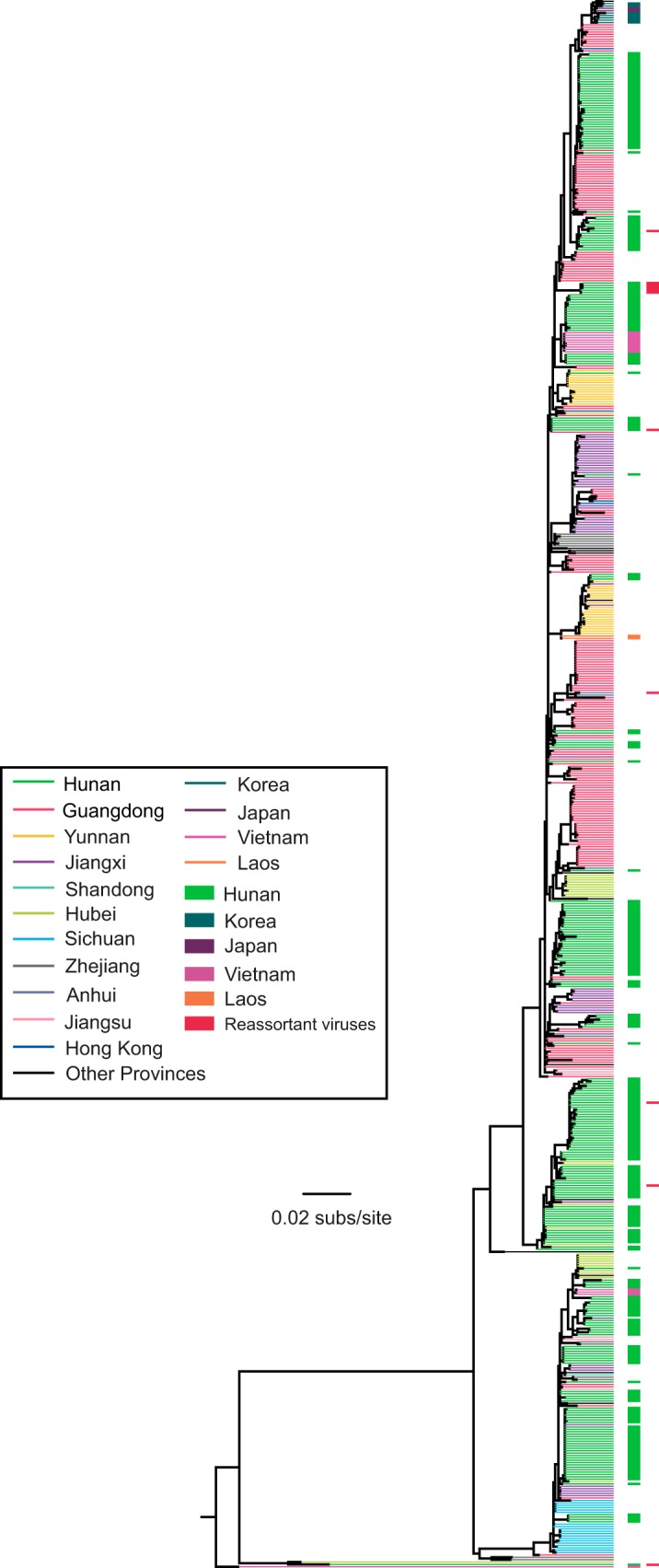
Global phylogenetic history and geographic distribution of H5N6 AIVs. Viruses sampled from different locations (n = 487 in total), including Wugang (i.e., Hunan), are distinguished by their colors, and all reassortant viruses detected in this study are also indicated. All branch lengths are scaled according to the number of substitutions per site (subs/site). The tree is rooted using A/duck/Vietnam/LBM360c1-4-1/2013 (H5N6), which was collected on 6 February 2013.
Evolutionary dynamics of H5N6.
An initial analysis of temporal structure by use of a root-to-tip regression provided evidence for some temporal (i.e., clock-like) structure for both the HA (n = 284; correlation coefficient = 0.71; R2 = 0.51) and NA (n = 284; correlation coefficient = 0.72; R2 = 0.52) gene segments sampled from China. However, there was also clearly rate variation, and when three clear outliers (Fig. 1B) were removed from the HA regression, the R2 value increased to 0.63. In addition, it is possible that evolutionary rates may be elevated to some extent by time dependency (15, 16), particularly given the short time span of sampling in this study. Hence, all evolutionary rates estimated here should be treated with caution. Using a Bayesian method, the rate of nucleotide substitution in the H5N6 viruses (for the same data sets as those described above) was estimated to be 4.28 × 10−3 nucleotide substitution/site/year (95% highest probability density [HPD] = 3.42 × 10−3 to 5.17 × 10−3 substitution/site/year) for the HA gene and 5.27 × 10−3 substitution/site/year (3.55 × 10−3 to 6.63 × 10−3 substitution/site/year) for the NA gene. Using the HA rate, we estimated that the times of origin (95% HPD) of the four major clusters (Fig. 1A) of viruses were as follows: WG1, March 2013 to June 2014; WG2, February 2015 to October 2015; WG3, November 2013 to May 2014; and WG4, November 2013 to June 2014. All these estimated dates are close to when (i.e., around 2014) the first human H5N6 case was reported in China.
DISCUSSION
As H5N1 avian influenza viruses pose a threat to public health, it is clearly significant that the H5N6 virus, which emerged in 2013, has internal genes derived from multiple subtypes, including H5, H6, H7, and H9 AIVs. This concern was heightened by a report on 21 November 2016 that a human death due to H5N6 had occurred in Wugang Prefecture, which was later confirmed by the WHO (http://www.who.int/csr/don/07-december-2016-ah5n6-china/en/). We therefore sought to examine the evolution and emergence of H5N6 AIV in the avian population in Wugang Prefecture prior to this human spillover event.
Our surveillance of H5N6 viruses in LPMs in Wugang Prefecture revealed that at least six types of reassortant H5N6 viruses have circulated in this region, covering all reassortant viruses reported to date and further illustrating the complexity of H5N6 evolution and the ability of H5N6 viruses to reassort with other AIV subtypes. In addition, all these reassortant strains were genetically close to genotypes previously reported in China, although sampling biases make it difficult to determine exactly where each reassortment event occurred.
As all currently reported H5N6 AIVs are members of H5N1 clade 2.3.4.4, our observation that one H5N6 virus seemingly carried an HA gene directly derived from H5N1 clade 2.3.2 is important. In particular, this demonstrates that earlier lineages of H5N1 AIV may still coinfect and reassort with more recently circulating H5N6 AIVs.
In sum, we have revealed a complex pattern of evolution and emergence of H5N6 viruses in a single locality in central China—Wugang Prefecture, Hunan Province—over a 2-year period. Clearly, H5N6 viruses are capable of receiving gene segments, even including the HA gene, from different subtypes and can act as the genomic source for other subtypes, such as H3N6 (17, 18). A careful monitoring of H5N6 viruses in LPMs and wild birds is evidently necessary to help prevent the virus from establishing itself in the human population.
MATERIALS AND METHODS
Sampling and virus isolation.
During 2014 and 2015, cloacal swabs and fecal samples from ducks and geese and environmental samples from LPMs in Wugang Prefecture, Hunan Province, were collected and screened for avian influenza virus. Samples were purified and propagated in 9- to 10-day-old specific-pathogen-free (SPF) embryonated hen eggs incubated at 37°C and were manually checked every 24 h until the eggs were sacrificed within 48 to 72 h. An HA inhibition assay was performed to determine the HA subtype of the isolated virus by using chicken anti-HA serum for each subtype. PCR was then performed to further identify HA and NA subtypes (conditions and primers are available on request). Allantoic fluids were collected and stored at −70°C until use.
RNA extraction, RT-PCR amplification, and sequencing.
Viral RNA was extracted from virus-infected allantoic fluid by use of a QIAamp viral RNA minikit (Qiagen). Viral cDNAs were synthesized from viral RNAs by reverse transcription (RT) using the primer Uni12, which is complementary to the conserved 3′ ends of all AIV RNA segments, and amplified by PCR with primers complementary to the conserved promoter and noncoding regions of each gene segment (primer sequences are available on request). The PCR products were purified using a gel extraction kit (Omega) according to the manufacturer's protocol and then sequenced using an ABI 3730 sequencer with BigDye Terminator cycle sequencing reagents (Applied Biosystems).
Phylogenetic analysis.
We analyzed all 175 H5N6 sequences generated in this study (see Table S1 in the supplemental material) together with sequences downloaded from the Influenza Virus Database in GenBank (NCBI) and the GISAID database that were released before 15 March 2017. All sequences were aligned using MUSCLE (version 3.8.31) (19), and each segment was analyzed independently. Phylogenetic relationships among all 487 H5N6 HA sequences were inferred using the maximum likelihood (ML) method available in PhyML (version 3.1) (20), utilizing subtree pruning and redrafting branch swapping and the GTR + Γ model of nucleotide substitution. The robustness of each node on the phylogenetic tree was determined using the Shimodaira-Hasegawa approximate likelihood ratio test (21).
Evolutionary dynamics of H5N6.
We next estimated rates of evolutionary change (i.e., nucleotide substitution) in the HA and NA gene segments of H5N6 viruses. For efficiency of analysis, we focused on H5N6 AIVs sampled in China alone (n = 284) and representative of the entire phylogenetic diversity of H5N6. To ensure that there was sufficient temporal structure in the HA and NA alignments for reliable rate estimation, we first performed a regression of root-to-tip genetic distances on the ML tree against exact sampling dates by using TempEst (22). This process revealed a small number of sequences (n = 3) that showed a clear deviation from clock-like behavior and that were removed from the analysis (see Results).
To obtain a more robust rate estimate, we used the Bayesian Markov chain Monte Carlo (MCMC) method implemented in the BEAST package (version 1.8.3) (23), employing the SRD06 nucleotide substitution model (24), an uncorrected lognormal relaxed molecular clock model, and a Bayesian Skyride coalescent model. Multiple runs of the MCMC method were computed and combined using LogCombiner (version 1.8.3) (http://beast.bio.ed.ac.uk/), utilizing 3.5 × 108 total steps for each data set, with sampling every 5,000 steps. Convergence (i.e., effective sample sizes of >200) of relevant parameters was assessed using Tracer (version 1.6) (http://beast.bio.ed.ac.uk/). The posterior distribution of trees obtained from the BEAST analysis (with 10% of the runs removed as burn-in) was also used to obtain the maximum clade credibility (MCC) tree for each segment.
Accession number(s).
All nucleotide sequences obtained in this study were compiled and edited using Lasergene (version 7.1) (DNASTAR). All viral genomes sequenced here have been deposited in the GISAID database (http://platform.gisaid.org/), and all accession numbers are listed in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the National Key Research and Development Program (grant 2016YFD0500201) and the CAS Pioneer Hundred Talents Program to J.C. E.C.H. was supported by an NHMRC Australia Fellowship (grant GNT1037231).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00143-17.
REFERENCES
- 1.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 2.Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, Zhang XL, Zhao D, Wang G, Feng Y, Ma J, Liu W, Wang J, Gao GF. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Shi J, Zhong G, Deng G, Tian G, Ge J, Zeng X, Song J, Zhao D, Liu L, Jiang Y, Guan Y, Bu Z, Chen H. 2010. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J Virol 84:8389–8397. doi: 10.1128/JVI.00413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong FY, Phommachanh P, Kalpravidh W, Chanthavisouk C, Gilbert J, Bingham J, Davies KR, Cooke J, Eagles D, Phiphakhavong S, Shan S, Stevens V, Williams DT, Bounma P, Khambounheuang B, Morrissy C, Douangngeun B, Morzaria S. 2015. Reassortant highly pathogenic influenza A(H5N6) virus in Laos. Emerg Infect Dis 21:511–516. doi: 10.3201/eid2103.141488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Lu R, Peng X, Xu L, Cheng L, Lu X, Jin C, Xie T, Yao H, Wu N. 2015. Novel reassortant highly pathogenic H5N6 avian influenza viruses in poultry in China. Infect Genet Evol 31:64–67. doi: 10.1016/j.meegid.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Fu Y, Yang J, Guo J, He J, Guo J, Weng S, Jia Y, Liu B, Li X, Zhu Q, Chen H. 2015. Genetic and biological characterization of two novel reassortant H5N6 swine influenza viruses in mice and chickens. Infect Genet Evol 36:462–466. doi: 10.1016/j.meegid.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Yang ZF, Mok CK, Peiris JS, Zhong NS. 2015. Human infection with a novel avian influenza A(H5N6) virus. N Engl J Med 373:487–489. doi: 10.1056/NEJMc1502983. [DOI] [PubMed] [Google Scholar]
- 9.Yuan R, Wang Z, Kang Y, Wu J, Zou L, Liang L, Song Y, Zhang X, Ni H, Lin J, Ke C. 2016. Continuing reassortant of H5N6 subtype highly pathogenic avian influenza virus in Guangdong. Front Microbiol 7:520. doi: 10.3389/fmicb.2016.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Y, Liu H, Xiong C, Liu D, Shi W, Li M, Liu S, Chen J, Chen G, Li Y, Yang G, Lei Y, Xiong Y, Lei F, Wang H, Chen Q, Chen J, Gao GF. 2016. Novel avian influenza A (H5N6) viruses isolated in migratory waterfowl before the first human case reported in China, 2014. Sci Rep 6:29888. doi: 10.1038/srep29888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Li X, Bai T, Zhao X, Zhao X, Zhang Y, Guo J, Li Z, Yang L, Wang D, Shu Y. 2016. A fatal case of infection with a further reassortant, highly pathogenic avian influenza (HPAI) H5N6 virus in Yunnan, China. Infect Genet Evol 40:63–66. doi: 10.1016/j.meegid.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Shen YY, Ke CW, Li Q, Yuan RY, Xiang D, Jia WX, Yu YD, Liu L, Huang C, Qi WB, Sikkema R, Wu J, Koopmans M, Liao M. 2016. Novel reassortant avian influenza A(H5N6) viruses in humans, Guangdong, China, 2015. Emerg Infect Dis 22:1507–1509. doi: 10.3201/eid2208.160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Li R, Jiang L, Xiong C, Chen Y, Zhao G, Jiang Q. 2016. The complexity of human infected AIV H5N6 isolated from China. BMC Infect Dis 16:600. doi: 10.1186/s12879-016-1932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Duchêne S, Holmes EC, Ho SY. 2014. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates. Proc Biol Sci 281:20140732. doi: 10.1098/rspb.2014.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes EC. 2016. Complexities of estimating evolutionary rates in viruses. J Virol 90:2155. doi: 10.1128/JVI.02570-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yang J, Liu B, Jia Y, Guo J, Gao X, Weng S, Yang M, Wang L, Wang LF, Cui J, Chen H, Zhu Q. 2016. Co-circulation of H5N6, H3N2, H3N8, and emergence of novel reassortant H3N6 in a local community in Hunan province in China. Sci Rep 6:25549. doi: 10.1038/srep25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Zhu W, Li X, Bo H, Zhang Y, Zou S, Gao R, Dong J, Zhao X, Chen W, Dong L, Zou X, Xing Y, Wang D, Shu Y. 2017. Genesis and dissemination of highly pathogenic H5N6 avian influenza viruses. J Virol 91:e02199-16. doi: 10.1128/JVI.02199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 21.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 22.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23:7–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.