ABSTRACT
Bats are natural reservoirs for many pathogenic viruses, and increasing evidence supports the notion that bats can also harbor group A rotaviruses (RVAs), important causative agents of diarrhea in children and young animals. Currently, 8 RVA strains possessing completely novel genotype constellations or genotypes possibly originating from other mammals have been identified from African and Chinese bats. However, all the data were mainly based on detection of RVA RNA, present only during acute infections, which does not permit assessment of the true exposure of a bat population to RVA. To systematically investigate the genetic diversity of RVAs, 547 bat anal swabs or gut samples along with 448 bat sera were collected from five South Chinese provinces. Specific reverse transcription-PCR (RT-PCR) screening found four RVA strains. Strain GLRL1 possessed a completely novel genotype constellation, whereas the other three possessed a constellation consistent with the MSLH14-like genotype, a newly characterized group of viruses widely prevalent in Chinese insectivorous bats. Among the latter, strain LZHP2 provided strong evidence of cross-species transmission of RVAs from bats to humans, whereas strains YSSK5 and BSTM70 were likely reassortants between typical MSLH14-like RVAs and human RVAs. RVA-specific antibodies were detected in 10.7% (48/448) of bat sera by an indirect immunofluorescence assay (IIFA). Bats in Guangxi and Yunnan had a higher RVA-specific antibody prevalence than those from Fujian and Zhejiang provinces. These observations provide evidence for cross-species transmission of MSLH14-like bat RVAs to humans, highlighting the impact of bats as reservoirs of RVAs on public health.
IMPORTANCE Bat viruses, such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), Ebola, Hendra, and Nipah viruses, are important pathogens causing outbreaks of severe emerging infectious diseases. However, little is known about bat viruses capable of causing gastroenteritis in humans, even though 8 group A viruses (RVAs) have been identified from bats so far. In this study, another 4 RVA strains were identified, with one providing strong evidence for zoonotic transmission from bats to humans. Serological investigation has also indicated that RVA infection in bats is far more prevalent than expected based on the detection of viral RNA.
KEYWORDS: bat, group A rotavirus, cross-species transmission, reassortment
INTRODUCTION
Group A rotaviruses (RVAs) are causative agents of severe diarrheal illness in infants and young children, reported to cause >200,000 deaths yearly of children younger than 5 years old, especially in the developing world (1). RVA is a member of the genus Rotavirus within the family Reoviridae. Inside the mature triple-layered particle, 11 segments of double-stranded RNA are encapsulated, with most of them encoding single polypeptides except for segment 11, which encodes 2 proteins (2). Due to the segmented nature of the RVA genome, reassortment is prone to occur when a cell is infected with more than one strain (2). A well-established nomenclature system has been developed with a sequence-based classification defining genotypes (G, P, I, R, C, M, A, N, T, E, and H genotypes) for each of the 11 gene segments (encoding VP7, VP4, VP6, VP1, VP2, VP3, NSP1, NSP2, NSP3, NSP4, and NSP5/6, respectively) based on a cutoff value of nucleotide identity (3–6). So far, 32 G, 47 P, 24 I, 18 R, 17 C, 17 M, 28 M, 18 N, 19 T, 24 E, and 19 H genotypes have been approved worldwide by the Rotavirus Classification Working Group (RCWG) (7). With the increasing amount of complete genomic data becoming available for RVA, sequence characterization of animals' RVA strains has suggested that most species possess RVA strains with particularly conserved genotype constellations (8, 9). Only RVA strains with 2 discrete genotype constellations are responsible for the vast majority of worldwide human RVA infections over a long period, which for the non-G, non-P genes were designated I1-R1-C1-M1-A1-N1-T1-E1-H1 and I2-R2-C2-M2-A2-N2-T2-E2-H2, referred to as Wa-like and DS-1-like, respectively (9). RVA strains from several other animal species show more or less conserved genotype constellations, such as I2-R2-C2-M2-A3/A13-N2-T6-E2-H3 for cattle (10), I1/I5-R1-M1-A1/A8-N1-T1/T7-E1-H1 for pigs (11), I2/I6-R2-C2-M3-A10-N2-T3-E2-H7 for horses (12), and I3-R3-C2-M3-A3/A9-N2-T3-E3-H3/H6 for cats and dogs (13).
In addition to infecting humans, RVAs are widely distributed among birds and young mammals, such as cats, dogs, pigs, cows, horses, rats, turkeys, pheasants, and pigeons (2). Mammals in the order Chiroptera, also known as bats and featuring the unique ability of flying, are divided into more than 900 species, being the second largest mammalian order (14). Bats are important virus reservoirs, and more than 200 viruses have been identified in them, including many that are highly pathogenic to humans (14, 15). With high-throughput next-generation sequencing technology, viral metagenomics has been successfully employed to uncover the bat virome, leading to discovery of many novel viruses from these creatures in the last a few years, such as coronaviruses, papillomaviruses, bunyaviruses, paramyxoviruses, hepatitis viruses, and even influenza A viruses (16–24). Recently, RVAs have also been sporadically reported to occur in bats from China and Africa (25–28). The first RVA strain, named RVA/Bat-wt/KEN/KE4852/2007/G25P[6], was detected in fruit bats (Eidolon helvum) in Kenya, which had the G25-P[6]-I15-Rx-C8-Mx-Ax-N8-T11-E2-H10 genotype constellation based on partial genomic sequences (25). Later, two bat RVAs were isolated and identified from two Chinese insectivorous bat species in locations more than 400 km apart; these were named RVA/Bat-tc/CHN/MSLH14/2012/G3P[3] and RVA/Bat-tc/CHN/MYAS33/2013/G3P[10] and had the same G3-P[x]-I8-R3-C3-M3-A9-N3-T3-E3-H6 genotype constellation, with the exception of their P genotypes, P[3] for MSLH14 and P[10] for MYAS33 (26, 27). Most recently, Yinda and colleagues identified 5 more RVA strains in bats from southwest Cameroon by using high-throughput sequencing (28). Four of them contained the genotype constellation Gx-P[x]-I22-R15-C15-M14-A25-N15-T17-E22-H17, whereas the last one possessed the G25-P[43]-I15-R16-C8-M15-A26-N8-T11-E23-H10 constellation, sharing six genotypes with that of Kenyan bat RVA strain KE4852 (28).
Although sequence information allows better understanding of the reassortment and evolution of bat RVAs, the lack of serological data on the prevalence of bat RVAs has prevented a full assessment of bat RVA infection status in bat populations. To understand the genomic diversities and seroprevalences of RVAs in bats, we collected bat fecal samples and tissues as well as sera in 5 provinces in South China. Serological investigation showed that more than 10% of bats were RVA seropositive, indicating a widely distributed exposure to RVAs. Viral detection and sequence analysis not only led to the identification of 4 RVA strains from 3 insectivorous bats and 1 frugivorous bat but also revealed cross-species transmission from bats to humans as well as reassortment events between human and bat RVAs.
RESULTS
Detection of RVA RNA in samples.
In total, 547 samples were collected (including 509 bat anal swabs from Guangxi, Fujian, and Zhejiang and 38 bat guts from Guangdong) and subjected to seminested reverse transcription-PCR (RT-PCR) screening. Four samples were positive for RVA (Table 1).These strains were detected from swabs of Scotophilus kuhlii in Beihai (1 out of 74 [1.4%]; strain YSSK5), Hipposideros pomona in Luzhai (1 out of 32 [3.1%]; strain LZHP2), and Taphozous melanopogon in Baise, Guangxi (1 out of 95 [1.1%]; strain BSTM70). The fourth strain was identified from gut tissue from a Rousettus leschenaultii in Luoding, Guangdong (1 out of 38 [2.6%]; strain GLRL1).
TABLE 1.
Details of anal swabs and sera from bats and number of RVA-positive bat samples detected by nested RT-PCR and IIFAa
| Province | Site | Bat species | Year | Diet | P/T (%) |
|
|---|---|---|---|---|---|---|
| PCR | IIFA | |||||
| Yunnan | Xishuangbanna | Rousettus leschenaultii | 2012 | F | ND | 30/142 (21.1) |
| Guangxi | Beihai | Scotophilus kuhlii | 2015 | I | 1/74 (1.4) | 13/50 (26.0) |
| Xing'an | Hipposideros larvatus | 2015 | I | 0/9 (0.0) | 0/9 (0.0) | |
| Wuming | H. larvatus | 2015 | I | 0/8 (0.0) | 0/15 (0.0) | |
| Long'an | H. larvatus | 2015 | I | 0/17 (0.0) | 0/17 (0.0) | |
| Lingshan | S. kuhlii | 2015 | I | 0/15 (0.0) | 0/15 (0.0) | |
| Miniopterus australis | 2015 | I | 0/15 (0.0) | 1/15 (6.7) | ||
| Luzhai | Hipposideros pomona | 2015 | I | 1/32 (3.1) | ND | |
| Baise | Taphozous melanopogon | 2015 | I | 1/95 (1.1) | ND | |
| Subtotal | 3/265 (1.1) | 14/121 (11.6) | ||||
| Guangdong | Luoding | R. leschenaultii | 2005 | F | 1/38 (2.6) | ND |
| Fujian | Xiamen | S. kuhlii | 2015 | I | 0/19 (0.0) | 2/19 (10.5) |
| Yanshi | Rhinolophus sinicus | 2016 | I | 0/48 (0.0) | 0/17 (0.0) | |
| Nanping | R. sinicus | 2016 | I | 0/1 (0.0) | 0/1 (0.0) | |
| Rhinolophus affinis | 2016 | I | 0/62 (0.0) | 1/60 (1.7) | ||
| Hipposideros armiger | 2016 | I | 0/10 (0.0) | 0/10 (0.0) | ||
| Shawu | R. affinis | 2016 | I | 0/21 (0.0) | 0/8 (0.0) | |
| Myotis horsfieldii | 2016 | I | 0/36 (0.0) | 0/23 (0.0) | ||
| Subtotal | 0/197 (0.0) | 3/138 (2.7) | ||||
| Zhejiang | Daishan | Rhinolophus ferrumequinum | 2016 | I | 0/47 (0.0) | 1/47 (2.1) |
| Total | 4/547 (0.7) | 48/448 (10.7) | ||||
Abbreviations: P/T (%), number positive/number tested (percentage); F, frugivorous; I, insectivorous; ND, not done.
Full-genome sequencing and genotype constellations.
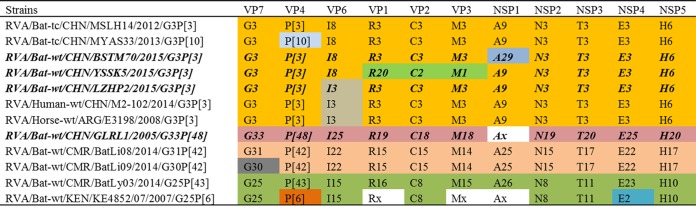
The combination of the contigs generated by viral metagenomics analysis and amplification using degenerate primers allowed us to obtain the near complete genomes of the four above-mentioned strains, except for the NSP1 gene of strain GLRL1. After genotyping using the RotaC online tool (4), NSP1 of BSTM70, VP1 of YSSK5, and the 10 sequenced segments of GLRL1 were proposed as new genotypes, which was further approved by the RCWG. The remaining gene segments fell into previously established genotypes. The complete genotype constellations of the 4 strains are shown in Fig. 1. The 3 strains identified from insectivorous bats in Guangxi Province shared a largely conserved genetic backbone of genotype constellations: G3-P[3]-I8-R3-C3-M3-A9-N3-T3-E3-H6, which is the same as that of strain MSLH14 isolated from a Rhinolophus hipposideros bat in 2012 in Dehong, Yunnan (26), although there were several variations for each strain (Fig. 1). Comparison of these genotype constellations with those of previously known bat RVAs revealed that 10 gene segments of RVA strain LZHP2 (except for VP6), 10 segments of BSTM70 (except for NSP1), and 8 segments of YSSK5 (except for VP1, VP2, and VP3) were shared with the genotype constellation of MSLH14 and with MYAS33 (except for VP4) isolated from an Aselliscus stoliczkanus bat in 2013 in Mengyuan, Yunnan (27). Notably, LZHP2 had the exact same genotype constellation as the human RVA strain M2-102, identified in a diarrheal infant from Sanjiang County, Guangxi, in 2014 (29), and with E3198, detected from feces of a 3-day-old diarrheic foal in 2008 in Argentina (30). However, the fruit bat-originated strain GLRL1, identified in Guangdong Province, did not share a single genotype with any other known RVA strain, representing a completely novel genotype constellation (Fig. 1). None of the strains in this study shared any genotypes with bat RVA strains detected from Cameroon and Kenya (Fig. 1).
FIG 1.
Genotype constellation comparisons of RVAs identified in this study and other known strains. Colors differentiate the individual genotypes, and the strains identified in this study are in bold italics.
Sequence comparison and phylogenetic relationship.
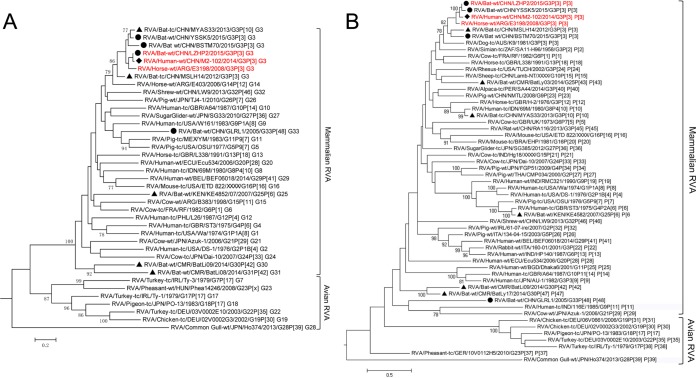
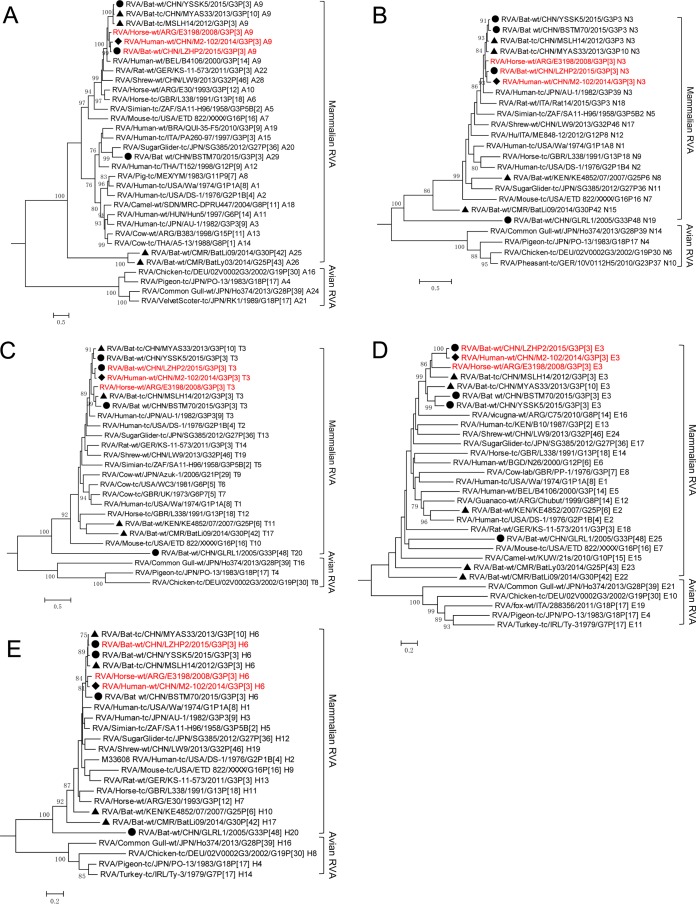
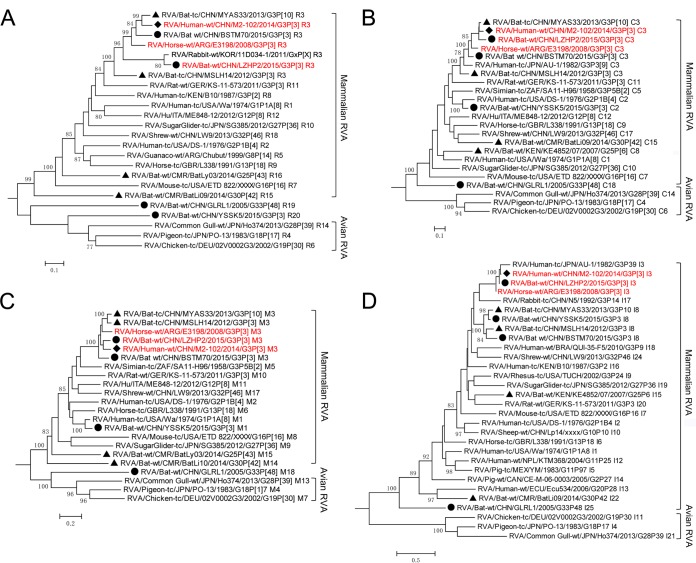
Representatives of all genotypes available in public databases were aligned with the sequences obtained in this study, and maximum likelihood trees of the 11 segments were generated. As previously established for each tree, a mammalian and avian RVA subcluster could be identified according to their hosts' hierarchical classes (Fig. 2 to 4). Generally, Guangdong strain GLRL1 was highly divergent from all currently known RVAs for the 10 obtained segments (NSP1 not obtained by any means) (Fig. 2 to 5). Phylogenetic analyses showed that all its segments except for VP7 and NSP4 were placed close to the roots of the evolutionary trees (Fig. 2 to 4). Intriguingly, VP3 and VP1 of GLRL1, along with VP1 of strain YSSK5, appeared more closely related to avian than to mammalian RVA strains (Fig. 3A and C). Meanwhile, NSP5 of GLRL1 had a larger open reading frame (ORF) than those of other known strains, due to an extra 70-nucleotide (nt) insert at the 3′ terminus.
FIG 2.
Phylogenetic analysis of VP7 (A) and VP4 (B) of RVAs identified in this study (circles), previously reported bat RVAs (triangles), and representatives of each genotype, including human RVA strain M2-102 (diamond). All strains were classified into two major phylogenetic clades representing mammalian and avian RVAs. The G/P genotype of each sequence is indicated at the last part of the strain name. LZHP2, M2-102, and E3198, sharing the same genotype constellation, are in red.
FIG 4.
Phylogenetic analysis of the NSP1 (A), NSP2 (B), NSP3 (C), NSP4 (D), and NSP5 (E) genes of RVAs identified in this study (circles), previously reported batRVAs (triangles), and representatives of each genotype, including human RVA strain M2-102 (diamond). All strains were classified into two major phylogenetic clades representing mammalian and avian RVA strains. The genotype of each sequence is indicated at the last part of the strain name. LZHP2, M2-102, and E3198, sharing the same genotype constellation, are in red.
FIG 5.
Nucleotide identity comparison of each segment of four bat strains in this study between each other and with two other Chinese bat strains (A), as well as with the human M2-102 and horse E3198 strains (B). The identities between strains are color coded according to the scale provided. Black cells indicate that no sequence was available (NA) for comparison.
FIG 3.
Phylogenetic analysis of the VP1 (A), VP2 (B), VP3 (C), and VP6 (D) genes of RVAs identified in this study (circles), previously reported bat RVAs (triangles), and representatives of each genotype, including human RVA strain M2-102 (diamond). All strains were classified into two major phylogenetic clades representing mammalian and avian RVA strains. The genotype of each sequence is indicated at the last part of the strain name. LZHP2, M2-102, and E3198, sharing the same genotype constellation, are in red.
Most segments of the remaining 3 Guangxi strains (BSTM70, YSSK5, and LZHP2), as shown in Fig. 2 to 4, clustered closely with two Yunnan bat strains (MSLH14 and MYAS33), the Guangxi human strain M2-102, and the horse strain E3198, found in Argentina. The only exceptions were VP1, VP2, and VP3 of YSSK5 and NSP1 of BSTM70. YSSK5 VP1 and BSTM70 NSP1 were differentiated as novel genotypes R20 and A29, respectively, and showed distant relationships with currently known RVAs (Fig. 2A and 3A), whereas the VP2 and VP3 genes of YSSK5 showed a close relationship (up to 93% nucleotide identities) with the typical human RVA genotypes C2 and M1, with DS-1 and Wa as their prototype strains (Fig. 3B and C). LZHP2 is a very interesting strain, not only because it has the same genotype constellation as the human strain M2-102 (Fig. 1) but also due to the fact that its VP7, VP4, VP6, NSP3, and NSP4 genes shared nucleotide identities as high as 95% to 98% to these genes of M2-102 (Fig. 5). Furthermore, its VP2 and NSP5 genes were 94% to 98% identical to the corresponding gene segments of bat strain MYAS33, while its VP1 gene shared the highest identity (96%) to that of strain 11D034-1, which is an unusual RVA detected in a rabbit stool in South Korea in 2011, with no other segment available in GenBank (Fig. 2 to 5). Although strain LZHP2 also shares the same genotype constellation as Argentinean horse strain E3198, their degree of genomic relationship was slightly lower than that between LZHP2 and M2-102 (Fig. 2 to 5).
Serological evidence for bat RVA infection.
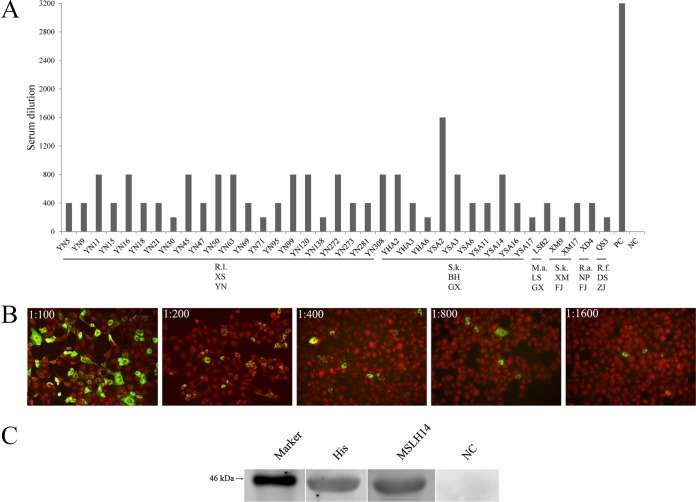
In total, 448 serum samples were collected from 4 provinces, including 142 from R. leschenaultii in Yunnan in 2012, 121 from insectivorous bats in Guangxi in 2015, and 138 and 47 from insectivorous bats in Fujian and Zhejiang in 2016 (Table 1). The indirect immunofluorescence assay (IIFA) screening showed that 48 of 448 sera (10.7%) were antibody (Ab) positive for RVA strain MSLH14; these were collected from 30 (21.1%) R. leschenaultii bats from Yunnan, 13 (26%) S. kuhlii bats from Beihai, 1 (6.7%) Miniopterus australis bat from Lingshan, Guangxi, 2 (10.5%) S. kuhlii bats from Xiamen, 1 (1.7%) Rhinolophus affinis bat from Nanping, Fujian, and 1 (2.1%) Rhinolophus ferrumequinum bat from Daishan, Zhejiang (Table 1). These prevalences were shown to be overall significantly different by χ2 test (χ2 = 30.3303; P < 0.0001). To compare the difference in seroprevalence between provinces, the significance level cutoff was adjusted to 0.0083 by the Bonferroni method with Fisher's exact test. Seroprevalence levels between bats in the western provinces Yunnan and Guangxi were not different (P = 0.0466 and P > 0.0083, respectively), whereas the rates in Yunnan and Guangxi were much higher than in the eastern provinces Fujian and Zhejiang (P < 0.00883). To test differences in Ab titers, we selected 22 positive sera from Yunnan, 11 from Guangxi (10 from Beihai and 1 from Lingshan), 3 from Fujian (2 from Xiamen and 1 from Nanping), and 1 from Daishan, Zhejiang, which had sufficient volumes to perform 2-fold endpoint dilution experiments. Compared to the 1:3,200 titer of the positive control (mouse hyperimmune serum against MSLH14), the titers observed in our study ranged between 1:200 and 1:1,600 (Fig. 6A and B), and only one sample from Beihai (sample code: YAS2) had a titer of 1:1,600 (Fig. 6B).
FIG 6.
(A) Endpoint titers of strain MSHL14-specific Abs in bat sera tested by IIFA. Sample codes are shown on the x axis. PC, positive control; NC, negative control; R.l., Rousettus lechenaultii; S.k., Scotophilus kuhlii; M.a., Miniopterus australis; R.a., Rhinolophus affinis; R.f., Rhinolophus ferrumequinum; XS, Xishuangbanna; BH, Beihai; LS, Lingshan; XM, Xiamen; NP, Nanping; DS, Daishan; YN, Yunnan; GX, Guangxi; FJ, Fujian. (B) A serum sample from Beihai, Guangxi (sample code YSA2), was applied in five different dilutions (range, 1:100 to 1:1,600) for the IIFA analyses. (C) Western blotting analyses of cross-reactivity of MSHL14-specific mouse hyperimmune sera against recombinant GLRL1 VP6. These lanes were taken from different gels. Marker, 46 kDa; His, anti-His tag monoclonal antibody; NC, negative serum control.
Antigenic characterization of VP6 of GLRL1.
RVAs with different G, P, and I genotypes can antigenically cross-react between each other (2). According to the topology of the VP7 phylogenetic trees, GLRL1 clustered among the mammalian RVA strains, whereas VP4 and VP6 were placed rather distinctly from most known mammalian and avian RVAs. Using Western blot (WB) analyses, anti-His antibody detected the GLRL1 VP6 fusion protein (∼46 kDa), indicating correct expression and effective transfer of the proteins to the membrane (Fig. 6C). Furthermore, mouse hyperimmune sera against MSLH14 reacted efficiently with VP6 of GLRL1, indicating significant cross-reactivity (Fig. 6C).
DISCUSSION
Increasing interest in bat RVAs has emerged recently. Based on viral RNA detection, 3 of 39 (7.7%) fecal swabs of E. helvum from Kenya and 5 of 24 pools containing 87 bat fecal samples in Cameroon were positive for RVA (25, 28). In China, 1.1% to 6.3% of fecal swabs or gut tissues of bats sampled in Yunnan, Guangxi, and Guangdong were positive for RVA RNA (26, 27), while samples collected from Fujian and Zhejiang were negative (Table 1). In this study, we conducted an investigation into the serology of RVA infections in bats using an IIFA, with MSLH14 as an antigenic target. Forty-eight out of 448 (10.7%) sera tested positive, which was significantly higher than the results of RNA detection in our study (total rate, 0.7% [Table 1]). As a diagnostic tool, therefore, it appears that RNA detection significantly underestimates the true prevalence of RVA in the bat population. Furthermore, all RNA-tested samples were from apparently health adult bats, which supposedly have robust immune competence and can clear viruses efficiently following infection (14). Unfortunately, no neonates were involved in this study, and therefore, we could not assess the outcomes and prevalence of RVA infection in young bats.
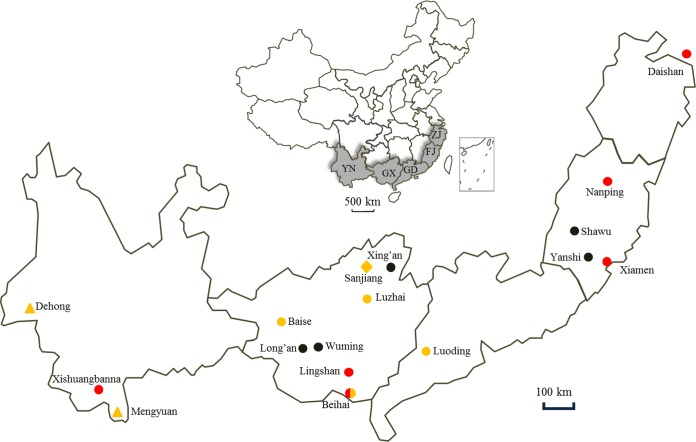
To further understand the difference in RVA seroprevalence in the investigated provinces, the serology data were compared; the IIFA results showed that RVA infections in bats vary in prevalence depending on the province. Infections appear to be more prevalent in Guangxi (11.6%) and Yunnan (21.1%) than in more eastern provinces (2.7% in Fujian and 2.1% in Zhejiang) (Table 1). However, a more reliable assessment of RVA infections in Yunnan bats will require further investigation over a wider geographical area as well as in a broader range of bat species, since only one species of bat was involved per site in this study.
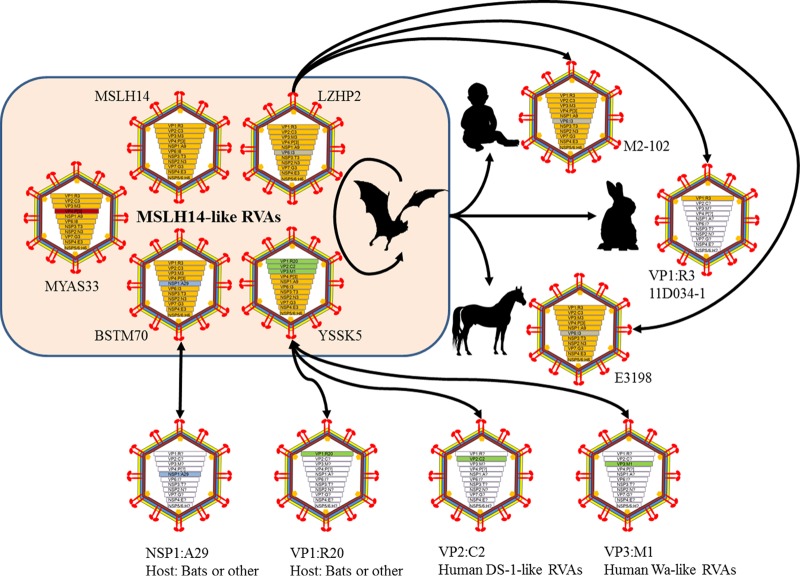
The present study identified another four novel RVAs in Chinese bats, which significantly improved our understanding of the genetic diversity of RVAs in Chinese bat populations as well as their relationship with human and other animal RVAs. Currently only five RVA strains have been identified in insectivorous bats in China: MSLH14 and MYAS33 in Yunnan and BSTM70, YSSK5, and LZHP2 in Guangxi (26, 27). Despite the fact that all these 5 strains were identified from locations at least 300 km apart (Fig. 7), they shared a similar genotype constellation, G3-P[3]-I8-R3-C3-M3-A9-N3-T3-E3-H6, referred to as “MSLH14-like” (Fig. 1), suggesting that MSLH14-like RVAs are true bat viruses widely distributed in insectivorous bat populations in this region.
FIG 7.
Map showing the locations of bats collected in this study (circles), previously reported bat RVAs (triangles), and human RVA strain M2-102 (diamond). Red circles indicate that the bats were positive by serological investigation, while the yellow ones indicate positivity for RVA RNA. Abbreviations for provinces: YN, Yunnan; GX, Guangxi; GD, Guangdong; FJ, Fujian; ZJ, Zhejiang.
One of the most striking findings in this study is that strain LZHP2, identified from an insectivorous bat sampled in a cave very close to a village, had exactly the same genotype constellations as the unique human strain M2-102, sharing very high nucleotide identities for most of their genome segments (Fig. 5). M2-102 was previously identified as the pathogen causing diarrhea in a 3-year-old boy in 2014 in Sanjiang, Guangxi, which is only 120 km away from the location (Luzhai) where LZHP2 was detected (Fig. 7) (29). Because strain M2-102 was highly divergent from all known human RVA strains, our data strongly suggest that this strain was the result of a cross-species transmission of an MSLH14-like bat RVA to the child. Our previous study also showed that MSLH14 was highly lethal to suckling mice, suggesting pathogenic properties in heterologous animal species (26). This finding has added bats to a long list of animal species for which it has been shown that their RVA strains are able to infect humans (Fig. 8) (31). Also very interesting is that the same genotype constellation was found in the Argentinean horse strain E3198, which, at that time, was shown to be distantly related to feline and canine RVA strains (30). However, our data suggest that the unusual equine RVA strain E3198 might also be the result of a cross-species transmission of an MSLH14-like bat RVA strain to a horse (Fig. 8). As strain E3198 was detected from a foal, without any history of travel, this would suggest that MSLH14-like bat RVA strains should also be present in South American bat populations. Regrettably, no complete genomes of South American bat RVA strains have been described to date. Our analyses also surprisingly showed that the VP1 gene of strain LZHP2 shared 96% identity with the corresponding gene of South Korean rabbit RVA strain 11D034-1, in comparison to only 90% or lower for other known RVAs (Fig. 3A). This might represent yet another example of a cross-species transmission event from an MSLH14-like bat RVA to a rabbit, although the rest of the Korean rabbit RVA genome should be determined to shed further light on this speculation (Fig. 8).
FIG 8.
Interspecies transmission and reassortment of MSLH14-like bat RVAs between bats and humans or other animals. Currently, 5 bat strains can be referred to as MSLH14-like, with MSLH14 as the prototype. The members of MSLH14-like bat RVAs can reassort between each other and cross-species transmit to humans (M2-102), rabbits (11D034-1), and horses (E3198) or reassort with human DS-1-like and Wa-like RVA strains. The NSP1 gene of BSTM70 and the VP1 gene of YSSK5 might suggest additional reassortments with other RVAs with unknown hosts. Genotypes within the virus diagrams are differentiated by color.
Bat RVA strains BSTM70 also possessed the MSLH14-like genotype constellation, with the only exception of a novel A29 genotype for NSP1, further expanding the genetic diversity for bat RVA strains in China (Fig. 1). Strain YSSK5 was identified from a palm-roosted bat in Beihai coast. It also contained 8 gene segments belonging to the MSLH14-like genotype constellations (Fig. 1). Its VP1 gene was shown to contain yet another novel genotype: R20. However, the observation that the VP2 and VP3 gene segments of YSSK5 were closely related to cogent genes of the typical human genotypes C2 and M1, respectively, was striking. Human strains possessing genotypes C2 and M1 are usually referred to as DS-1-like and Wa-like RVAs, respectively, and have been successful in sustaining infection of humans worldwide for as long as scientists have studied human RVAs (9). These genotypes have also been found in Chinese children (32, 33), suggesting the occurrence of one or more reassortment events between human and bat RVAs, which must have been preceded by additional interspecies transmission events in the past (Fig. 8). This finding adds further evidence for the role bats might play as a reservoir for pathogenic human RVA strains (Fig. 8).
As the only RVA strain from Chinese fruit bats known to date, GLRL1 possessed a genotype constellation distinct from that of any known RVA and was therefore assigned as the prototype species for multiple novel genotypes (Fig. 1). Regrettably, we were unable to obtain the NSP1 gene segment of this strain. Eight (VP7, VP4, VP2, VP6, and NSP2 to -5) of the obtained 10 segments of GLRL1 were more closely related to the cluster of mammalian RVA strains (Fig. 2 to 4), and the VP4, VP2, VP6, NSP2, NSP3, and NSP5 genes were located at the root of the mammalian RVA cluster (Fig. 2 to 4), a feature which was also observed for the VP7, VP1, VP3, NSP1, and NSP4 gene segments of the Cameroonian bat RVA strains BatLy03 and BatLi10 (28) (Fig. 2 to 4). This observation suggests that some of the bat RVA strains might have diverged from other mammalian RVAs a very long time ago. Of note is that the VP1 segments of GLRL1 and YSSK5 and VP3 of GLRL1 were phylogenetically more closely related to avian RVAs than to mammalian RVA strains (Fig. 3A and C), suggesting a complicated evolutionary history of bat RVAs in China and the rest of the world. However, due to the vast amount of known bat species and the very limited number of locations and species investigated, the currently observed diversity represents without a doubt only the tip of the iceberg. Despite the genetic difference of strain GLRL1 from known RVA strains, WB analysis showed cross-reactivity of MSLH14-specific Ab against VP6 of GLRL1, suggesting that GLRL1 is a true member of the rotavirus species A, since serological cross-reactivity using either polyclonal sera or monoclonal antibodies against VP6 is an established criterion for rotavirus species demarcation (34) (Fig. 6C).
RVA infections of children in China and the rest of the world pose a huge health and financial burden on their families and the communities (35, 36). Surveillance of RVA diarrhea in China showed that RVA was the dominant pathogen causing diarrhea in young children, accounting for nearly 50% of the cases, with the G3 genotype being most commonly identified (37). Other studies in China found that RVAs were prevalent in animals such as pigs, rabbits, and rodents (38–41) and animal-associated RVAs in humans (32, 42). Evidence for interspecies transmission and for genetic reassortment between human and animal rotaviruses was illustrated in recent years, and in particular, some animal species (cows, pigs, cats, and dogs) appear to contribute frequently to the antigenic/genetic diversity found in human RVAs (6, 13, 32, 42–46), presumably because of the close interactions between animals and their physical proximity. The present study highlights the role of bats as reservoirs of RVAs and their bat-to-human cross-species transmission as well as interspecies reassortment (Fig. 8). Continuous epidemiological surveillance is critical for understanding the short- and long-term effects as well as control and prevention of the spread of bat RVAs to humans.
MATERIALS AND METHODS
Ethics statement.
The procedures for sampling of bats in this study were reviewed and approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary, Academy of Military Medical Sciences, China (Laboratory Animal Care and Use Committee Authorization, permit number JSY-DW-2015-01). All live bats were maintained and handled according to the principles and guidelines for laboratory animal medicine (2006) of the Ministry of Science and Technology, China.
Sample collection and preparation.
Adult bats were live-captured with nets near or in human-inhabited communities between 2005 and 2016 in 5 provinces of South China (Fig. 7). All bats were apparently healthy at capture. About 100 μl of blood from every bat was sampled by wing vein puncture, and serum was collected after blood clotting. Anal specimens were collected using sterile swabs and immediately transferred to viral transport medium (VTM; Earle's balanced salt solution, 0.2% sodium bicarbonate, 0.5% bovine serum albumin, 18 μg/liter of amikacin, 200 μg/liter of vancomycin, 160 U/liter of nystatin) and stored in liquid nitrogen prior to transport to the laboratory, where they were stored at −80°C. All captured bats were released after sample collection, with the exception of 38 bats from Guangdong in 2005, which were euthanized humanely, with their guts (along with the contents) and lungs collected and transported to the laboratory as described above. Bat sample details are listed in Table 1.
RVA RNA screening.
Anal specimens as well as gut contents of bats from each location were pooled and subjected to viral metagenomic analysis as per our published method (22). Seminested RT-PCR primers targeting the entire NSP5 segment were designed based on all known genotypes of NSP5. First-round PCR was carried out with a mixture of forward primer 5′-AAAGCRCTACMGTGATGTC-3′ and reverse primer 5′-GGTCACAAAMGGRAGTGG-3′; seminested PCR was carried out with a mixture of forward primer 5′-CTGGAAAATCTRTTRGTAGG-3′ and the same reverse primer as used in the first-round PCR. Total RNA of each anal swab and gut was extracted automatically using the RNeasy minikit (Qiagen) in a QIAcube (Qiagen). Reverse transcription was performed with the 1stcDNA synthesis kit (TaKaRa) according to the manufacturer's protocol. The cDNA was amplified using PCR master mix (Tiangen) with the following PCR programs: 30 cycles (outer PCR) or 35 cycles (inner PCR) of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 40 s, with double-distilled water (ddH2O) as a negative control. Positive PCR amplicons were ligated into pMD18T vectors (TaKaRa) and used to transfect competent Escherichia coli DH5α cells (Tiangen). Five clones of each amplicon were randomly picked for sequencing by the Sanger method on an ABI 3730 sequencer (ComateBio).
Full-genome sequencing.
To obtain the full genomes of the identified RVA-positive samples, the RVA contigs generated by viral metagenomic sequencing were further confirmed by Sanger sequencing. The previously developed degenerate PCR primer pairs used to amplify MSLH14 and MYAS33 were slightly modified to target the near full lengths of all 11 segments in this study (26, 27). Viral cDNA was prepared as described above directly from positive samples and amplified using the Fast HiFidelity PCR kit (Tiangen). The amplicons were sequenced after blunt ligation into the pZeroBack vector (Tiangen). Overlapping amplicons were assembled with SeqMan v.7.0 into full genomic sequences.
Nucleotide sequence and maximum likelihood phylogenetic analysis.
To identify the genotypes of different segments of these viruses, all segments were genotyped using the online tool RotaC or after consultation with the RCWG (4, 5, 7). Representatives of each genotype of 11 segments were retrieved from GenBank and aligned with sequences in this study using ClustalW v.2.0, and nucleotide identities were calculated using MegAlign. The best-fit nucleotide substitution models were determined using Model Generator. For VP1, VP2, VP3, and NSP1, the HKY+ Γ+ I model was selected. For VP4, VP6, NSP2, and NSP3, the GTR+ Γ model was selected, whereasVP7, NSP4, and NSP5 were analyzed using the HKY+ Γ, the HKY, and the GTR+ Γ+ I models, respectively. Phylogenetic reconstructions were performed using MEGA v.6 and the maximum likelihood method with 1,000 bootstrap replicates (47).
Serological analyses.
To detect bat antibodies (Abs) against RVA, an indirect immunofluorescence assay (IIFA) was performed according to a previous protocol, with slight modifications (26). Briefly, Marc-145 cells (embryonic rhesus monkey kidney cells) were infected with 200 50% tissue culture infective doses (TCID50) of RVA strain MSLH14 and washed three times with sterile phosphate-buffered saline (PBS) after 18 h. Following fixation with cold acetone, cells were incubated with bat serum at a 1:100 dilution at 37°C for 1 h, followed by three washes with PBS-Tween (PBST). Fluorescein isothiocyanate (FITC)-labeled proteins A and G (Abcam) equally mixed at a 1:1,000 dilution were used to detect the primary antibodies. Positive and negative mouse sera against MSLH14 were used as controls. Analysis was performed with an immunofluorescence microscope (Zeiss). The Ab titers of the bat sera with a sufficient volume were determined by endpoint titrations using serial 2-fold dilutions (1:100 to 1:3,200).
To determine antigenic cross-reactivity of the novel bat RVA strain GLRL1 with other RVAs, its VP6 entire open reading frame (subgroup antigen) was amplified using HiFidelity polymerase (Tiangen) and ligated into pET-28a with a His tag at its C terminus. The fusion protein was expressed in E. coli BL21 cells, preliminarily purified using high-affinity nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen), and subsequently subjected to Western blot (WB) analysis using mouse hyperimmune serum against strain MSLH14 (26). A total of 10 μg of protein was boiled in 2× loading buffer (Tiangen) for 10 min, separated by 12% SDS-PAGE, and transferred onto a nitrocellulose membrane (Millipore). The blocked membrane was then incubated with mouse antibodies against MLSH14 and His at a 1:1,000 dilution, followed by IRDye 800CW goat anti-mouse secondary antibody (1:5,000). The washed membrane was then scanned in an Odyssey infrared imaging system (LI-COR Bioscience) at 700-nm and 800-nm wavelengths to detect target proteins.
Statistical analyses.
A χ2 test was used to generally analyze the seroprevalence results from IIFA. Fisher's exact test was employed to compare differences between provinces, with significance level adjusted by the Bonferroni method. The serum titers were not normally distributed, so the differences between them were compared using a Wilcoxon rank sum test. All data processes were conducted using Statistical Analysis System v.9.2 (SAS).
Accession number(s).
All sequences generated in this study have been deposited in GenBank under accession numbers KX814921 to KX814963.
ACKNOWLEDGMENTS
This study was supported by the General Program of the National Natural Science Foundation of China (31572529), the National Key Basic Research and Development Program of China (2016YFC1200100), the Youth Innovation Fund of the Academy of Military Medical Sciences (2015CXJJ28), and Post-doctoral Special Assistance of China (2016T91011).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, Agocs M, Serhan F, de Oliveira L, Mwenda JM, Mihigo R, Ranjan Wijesinghe P, Abeysinghe N, Fox K, Paladin F. 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 62:S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 2.Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maes P, Matthijnssens J, Rahman M, Van Ranst M. 2009. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.RCWG. 2016. Newly assigned genotypes. https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg.
- 8.Ghosh S, Kobayashi N. 2011. Whole-genomic analysis of rotavirus strains: current status and future prospects. Future Microbiol 6:1049–1065. doi: 10.2217/fmb.11.90. [DOI] [PubMed] [Google Scholar]
- 9.Matthijnssens J, Van Ranst M. 2012. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr Opin Virol 2:426–433. doi: 10.1016/j.coviro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Matthijnssens J, Potgieter CA, Ciarlet M, Parreno V, Martella V, Banyai K, Garaicoechea L, Palombo EA, Novo L, Zeller M, Arista S, Gerna G, Rahman M, Van Ranst M. 2009. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J Virol 83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HH, Matthijnssens J, Kim HJ, Kwon HJ, Park JG, Son KY, Ryu EH, Kim DS, Lee WS, Kang MI, Yang DK, Hyun BH, Park SI, Park SJ, Cho KO. 2012. Full-length genomic analysis of porcine G9P[23] and G9P[7] rotavirus strains isolated from pigs with diarrhea in South Korea. Infect Genet Evol 12:1427–1435. doi: 10.1016/j.meegid.2012.04.028. [DOI] [PubMed] [Google Scholar]
- 12.Matthijnssens J, Mino S, Papp H, Potgieter C, Novo L, Heylen E, Zeller M, Garaicoechea L, Badaracco A, Lengyel G, Kisfali P, Cullinane A, Collins PJ, Ciarlet M, O'Shea H, Parreno V, Banyai K, Barrandeguy M, Van Ranst M. 2012. Complete molecular genome analyses of equine rotavirus A strains from different continents reveal several novel genotypes and a largely conserved genotype constellation. J Gen Virol 93:866–875. doi: 10.1099/vir.0.039255-0. [DOI] [PubMed] [Google Scholar]
- 13.Matthijnssens J, De Grazia S, Piessens J, Heylen E, Zeller M, Giammanco GM, Banyai K, Buonavoglia C, Ciarlet M, Martella V, Van Ranst M. 2011. Multiple reassortment and interspecies transmission events contribute to the diversity of feline, canine and feline/canine-like human group A rotavirus strains. Infect Genet Evol 11:1396–1406. doi: 10.1016/j.meegid.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moratelli R, Calisher CH. 2015. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem Inst Oswaldo Cruz 110:1–22. doi: 10.1590/0074-02760150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol 84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shi Z. 2012. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol 86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, Qian Z, Dong J, Sun L, Zhu Y, Du J, Yang F, Zhang S, Jin Q. 2015. Deciphering the bat virome catalog to better understand tha ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 2015:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B, Fan Q, Yang F, Hu T, Qiu W, Feng Y, Li Z, Li Y, Zhang F, Guo H, Zou X, Tu C. 2013. Hepatitis virus in long-fingered bats, Myanmar. Emerg Infect Dis 19:638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, Li Y, Wang Y, Su N, Zhang F, Fan Q, Tu C. 2013. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS One 8:e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen L-M, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esona MD, Mijatovic-Rustempasic S, Conrardy C, Tong S, Kuzmin IV, Agwanda B, Breiman RF, Banyai K, Niezgoda M, Rupprecht CE, Gentsch JR, Bowen MD. 2010. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis 16:1844–1852. doi: 10.3201/eid1612.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He B, Yang F, Yang W, Zhang Y, Feng Y, Zhou J, Xie J, Feng Y, Bao X, Guo H, Li Y, Xia L, Li N, Matthijnssens J, Zhang H, Tu C. 2013. Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses. J Virol 87:12357–12366. doi: 10.1128/JVI.02013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia L, Fan Q, He B, Xu L, Zhang F, Hu T, Wang Y, Li N, Qiu W, Zheng Y, Matthijnssens J, Tu C. 2014. The complete genome sequence of a G3P[10] Chinese bat rotavirus suggests multiple bat rotavirus inter-host species transmission events. Infect Genet Evol 28:1–4. doi: 10.1016/j.meegid.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Yinda CK, Zeller M, Conceição-Neto N, Maes P, Deboutte W, Beller L, Heylen E, Ghogomu SM, Van Ranst M, Matthijnssens J. 2016. Novel highly divergent reassortant bat rotaviruses from bats in Cameroon, without evidence of zoonosis. Sci Rep 6:34209. doi: 10.1038/srep34209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Qian Y, Nong Y, Zhang Y, Mo Z, Li R. 2016. Genomic characterization of an unusual human G3P[3] rotavirus with multiple cross-species reassortment. Chin J Virol 32:129–140. [PubMed] [Google Scholar]
- 30.Miño S, Matthijnssens J, Badaracco A, Garaicoechea L, Zeller M, Heylen E, Van Ranst M, Barrandeguy M, Parreño V. 2013. Equine G3P[3] rotavirus strain E3198 related to simian RRV and feline/canine-like rotaviruses based on complete genome analyses. Vet Microbiol 161:239–246. doi: 10.1016/j.vetmic.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh S, Kobayashi N. 2014. Exotic rotaviruses in animals and rotaviruses in exotic animals. Virus Dis 25:158–172. doi: 10.1007/s13337-014-0194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong H-J, Qian Y, Huang T, Zhu R-N, Zhao L-Q, Zhang Y, Li R-C, Li Y-P. 2013. Identification of circulating porcine-human reassortant G4P[6] rotavirus from children with acute diarrhea in China by whole genome analyses. Infect Genet Evol 20:155–162. doi: 10.1016/j.meegid.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Wen Y, Liu X, Xiong X, Cao Z, Zhao Q, Yu Y, Yin X, Li C, Fan Y. 2008. Full genomic analysis of human rotavirus strain TB-Chen isolated in China. Virology 375:361–373. doi: 10.1016/j.virol.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, Dermody TS, Dormitzer PR, Duncan R, Fang Q, Graham R, Grulielmi KM, Harding RM, Hillman B, Makkey A, Marzachi C, Matthijinssens J, Milne RG, Mohd Jaafar F, Mori H, Noordeloos AA, Omura T, Patton JT, Rao S, Maan M, Stoltz D, Suzuki N, Upadhyaya NM, Wei C, Zhou H. 2012. Family Reoviridae, p 541–637. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom. [Google Scholar]
- 35.Wu D, Yen C, Yin Z-D, Li Y-X, Liu N, Liu Y-M, Wang H-Q, Cui F-Q, Gregory CJ, Tate JE, Parashar UD, Yin D-P, Li L. 2016. The public health burden of rotavirus disease in children younger than five years and considerations for rotavirus vaccine introduction in China. Pediatr Infect Dis J 35:e392–. doi: 10.1097/INF.0000000000001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natchu UCM, Bhatnagar S. 2013. Diarrhoea in children: identifying the cause and burden. Lancet 382:184–186. doi: 10.1016/S0140-6736(13)60941-1. [DOI] [PubMed] [Google Scholar]
- 37.Duan ZJ, Liu N, Yang SH, Zhang J, Sun LW, Tang JY, Jin Y, Du ZQ, Xu J, Wu QB, Tong ZL, Gong ST, Qian Y, Ma JM, Liao XC, Widdowson MA, Jiang B, Fang ZY. 2009. Hospital-based surveillance of rotavirus diarrhea in the People's Republic of China, August 2003–July 2007. J Infect Dis 200(Suppl 1):S167–S173. doi: 10.1086/605039. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Hu R, Tang X, Wu C, He Q, Zhao Z, Chen H, Wu B. 2013. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch Virol 158:1631–1636. doi: 10.1007/s00705-013-1659-x. [DOI] [PubMed] [Google Scholar]
- 39.Shi H, Chen J, Li H, Sun D, Wang C, Feng L. 2012. Molecular characterization of a rare G9P[23] porcine rotavirus isolate from China. Arch Virol 157:1897–1903. doi: 10.1007/s00705-012-1363-2. [DOI] [PubMed] [Google Scholar]
- 40.Guo D, Liu J, Lu Y, Sun Y, Yuan D, Jiang Q, Lin H, Li C, Si C, Qu L. 2012. Full genomic analysis of rabbit rotavirus G3P[14] strain N5 in China: identification of a novel VP6 genotype. Infect Genet Evol 12:1567–1576. doi: 10.1016/j.meegid.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Li K, Lin X-D, Huang K-Y, Zhang B, Shi M, Guo W-P, Wang M-R, Wang W, Xing J-G, Li M-H, Hong W-S, Holmes EC, Zhang Y-Z. 2016. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology 494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YH, Pang BB, Zhou X, Ghosh S, Tang WF, Peng JS, Hu Q, Zhou DJ, Kobayashi N. 2013. Complex evolutionary patterns of two rare human G3P[9] rotavirus strains possessing a feline/canine-like H6 genotype on an AU-1-like genotype constellation. Infect Genet Evol 16:103–112. doi: 10.1016/j.meegid.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee A, Dutta D, Ghosh S, Bagchi P, Chattopadhyay S, Nagashima S, Kobayashi N, Dutta P, Krishnan T, Naik TN, Chawla-Sarkar M. 2009. Full genomic analysis of a human group A rotavirus G9P[6] strain from Eastern India provides evidence for porcine-to-human interspecies transmission. Arch Virol 154:733–746. doi: 10.1007/s00705-009-0363-3. [DOI] [PubMed] [Google Scholar]
- 44.Tsugawa T, Hoshino Y. 2008. Whole genome sequence and phylogenetic analyses reveal human rotavirus G3P[3] strains Ro1845 and HCR3A are examples of direct virion transmission of canine/feline rotaviruses to humans. Virology 380:344–353. doi: 10.1016/j.virol.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant L, Esona M, Gentsch J, Watt J, Reid R, Weatherholtz R, Santosham M, Parashar U, O'Brien K. 2011. Detection of G3P[3] and G3P[9] rotavirus strains in American Indian children with evidence of gene reassortment between human and animal rotaviruses. J Med Virol 83:1288–1299. doi: 10.1002/jmv.22076. [DOI] [PubMed] [Google Scholar]
- 46.Ben Hadj Fredj M, Heylen E, Zeller M, Fodha I, Benhamida-Rebai M, Van Ranst M, Matthijnssens J, Trabelsi A. 2013. Feline origin of rotavirus strain, Tunisia, 2008. Emerg Infect Dis 19:630–634. doi: 10.3201/eid1904.121383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]