ABSTRACT
Although a varicella-zoster virus (VZV) vaccine has been used for many years, the neuropathy caused by VZV infection is still a major health concern. Open reading frame 7 (ORF7) of VZV has been recognized as a neurotropic gene in vivo, but its neurovirulent role remains unclear. In the present study, we investigated the effect of ORF7 deletion on VZV replication cycle at virus entry, genome replication, gene expression, capsid assembly and cytoplasmic envelopment, and transcellular transmission in differentiated neural progenitor cells (dNPCs) and neuroblastoma SH-SY5Y (dSY5Y) cells. Our results demonstrate that the ORF7 protein is a component of the tegument layer of VZV virions. Deleting ORF7 did not affect viral entry, viral genome replication, or the expression of typical viral genes but clearly impacted cytoplasmic envelopment of VZV capsids, resulting in a dramatic increase of envelope-defective particles and a decrease in intact virions. The defect was more severe in differentiated neuronal cells of dNPCs and dSY5Y. ORF7 deletion also impaired transmission of ORF7-deficient virus among the neuronal cells. These results indicate that ORF7 is required for cytoplasmic envelopment of VZV capsids, virus transmission among neuronal cells, and probably the neuropathy induced by VZV infection.
IMPORTANCE The neurological damage caused by varicella-zoster virus (VZV) reactivation is commonly manifested as clinical problems. Thus, identifying viral neurovirulent genes and characterizing their functions are important for relieving VZV related neurological complications. ORF7 has been previously identified as a potential neurotropic gene, but its involvement in VZV replication is unclear. In this study, we found that ORF7 is required for VZV cytoplasmic envelopment in differentiated neuronal cells, and the envelopment deficiency caused by ORF7 deletion results in poor dissemination of VZV among neuronal cells. These findings imply that ORF7 plays a role in neuropathy, highlighting a potential strategy to develop a neurovirulence-attenuated vaccine against chickenpox and herpes zoster and providing a new target for intervention of neuropathy induced by VZV.
KEYWORDS: varicella-zoster virus, ORF7, differentiated neural progenitor cells, differentiated SH-SY5Y cells, cytoplasmic envelopment
INTRODUCTION
Varicella-zoster virus (VZV) has the smallest double-stranded DNA genome among the members of human herpesvirus family and encodes at least 70 open reading frames (ORFs) (1). This highly cell-associated virus follows various infection processes and induces differential pathological reactions in distinct tissues (2). During primary infection, VZV progenies are released into the blood circulation system, where T cells serve as the vector, causing chickenpox (3). The viral particles are then retrogradely transported from the sensory nerve terminus to cell bodies in the sensory ganglia, where VZV establishes lifelong persistence/latency (4). Conditionally, the latent VZV reactivates in ganglia, anterogradely spreads to the innervated skin, causing herpes zoster (4). Although productive infection of VZV in skin and circulation system can be controlled by antiviral medications, no effective therapy has been developed to treat complications related to VZV neuronal infection, particularly postherpetic neuralgia (4–6). Understanding the functions of VZV neurotropic gene(s) can facilitate an understanding of the mechanisms of VZV-induced neuropathy, leading to the development of interventional strategies. A previous study identified VZV ORF7 as a neurotropic factor required for neuronal infection in human dorsal root ganglia (DRG), the most common site for VZV latency and reactivation (7). However, the role of the ORF7 protein (pORF7) in VZV replication remains unknown.
A typical VZV replication cycle consists of the following steps: virus entry and uncoating, viral gene expression and genome replication, nucleocapsid assembly and egress from nucleus to cytoplasm, virion assembly in the Golgi apparatus, and virion release and transcellular transmission from host cell to neighboring cells (8–10). The VZV genome encodes at least 20 tegument proteins with regulatory function or tegumentation (11). The functions of these tegument proteins are normally relevant to their subcellular localization. For instance, pORF62, pORF63, and pORF10 are immediate early regulatory proteins locating within nuclei (12, 13); pORF11 presents in both the nuclei and cytoplasm, defined as an RNA binding protein, and deletion of ORF11 causes lower expression of several immediate early proteins (14); pORF12 localizes in the cytoplasm and indirectly affects cell cycle through the phosphatidylinositol 3-kinase/Akt pathway (15); and phosphorylation of pORF9 is critical for the egress and maturation of VZV particles (16, 17). pORF7 is a putative tegument protein localizing in the Golgi apparatus (7). Homologues of pORF7 include the herpes simplex virus 1 (HSV-1) protein UL51 (pUL51) and pseudorabies virus (PRV) pUL51. HSV-1 pUL51 is a phosphoprotein with a molecular mass of ∼30 kDa and presents N-terminal palmitoylation-dependent Golgi localization (18). The UL51 deletion HSV-1 mutant shows defective nucleocapsid egress and cytoplasmic envelopment (19). Defective cytoplasmic envelopment is also observed in UL51 deficient PRV-infected kidney cells (20).
In this study, we used distinct cells—including epithelium ARPE-19 cells, neural progenitor cells (NPCs), differentiated NPCs (dNPCs), neuroblastoma SH-SY5Y cells (SY5Y), and differentiated SH-SY5Y cells (dSY5Y)—to decode the function of pORF7. We analyzed the effect of ORF7 deletion on viral replication processes, including viral entry, viral gene expression and genome replication, viral particle assembly and envelopment, and intercellular viral spread. The results showed that pORF7 participates in VZV cytoplasmic envelopment in neuronal cells which could be a target for the attenuation of VZV neurovirulence.
RESULTS
ORF7 deletion attenuates VZV replication.
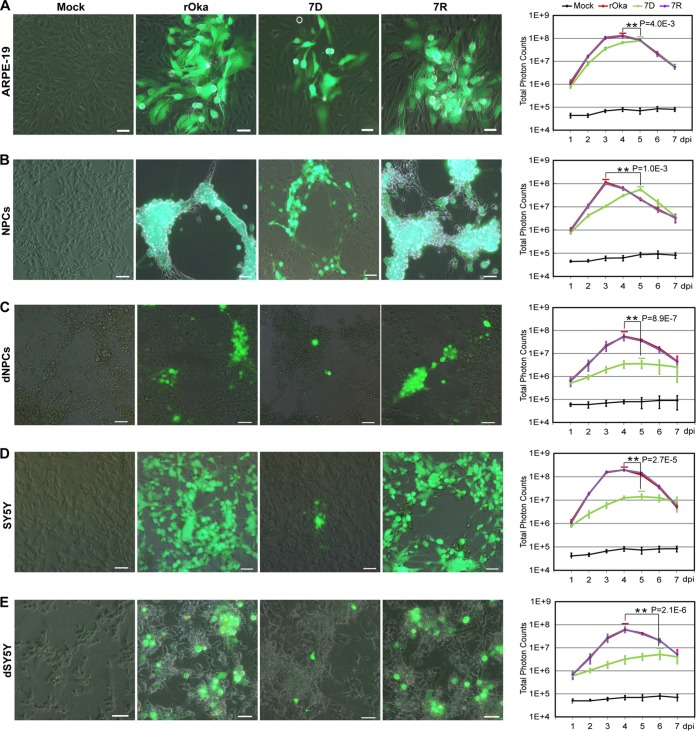
To comprehensively evaluate the role of ORF7 in VZV replication, the growth curves of VZV-rOka (rOka), VZV-7D (7D), and VZV-7R (7R) in ARPE-19 cells, NPCs, dNPCs, SY5Y cells, and dSY5Y cells were determined based on luciferase activity (Fig. 1). In the infected ARPE-19 cells, rOka and 7R displayed identical growth kinetics with the same peak value, whereas 7D showed a significantly lower peak level after a 24-h delay (Fig. 1A). NPCs and dNPCs were fully permissive for VZV infection, and the replication levels of rOka and 7R in both cells were similar. 7D replicated in NPCs, reaching a lower peak with a 48-h delay (Fig. 1B), but 7D replication was significantly impaired in dNPCs (Fig. 1C). SY5Y and dSY5Y cells fully supported rOka and 7R infection, but 7D presented a significantly lower and lagged replication without a clear peak (Fig. 1D), and the replication deficiency was more severe in dSY5Y than in SY5Y (Fig. 1E). Taken together, rOka and 7R presented almost identical growth kinetics in all of the cells tested, replication of 7D was delayed or attenuated, and more significant effects were observed in dNPCs and dSY5Y. These data confirmed a neurotropic character of ORF7 and indicated that certain steps in the VZV replication cycle are influenced by ORF7 deletion, especially in differentiated neuronal cells.
FIG 1.
Growth of rOka, 7D, and 7R in different cell types. (A to E) Growth curves in ARPE-19 cells (A), NPCs (B), dNPCs (C), SY5Y cells (D), and differentiated SH-SY5Y (dSY5Y) cells (E). Cultured cells (3.5 × 105 cells per 35-mm dish) were infected with cell-free rOka, 7D, and 7R, respectively, at an MOI of 0.001. Images of infected cells were captured at 3 dpi are shown (left panel). Scale bar, 50 μm. Growth curves of rOka, 7D, and 7R were generated based on the total photon counts of luciferase bioluminescence. The virus replication levels were measured every 24 h by adding d-luciferin into the dishes and recording the bioluminescence signal with an in vivo fluorescence imaging system (IVIS) system. The total photon count values (photons/s/cm2/steradian) for each group at the indicated time points were collected, and the average was obtained from three independent experiments. The results are represented as averages ± the SD. Significant differences in the replication peak levels between rOka and 7D were observed in all cell groups (**, P < 0.01).
VZV is a highly cell-associated virus, and its spread mainly depends on cell-cell transmission (8). At 3 days postinfection (dpi), the plaques and cytopathic effects (CPEs) were similar for rOka and 7R, but 7D formed smaller plaques with fewer green fluorescent protein (GFP)-positive cells in ARPE-19 cells, NPCs, and SY5Y cells compared to rOka. Since the cell morphologies of ARPE-19 cells, NPCs, and SY5Y cells are different, and the virus growth is also various, distinct plaques and CPEs induced by rOka, 7R, and 7D were therefore observed (Fig. 1A, B, and D). More interestingly, there were no distinct plaques and CPEs appearing in 7D-infected dNPCs and dSY5Y cells compared to the rOka infection (Fig. 1 C and E). These data indicated that ORF7 deletion clearly affects virus transmission in differentiated neuronal cells.
ORF7 deletion impairs VZV transportation in differentiated neuronal cells.
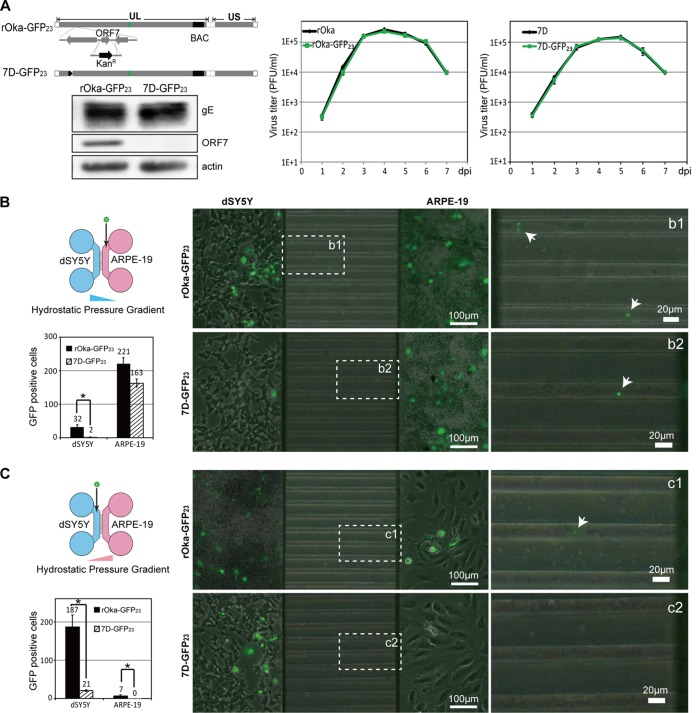
To visualize the transportation of viral particles and identify the effect of ORF7 deletion on VZV transmission, viruses with small capsid protein ORF23 fused with GFP were applied. 7D-GFP23 (an ORF7 deletion mutant) was generated from VZV GFP-ORF23 (designated rOka-GFP23) (Fig. 2A, left upper panel), and the absence of pORF7 in 7D-GFP23 was verified by Western blotting (Fig. 2A, left lower panels). The growth of rOka, rOka-GFP23, 7D, and 7D-GFP23 was determined by plaque-forming assay, but no significant differences in growth kinetics were observed between rOka-GFP23 and rOka or between 7D-GFP23 and 7D (Fig. 2A, right panel).
FIG 2.
Transcellular transmission of VZV. (A) Construction and growth evaluation of 7D-GFP23. The entire ORF7 of rOka-GFP23 was replaced by kanamycin-resistant (Kanr) gene via homologous recombination in E. coli DY380. The absence of pORF7 in 7D-GFP23 was confirmed by Western blotting (left lower panel). The growth curves suggest that the growth profiles of rOka-GFP23 and rOka were identical, as well as the growth curves of 7D-GFP23 and 7D. (B) Virus transmission from ARPE-19 cells to dSY5Y. A diagram of the cell-seeding and virus-inoculating schema is shown (left upper panel); hydrostatic pressure was generated from the difference in medium height (higher in the left chamber). ARPE-19 cells (5 × 104 cells seeded, right chamber) were infected with 5,000 PFU of rOka-GFP23 (right upper panel) or 7D-GFP23 (right lower panel), and virus transmission and infection signals in dSY5Y cells (2 × 105 cells seeded, left chamber) were examined at 7 dpi. The green viral particles within the microchannels are indicated by dashed squares and are displayed at higher magnifications (b1 for the rOka-GFP23 particle and b2 for the 7D-GFP23 particle). The virus particles are indicated by the white arrows. The GFP-positive cells in both chambers were counted and are shown (left lower panel). (C) Virus transmission from dSY5Y to ARPE-19 cells. The cells were similarly seeded, the rOka-GFP23 and 7D-GFP23 viruses were inoculated into the left chamber, and transmissions from dSY5Y to ARPE-19 cells were examined at 7 dpi. The GFP-positive cells in both chambers were quantified and are shown.
rOka-GFP23 and 7D-GFP23 were further used to investigate the differences in viral transmission between ARPE-19 and dSY5Y cells within the microfluidic devices (21, 22). SY5Y and ARPE-19 cells were sequentially seeded into the microfluidic chambers (23) and infected with rOka-GFP23 or 7D-GFP23 at the indicated times. The results at 7 dpi are shown in Fig. 2B. Prior to virus inoculation, the neuronal terminals of dSY5Y cells already passed through the microchannel (450-μm length, 10-μm width, and 4-μm depth), reaching the right chamber, where ARPE-19 cells were cultured. During viral transmission from ARPE-19 to dSY5Y, the offspring viral particles of rOka-GFP23 and 7D-GFP23 produced in ARPE-19 cells were transported retrogradely to dSY5Y cells. The invasive rOka-GFP23 particles further replicated in dSY5Y, transmitted to and labeled adjacent dSY5Y cells with GFP (GFP-positive cells). 7D-GFP23 infection resulted in a slightly smaller number of GFP-positive ARPE-19 cells compared to rOka-GFP23 infection at 7 dpi (163 ± 12 versus 221 ± 18); however, significantly fewer GFP-positive cells were observed among dSY5Y cells (2 ± 1 versus 32 ± 7) (Fig. 2B). During viral transmission from dSY5Y to ARPE-19, rOka-GFP23 infection resulted in more GFP-positive dSY5Y cells (187 ± 31) and more anterogradely labeled GFP-positive ARPE-19 cells (7 ± 3). 7D-GFP23 infection resulted in significantly fewer GFP-positive dSY5Y cells (21 ± 2), and no 7D-GFP23 particles transported from infected dSY5Y to ARPE-19; thus, no GFP-positive ARPE-19 cells were observed. In addition, the damage to dSY5Y cells caused by 7D-GFP23 infection was milder than that observed for rOka-GFP23 infection (Fig. 2C). These data demonstrated that ORF7 deletion resulted in attenuated viral replication and deficient virus transportation in dSY5Y cells, indicating that ORF7 plays an important role in VZV replication, transportation, and pathogenesis in neuronal cells.
pORF7 resides in the tegument layer of virion.
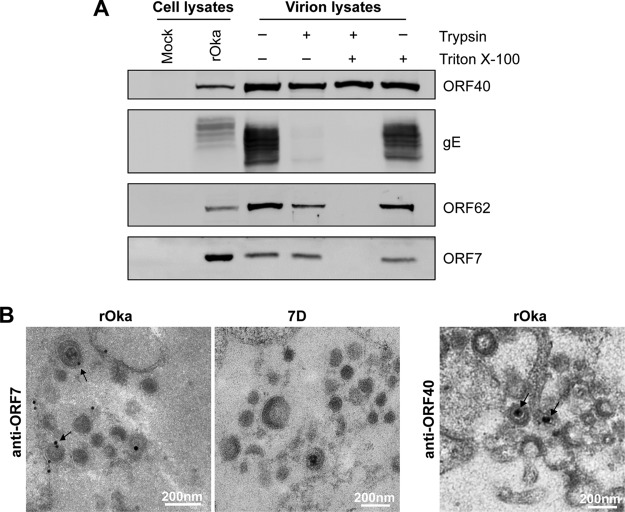
We have previously identified pORF7 as a virion component (7). ORF7 of VZV, a homologue of UL51 from HSV-1, potentially encodes a tegument protein (24). However, whether pORF7 resides in the tegument layer of the virion is unclear. To investigate the localization of ORF7, cell-free rOka virions were purified and then treated with trypsin with or without detergent. Samples were analyzed by Western blotting. As expected, the major capsid protein pORF40 was resistant to trypsin even in the presence of detergent, which is consistent with the features of a HSV-1 capsid protein (25, 26). Envelope protein gE, which is located in the exterior of virion, was sensitive to trypsin in the presence or absence of detergent. Conversely, pORF62, a known tegument protein, was resistant to trypsin only in the absence of detergent but sensitive to trypsin in the presence of detergent. pORF7 had a profile similar to that of pORF62 (Fig. 3A), which is more resistant to trypsin than envelope protein gE but less than capsid protein pORF40. These data indicate that location of pORF7 is similar to pORF62 in the tegument layer, between the envelope and capsid.
FIG 3.
Localization of ORF7 in VZV virions. (A) Localization analysis of pORF7 in purified VZV virions. Purified rOka virions were treated with trypsin in either the presence (+) or the absence (−) of Triton X-100. Equivalent amounts of cell lysates and virion samples were analyzed by Western blotting for the indicated viral proteins. (B) Localization of pORF7 in virions examined by immuno-EM. rOka- and 7D-infected ARPE-19 cells were fixed and subjected to a standard immuno-EM procedure. Images were obtained using a Hitachi H-7000FA transmission electron microscope. The colloidal gold signals of pORF7 in viral particles in the cytoplasm of infected cells are indicated by black arrows. No colloidal gold signal was observed in 7D-infected cells. The capsid protein ORF40 was also stained to clarify the location. Colloidal gold is indicated by black arrows. Scale bar, 200 nm.
The location of pORF7 in virion was also analyzed by immuno-electron microscopy (immuno-EM). As shown in Fig. 3B, pORF7 appeared in the tegument layer of rOka virions (black arrows) and in the cytoplasm of rOka-infected cells. In contrast, no pORF7 was observed in 7D viral particles and in the cytoplasm of 7D-infected cells. The capsid protein ORF40 is a reference for the capsid location, which is closer to the center of virion than pORF7. These data further confirmed that pORF7 resides in the tegument layer of VZV virion.
ORF7 deletion does not affect viral entry, genome replication, or expression of typical viral genes.
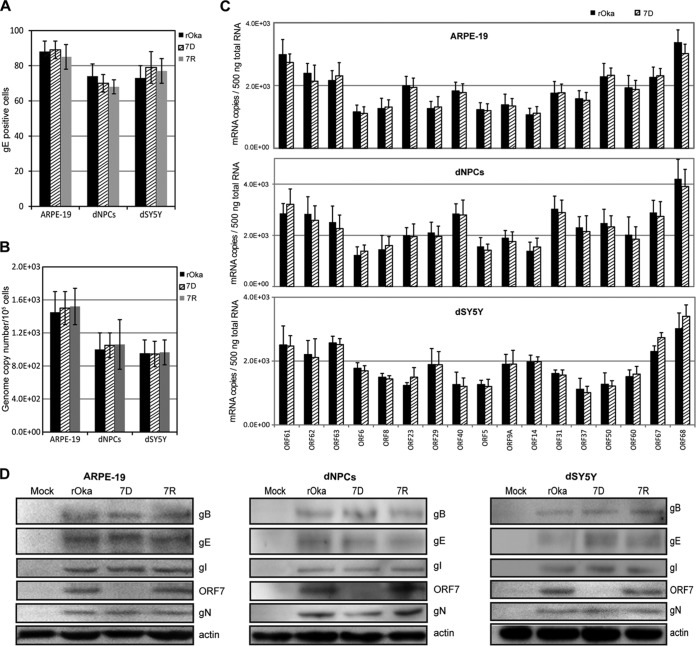
ARPE-19 cells, dNPCs, and dSY5Y cells were infected with cell-free rOka, 7D, and 7R viruses, respectively, at a multiplicity of infection (MOI) of 0.001. To synchronize viral entry, the cells were placed at 4°C, infected with virus, and incubated for 30 min. After being washed with phosphate-buffered saline (PBS), the cells were incubated at 37°C and harvested at 10 h postinfection (hpi). The determinations of viral entry, genome replication, and expression of typical viral genes were conducted by using immunofluorescence assay (IFA), quantitative PCR (qPCR), quantitative reverse transcription-PCR (qRT-PCR), and Western blotting. First, gE staining by IFA was used to indicate viral entry, and the gE-positive cells were counted. No significant differences in viral entry were observed among the infections of rOka, 7D, and 7R viruses, indicating that ORF7 deletion has no influence on viral entry (Fig. 4A). Next, the viral genome replication levels were investigated and found to be indistinguishable among rOka, 7D, and 7R infections within the first viral replication cycle (Fig. 4B). Similarly, no significant differences in the transcription levels of the 17 investigated viral genes were observed among the different infections. The 17 viral genes examined were immediate early genes (ORF62, ORF63, and ORF61), early genes encoding viral DNA replication associated proteins (ORF29, ORF6, and ORF8), late genes encoding capsid proteins (ORF23 and ORF40), and glycoproteins (ORF5, ORF9A, ORF14, ORF31, ORF37, ORF50, ORF60, ORF67, and ORF68) (27–31) (Fig. 4C). The data from 7R were similar to the data for rOka and 7D (not shown).
FIG 4.
Role of ORF7 in viral entry, genome replication, and expression of typical viral genes. ARPE-19 cells, dNPCs, and dSY5Y cells were infected with cell-free rOka, 7D, and 7R viruses, respectively, at an MOI of 0.001. The cells were harvested at 10 hpi. (A) Entry of the viruses. The harvested cells were fixed and subjected to gE IFA staining. The gE-positive cells are counted as infected cells with viral entry. (B) Viral genome replication. DNA was extracted from the cells infected with VZV and the mutants, and viral genome copy numbers were determined by qPCR. (C) Transcription of typical viral genes. Harvested cells were subjected to RNA extraction, followed by quantitation of the mRNA level of viral genes by qRT-PCR. (D) Expression of typical glycoproteins. Cell lysates were prepared from the harvested cells. Four representative glycoproteins and pORF7 were detected by Western blotting. All of the results were obtained from three independent experiments and are represented as averages ± the SD or else representative images are shown.
Finally, certain tegument proteins can conjugate with glycoproteins and be packed into viral particles in the Golgi apparatus (32–34). The levels of glycoproteins (gE, gI, gB, and gN) in rOka-, 7D-, and 7R-infected cells were determined by Western blotting. pORF7 was absent in 7D-infected cells, and no significant change of the examined glycoproteins was observed (Fig. 4D). These data demonstrated that ORF7 deletion does not affect viral entry, the first round of viral genome replication, and the transcription and expression of typical viral genes.
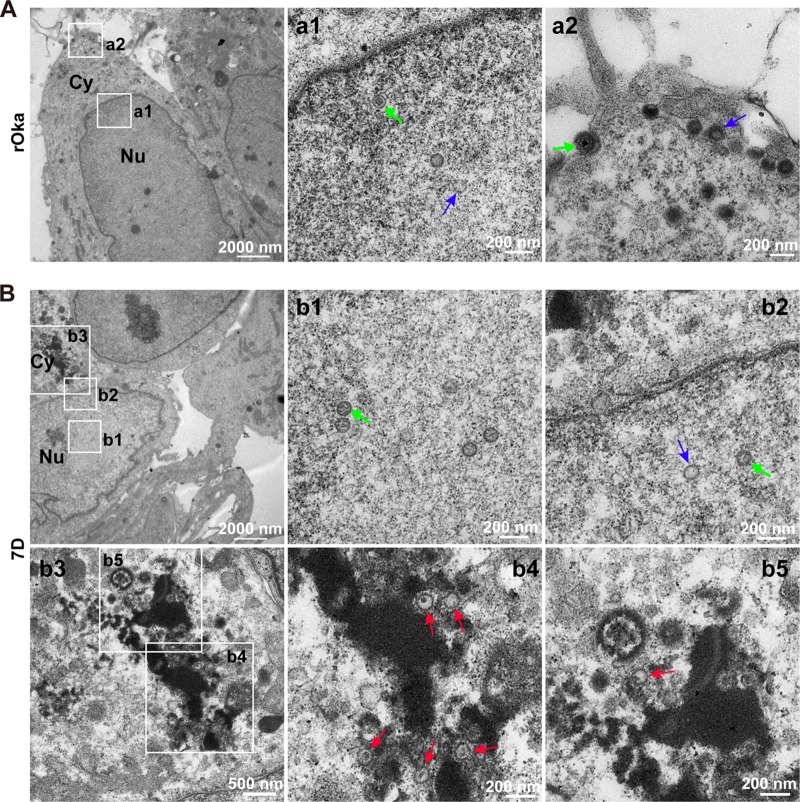
ORF7 deletion impairs cytoplasmic envelopment in dNPCs.
Deletion of ORF7 does not affect viral entry, viral genome replication, and gene expression, but typical plaque and CPEs are absent in the 7D-infected dNPCs and dSY5Y cells (Fig. 1). This prompted us to consider whether deletion of ORF7 impacts the late stage of viral replication, including nucleocapsid formation, nucleocapsid egress, and cytoplasmic envelopment, which may in turn result in defective viral particles and affect virus spread. To address this question, viral particles in rOka- and 7D-infected ARPE-19 cells, NPCs, dNPCs, and dSY5Y cells were observed by electron microscopy (EM), and the viral particles were classified and counted.
In the nuclei of rOka-infected dNPCs (Fig. 5A), intact nucleocapsids (Fig. 5A, a1, green arrow) and empty nucleocapsids (without viral genome) (Fig. 5A, a1, blue arrow) were observed; in the cytoplasm and cell surface, mature virions (Fig. 5A, a2, green arrow) and hollow particles without nucleocapsids (Fig. 5A, a2, blue arrow) were observed. Similarly, in the nuclei of 7D-infected dNPCs (Fig. 5B), intact (green arrow) and empty nucleocapsids (blue arrow) were observed, and the ratio of intact to empty nucleocapsids was consistent with that in nuclei of rOka-infected dNPCs (Fig. 5B, b1 and b2), indicating that nucleocapsid assembly in nuclei was not affected by ORF7 deletion. However, no mature virion was definitely observed in the cytoplasm, and almost all the viral particles were envelope defective (red arrows) (Fig. 5B, b3, b4, and b5). These data indicated that pORF7 involves in cytoplasmic envelopment in dNPCs.
FIG 5.
Viral particles in rOka- and 7D-infected dNPCs. (A) rOka-infected dNPCs. Normal VZV particles were observed in the nucleus (a1, capsids with genome, green arrow; capsids without genome, blue arrow) and in the cytoplasm and on the cell surface (a2, intact virion, green arrow; hollow particle, blue arrow), respectively. (B) 7D-infected dNPCs. 7D nucleocapsids were observed in the nucleus (b1 and b2, capsids with genome, green arrow; capsids without genome, blue arrow). Envelope-defective particles (red arrows) were captured in the cytoplasm (b3-b5). “Nu” and “Cy” indicate the nucleus and cytoplasm, respectively.
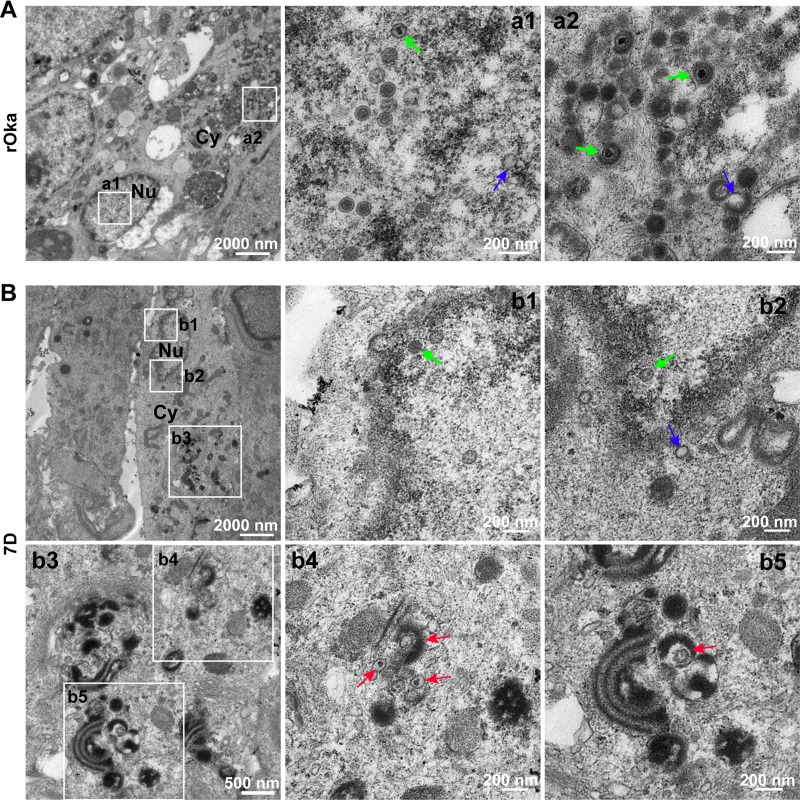
ORF7 deletion impairs cytoplasmic envelopment in dSY5Y.
Consistent with the observations for the rOka-infected dNPCs, intact and empty nucleocapsids in nuclei, mature virions, and hollow particles in the cytoplasm were observed in rOka-infected dSY5Y cells (Fig. 6A, a1 and a2). In 7D-infected dSY5Y cells, nucleocapsids in the nuclei were similar to those in rOka-infected dSY5Y cells and dNPCs, and no nucleocapsid accumulation in nuclei and around the nuclear membrane was observed (Fig. 6B, b1 and b2). However, cytoplasmic envelopment of nucleocapsids in 7D-infected dSY5Y cells was severely impaired, and no intact virion was observed, whereas an increased number of envelope-defective particles was clearly observed (Fig. 6B, b3, b4, and b5, red arrows). These results suggested that cytoplasmic envelopment is impaired by ORF7 deletion in dSY5Y cells.
FIG 6.
Viral particles in rOka- and 7D-infected dSY5Y cells. (A) rOka-infected dSY5Y cells. Normal VZV particles were observed in the nucleus (a1, capsids with genome, green arrow; capsids without genome, blue arrow), and in the cytoplasm (a2, intact virion, green arrow; hollow particle, blue arrow), respectively. (B) 7D-infected dSY5Y cells. 7D nucleocapsids were observed in the nucleus (b1 and b2, capsids with genome, green arrow; capsids without genome, blue arrow). Envelope-defective particles (red arrows) were captured in the cytoplasm (b3 to b5). “Nu” and “Cy” indicate the nucleus and the cytoplasm, respectively.
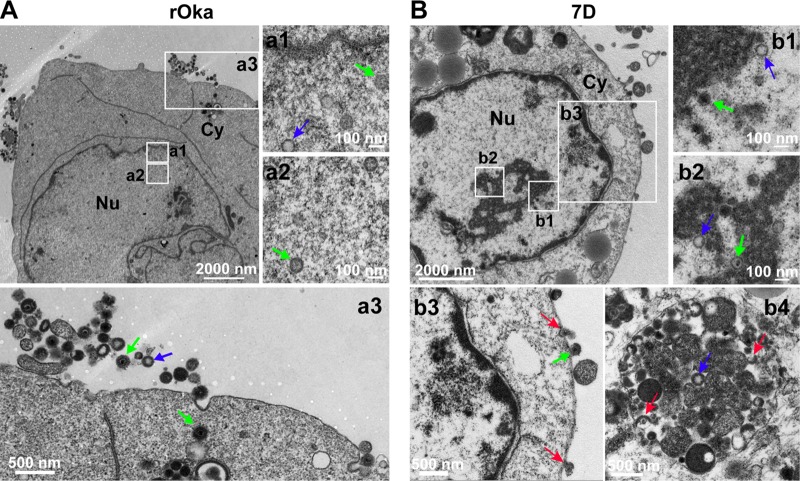
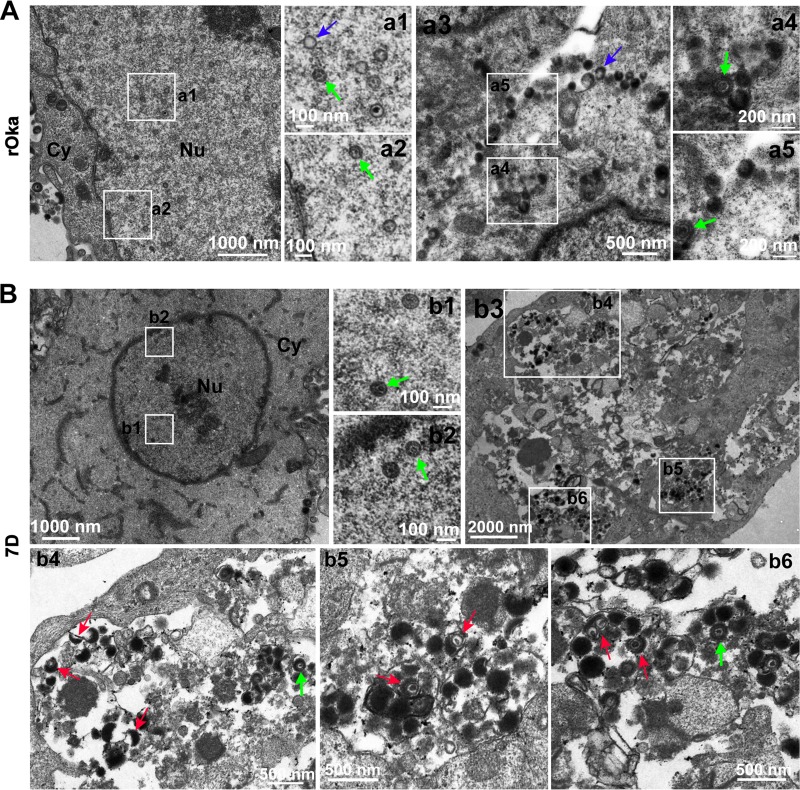
Viral particles in rOka- and 7D-infected ARPE-19 cells (Fig. 7) and NPCs (Fig. 8) were also analyzed. The results from rOka- and 7D-infected ARPE-19 cells were similar to that from rOka- and 7D-infected NPCs. ORF7 deletion also results in cytoplasmic envelopment deficiency in ARPE-19 cells and NPCs, but it is not as severe as that in dNPCs and dSY5Y cells, since mature virions appeared in the cytoplasm of 7D-infected ARPE-19 cells (Fig. 7B, b3 and b4) and NPCs (Fig. 8B, b4 to b6).
FIG 7.
Viral particles in rOka- and 7D-infected ARPE-19 cells. (A) rOka-infected ARPE-19 cells. Normal VZV particles were observed in the nucleus (a1 and a2, capsids with genome, green arrow; capsids without genome, blue arrow) and in the cytoplasm and on the cell surface (a3 and a4, intact virion, green arrow; hollow particle, blue arrow), respectively. (B) 7D-infected ARPE-19 cells. 7D nucleocapsids were observed in the nucleus (b1 and b2, capsids with genome, green arrow; capsids without genome, blue arrow). Envelope-defective particles (red arrows) were captured in the cytoplasm and on the cell surface (b3 and b4). “Nu” and “Cy” indicate the nucleus and the cytoplasm, respectively.
FIG 8.
Viral particles in rOka- and 7D-infected NPCs. (A) rOka-infected NPCs. Normal VZV particles were observed in the nucleus (a1 and a2, capsids with genome, green arrow; capsids without genome, blue arrow) and in the cytoplasm and on the cell surface (a3 to a5, intact virion, green arrow; hollow particle, blue arrow), respectively. (B) 7D-infected NPCs. 7D nucleocapsids were observed in the nucleus (b1 and b2, capsids with genome, green arrow). Envelope-defective particles (red arrows) were captured in the cytoplasm (b3 to b6). “Nu” and “Cy” indicate the nucleus and the cytoplasm, respectively.
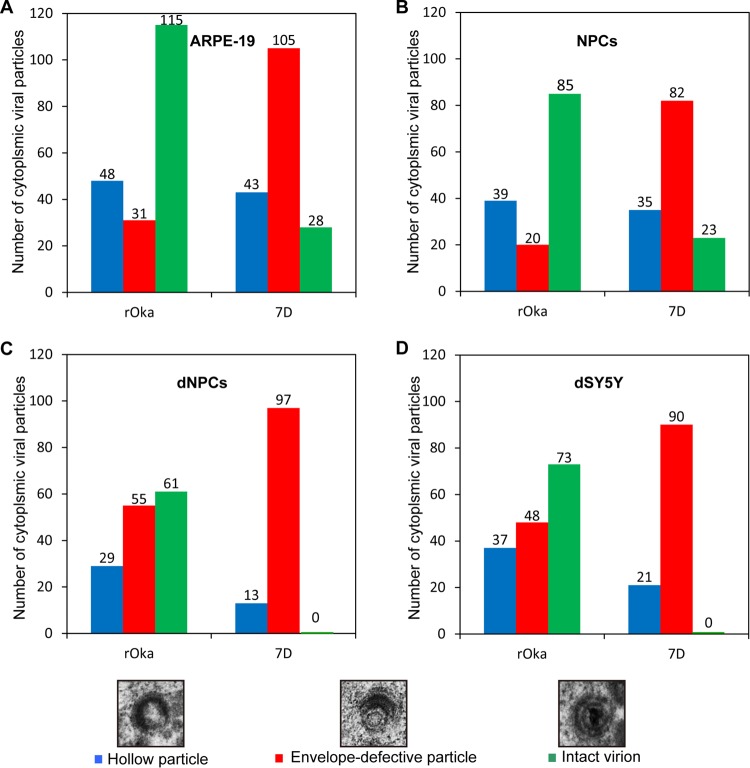
To further demonstrate the cytoplasmic envelopment deficiency induced by ORF7 deletion, viral particles in cytoplasm were analyzed according to composition of viral particles. There were three major types of viral particles observed in the cytoplasm: hollow particles, envelopes without nucleocapsid particles (blue column), and envelope-defective particles with nucleocapsids (red column) and virions (green column). Particles were counted in five cells randomly selected from the three independent experiments, and the results from rOka- or 7D-infected ARPE-19 cells, NPCs, dNPCs, and dSY5Y cells are shown in Fig. 9A, B, C, and D, respectively. A dramatic increase in the number of envelope-defective particles and decrease in virion number were clearly observed in 7D-infected cells compared to rOka-infected cells; notably, no virions were observed in 7D-infected dNPCs and dSY5Y cells. The severity of cytoplasmic envelopment deficiency in 7D-infected dSY5Y cells and in dNPCs was similar. These results indicate that ORF7 plays an important role in cytoplasmic envelopment in neuronal cells, clarifying the underlying mechanisms associated with the in vivo neurotropic characteristics of ORF7.
FIG 9.
Particles in cytoplasm of rOka- and 7D-infected cells. ARPE-19 cells (A), NPCs (B), dNPCs (C), and dSY5Y cells (D) were infected with rOka and 7D. Cytoplasmic viral particles were counted from five random infected cells from three independent experiments. Hollow particles, particles with a viral envelope but no capsid core (blue column); envelope-defective particles, particles with a capsid core but without complete envelope (red column); intact virions, particles with complete envelope, tegument layer, and capsid core (green column). No intact virions were observed in 7D-infected dNPCs and dSY5Y cells.
DISCUSSION
VZV infection-associated neuropathy is an important clinical complication. Shingles is the most common clinical manifestation, resulting from reactivation of latent/persistent VZV in neuronal cells, and relies on viral replication within neurons spreading to neighboring neurons or innervated nonneuronal cells (e.g., epithelial cells). Vaccination in childhood cannot prevent shingles, since the vaccinated virus still can potentially establish latency in DRG (2). Therefore, intervention by selectively impairing VZV replication in neuronal cells and the ability to spread to adjacent neurons can be a novel strategy for the treatment of VZV-associated neuropathy.
ORF7 of VZV has been identified as an in vivo neurotropic factor whose mutation has the potential to limit the neurological damage (7), but its function is unclear. In the present study, the role of pORF7 in the VZV replication cycle has been comprehensively characterized in ARPE-19 cells, NPCs, and SY5Y cells and in differentiated neuronal dNPCs and dSY5Y cells. A delayed or attenuated growth of 7D was observed in all the cells examined, and a severe impairment in transcellular spread of 7D in differentiated neuronal cells was found. Moreover, we demonstrated that ORF7 deletion does not affect viral entry, genome replication, and the expression of typical viral genes, indicating that, as an identified in vivo neurotropic factor, ORF7 is not a ligand that is used for viral entry to neuronal cells and thus is not associated with in vitro cell tropism.
If ORF7 does not play a vital role in regulating viral genome replication and gene expression, could ORF7 affect the late stage of replication and VZV-induced neuropathy? We found that pORF7 is located in the tegument layer, indicating that it is a structural protein similar to pUL51 of HSV-1 and PRV (18–20, 35, 36). Normally, when viral particles within vesicles have completed the secondary envelopment in the Golgi apparatus, they are transported to the cell surface and neuronal terminals to invade neighboring cells (8, 37, 38). 7D virus infection causes smaller plaques in ARPE-19 cells, NPCs, and SY5Y cells; no distinct plaque was observed in differentiated neuronal cells of dNPCs and dSY5Y compared to the rOka virus infection, and 7D transmission from dSY5Y cells to ARPE-19 cells was severely impaired. All of these data indicate that the deletion of ORF7 affects virus trafficking and spread, which is consistent with the tegument proteins of other herpesviruses. For example, PRV-UL37 and HSV-1 UL37 play an important role in virus trafficking to cell junctions for cell-to-cell spread (39, 40). UL37 interacts with UL36 to recruit capsids (41), associating with the host protein dystonin/BPAG1 (42) for microtubule-based transportation (43). How ORF7 affects trafficking of viral particles needs to be further studied.
Tegument proteins also affect morphogenesis at the late stage of infection, which in turn impacts the integrity of viral particle structure and the stability of viral particles (18–20, 35, 36). In the nuclei of 7D-infected cells nucleocapsids were not affected, but in the cytoplasm of 7D-infected cells the number of defective viral particles, namely, envelope-defective particles, increased dramatically. This indicates that pORF7 is involved in secondary envelopment, probably by conjugating with other tegument proteins or glycoproteins to facilitate the formation of a tegument-glycoprotein complex to aid in anchoring the envelope layer to the capsid outer surface (35, 36). Interestingly, the severity of envelopment deficiency caused by ORF7 deletion is different among the various cells used. Envelopment deficiency was more severe in dNPCs and dSY5Y cells than in ARPE-19 cells and NPCs. In the absence of pORF7, 7D might hijack other cellular machinery in certain cells to compensate for the loss of this tegument protein, as observed in other herpesviruses. For instance, in the absence of tegument proteins US3, UL47, or UL49, cellular F-actin is incorporated into the recombinant PRV particles to produce mature virions (44, 45). HSV-1 pUL37 recruits pUL36 (41, 46, 47) and interacts with glycoprotein K and membrane protein UL20 for efficient cytoplasmic envelopment (48). Intact virions were produced in 7D-infected ARPE-19 cells and NPCs, but not in the differentiated neuronal cells of dNPCs and dSY5Y. The difference in the production of virions between 7D-infected ARPE-19 cells/NPCs and differentiated neuronal cells may be related to the cellular environment. Certain cellular factor(s) may also be involved in the envelopment of VZV, but how ORF7 mediates VZV cytoplasmic envelopment and the interaction with cellular factor(s) needs to be specified.
In summary, ORF7 deletion attenuates the VZV replication in ARPE-19 cells and NPCs and severely impairs viral replication and neurovirulence in differentiated neuronal cells. 7D shows impaired transneuronal transmission and an impaired capacity for producing intact virions in neurons and for spreading to neighboring neurons or innervated nonneuronal cells. These results suggest that 7D could be a basis for a safer vaccine.
MATERIALS AND METHODS
Ethics statement.
The Wuhan Institute of Virology Institutional Review Board approved (WIVH10201202) the isolation of primary human NPCs from postmortem fetal embryonic tissue and waived the need for consent. The isolated NPCs are maintained in our laboratory and complied with the rule that NPCs must be less than nine passages (49).
Cells and cell culture.
ARPE-19 cells (ATCC, CRL-2302) were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin (100 U/ml and 100 μg/ml), all from Gibco/Life Technology. NPCs were isolated from the postmortem neonate brain tissue and cultured as described previously (21). To differentiate NPCs toward neurons, monolayer NPCs were cultured in the presence of 25 ng/ml human basic fibroblast growth factor 2 (FGF-2), 20 ng/ml nerve growth factor (NGF), and 10 ng/ml brain-derived neurotrophic factor (BDNF), all from Prospec; 100 μM dibutryl cyclic AMP (Selleck); and 1 μM retinoic acid (Sigma) for 10 days as described previously (50). The differentiated NPCs were designated dNPCs. SH-SY5Y (designated SY5Y, ATCC, CRL-2266) cells were maintained in DMEM-F12 containing 10% FBS. To differentiate SY5Y cells toward neurons, cells were treated with 50 μM retinoic acid for 5 days, followed by treatment with neurotropic growth factors (100 nM NGF and 50 nM BDNF) for 7 days (23). The differentiated SY5Y cells were designated dSY5Y.
Viruses and infection.
rOka, 7D, and 7R were all derived from the parental wild-type Oka strain (7). rOka contains a GFP coding gene and a luciferase gene in the genome and has been shown to have growth kinetics similar to those of the parental Oka strain (51). 7D is constructed by deleting the ORF7 gene from the rOka genome, and 7R is a revertant virus of 7D (7).
Generation of 7D-GFP23.
VZV GFP-ORF23 (rOka-GFP23), a gift from Paul Kinchington (University of Pittsburgh), is a recombinant virus with GFP fused to the N-terminal end of the small capsid protein encoded by ORF23 (pORF23) (52). The entire ORF7 gene was deleted from the rOka-GFP23 genome via homologous recombination in Escherichia coli DY380, and the corresponding recombinant virus was obtained by transfecting the ARPE-19 cells with the generated construct using SuperFect transfection reagent (Qiagen) as described previously (53). The resulting ORF7-deficient GFP-tagged virus was designated 7D-GFP23.
Preparation of cell-free virus.
Cell-free viruses of rOka, 7D, 7R, rOka-GFP23, and 7D-GFP23 were prepared as described previously, with slight modifications (54). Briefly, the viruses were propagated in ARPE-19 cells and harvested when >80% of the cells displayed severe CPEs. Supernatant (supernatant-A) and cells were collected separately. Cells collected from five T-175 flasks were resuspended in 10 ml of DMEM and sequentially treated by ultrasonication (Sonics Vibra-Cell; 20 kHz, 45% amplitude, 15 s) to release viral particles and low-speed centrifugation (Eppendorf rotor F-34-6-38; 1,000 × g, 5 min, 4°C) to remove cell debris. The obtained supernatant was combined with supernatant-A and subjected to high-speed centrifugation (Beckman rotor SW32; 80,000 × g, 3 h, 4°C) to concentrate the viruses. Obtained sediments were resuspended in small volume DMEM. Virus titers were determined by a plaque-forming assay in ARPE-19 cells.
Virus infections were performed with prepared cell-free viruses at an MOI of 0.001. Virus entry was synchronized by placing cells at 4°C for 30 min; the cells were then infected with viruses, followed by incubation for 30 min. After being washed with PBS, the cells were switched to a 37°C incubator for further culture.
Virus growth curve.
ARPE-19 cells, NPCs, dNPCs, SY5Y cells, and dSY5Y cells were cultured in dishes (3 × 105 cells per 35-mm dish), which were then divided into four groups, comprising infection (at an MOI of 0.001) with rOka, 7D, or 7R or mock infection (isovolumetric culture medium). rOka, 7D, and 7R viruses have luciferase as a reporter. The luciferase activity was measured daily by adding fresh medium containing 150 μg/ml d-luciferin into each dish, followed by incubation for 10 min at 37°C. The luminescent signal was recorded using a Maestro in vivo fluorescence imaging system (IVIS; CRi). Luciferin-containing medium was replaced by fresh medium after each measurement (7).
Similarly, to evaluate the growth features of rOka-GFP23 and 7D-GFP23 and to compare them with rOka and 7D, ARPE-19 cells were infected with rOka, rOka-GFP23, 7D, and 7D-GFP23, respectively, at an MOI of 0.001. Samples were collected daily. Since rOka-GFP23 and 7D-GFP23 do not have a luciferase reporter, virus titers were determined by a plaque-forming assay, and the average titers were taken from three independent experiments.
Immunofluorescence assay.
To determine the entry of VZV particles, 35-mm coverglass-bottom dishes (Nest Biotechnology) containing ARPE-19 cells, dNPCs, and dSY5Y cells (3 × 105 cells per dish) were placed at 4°C and infected with rOka, 7D, or 7R, respectively, at an MOI of 0.001. To synchronize virus entry, the cells were maintained 4°C for 30 min and then switched to 37°C for 1 h to allow virus entry. After incubation, the cells were washed with fresh medium three times to remove free virus; the cultures were then continued at 37°C until harvesting. At 10 hpi, the cells were fixed with 4% paraformaldehyde (PFA), permeabilized, and subsequently incubated with mouse anti-gE antibody (9H8; Abcam) and Alexa Fluor 594-conjugated goat anti-mouse IgG antibody (A11032; Invitrogen) as described previously (49). Cell nuclei were counterstained with Hoechst 33342 (catalog no. 62249; Thermo Fisher Scientific). Samples were observed under a Nikon Eclipse 80i fluorescence microscope. All of the gE-positive cells were counted.
Virus transmission in microfluidic device.
The microfluidic culture platform was fabricated according to a previously described protocol (21). The microchannel mask is designed to generate a microchannel 450 μm in length, 10 μm in width, and 4 μm in depth (Nanjing Microclear Electronics Technology, China). In addition, the photoresist products SU-8 GM 1050 and GM 1075 (Gersteltec Sarl), propylene glycol methyl ether acetate (Sigma), a chamber mask (Nanjing Microclear Electronics Technology), and polydimethylsiloxane (Sylgard 184; Dow Corning) were used to fabricate the microfluidic devices. SY5Y cells (2 × 105) were seeded into the left chamber and differentiated as described above, and axonal fibers of dSY5Y cells were allowed to pass through the microchannels. Five days after the SY5Y cells were seeded, ARPE-19 cells (5 × 104) were seeded into the right chamber. rOka-GFP23 and 7D-GFP23 viruses (5,000 PFU), whose capsid is labeled by fused GFP, were inoculated into the chamber containing dSY5Y or ARPE-19 cells at 12 days after seeding with SY5Y cells. To generate and maintain hydrostatic pressure, the medium was refreshed daily, and the volume in the chambers was maintained at 200 μl in the virus inoculation chamber and at 400 μl in the opposite chamber. To prevent virus flow from the inoculation chamber to the opposite one, the medium volume in the virus inoculation chamber was always maintained at a lower value relative to the opposite chamber (22).
Analysis of ORF7 localization in purified VZV virions.
rOka virions were purified by a procedure modified from a previously described method (55). Briefly, cytoplasmic extract was prepared from rOka-infected ARPE-19 cells when 90% of the cells showed CPEs. The cells were homogenized using a rotary homogenizer. The clarified cytoplasmic extract was centrifuged through a linear 20 to 50% sucrose gradient. The band at the 40 to 50% sucrose interface, which contains enveloped virions, was harvested. The collected band was washed with PBS and further purified by centrifugation (Beckman rotor SW28; 71,934 × g, 24 h, 15°C) with linear 30 to 0% reverse glycerol/0 to 50% potassium tartrate gradients. The further purified virions were treated with trypsin (0.15 mg/ml) either in the presence or in the absence of 1% Triton X-100 for 10 min at 37°C. The proteolysis reaction was terminated by adding trypsin inhibitor (1.5 mg/ml; T2011; Sigma). Equivalent amounts of cell lysates and virion samples were boiled in lysis buffer containing 0.5% β-mercaptoethanol (Sigma) and processed for Western blotting (as described below). Mouse monoclonal antibodies against gE (clone 4A2), ORF7 (clone 8H3), ORF40 (clone 10F2), or ORF62 (clone 1B7) (all made in the lab) were used as primary antibodies (56, 57).
Western blotting.
To determine the levels of viral proteins during the first round of infection in ARPE-19 cells, dNPCs, and dSY5Y cells, cells (3 × 106 per 100-mm dish) were infected with rOka, 7D, and 7R (MOI = 0.001), respectively, and collected at 10 hpi. To verify the absence of ORF7 in 7D-GFP23, ARPE-19 cells were infected with rOka-GFP23 and 7D-GFP23 viruses (MOI = 0.001) in parallel and harvested at 72 hpi. Cell pellets were snap-frozen in liquid nitrogen and stored at −80°C. Cell lysates were prepared, and the protein concentration was quantitated as described previously (49, 58). Equal amounts of protein from each sample were subjected to SDS–8% PAGE, transferred to polyvinylidene difluoride membrane (Millipore), and blotted sequentially with primary anti-ORF7 (clone 8H3), anti-gE (clone 4A2), anti-gI (clone 8C4), anti-gB (clone 10E10), or anti-gN (clone 12E10) (all made in the lab) or anti-actin (sc-8432; Santa Cruz Biotechnology) antibodies and with horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (Santa Cruz Biotechnology).
qPCR and qRT-PCR.
Infected cells were collected at 10 hpi and processed for extraction of DNA and RNA using a TIANamp virus DNA/RNA kit (Tiangen) according to the manufacturer's instructions. Total RNA was extracted by using TRIzol Reagent (TaKaRa), followed by treatment with 10 U of DNase (TaKaRa). RNA was reverse transcribed using an RT master mix Perfect-Real-Time kit (PrimeScript; TaKaRa). qPCR was performed on a real-time thermocycler (Bio-Rad; Connect) using SYBR green PCR master mix (Applied Biosystems) in a 20-μl reaction for 40 PCR cycles as described previously (49, 58, 59). DNA samples were amplified, along with standard rOka genome DNA. cDNA samples were amplified, along with GAPDH plasmid. The VZV genome copy number was calculated according to the quantity of ORF40 and ORF62. All primers used in qPCR analysis were listed in Table 1.
TABLE 1.
Primers used for qPCR and qRT-PCR
| Gene | Orientationa | Sequence (5′-3′) | Product size (bp) |
|---|---|---|---|
| ORF61 | F | ACATCCCTGCGTTGTCTTT | 172 |
| R | TTGAGGTGGTTTCTGGTCTTA | ||
| ORF62 | F | ATGTGGTTTCCAAGGCCAAGAG | 101 |
| R | TCCGTCAAGTGGCATCGTTATT | ||
| ORF63 | F | CTTCAACCCACCCAGACGC | 206 |
| R | GAATCCCGAAATTCAATTACATCC | ||
| ORF5 | F | GGGATATTAACTCACAGG | 192 |
| R | AAACCGCTATTTCTACAC | ||
| ORF6 | F | GAGGGCTTCTAATGGAGTT | 129 |
| R | CAAAGTGGTCGTACTGGTT | ||
| ORF8 | F | ATTTGACGGCGAGTTTGAT | 215 |
| R | CCAACGGAGTTATTGATGC | ||
| ORF9A | F | AACTTTGCAGAACGGAATT | 130 |
| R | CTAACGAAATAAGGGCTAC | ||
| ORF14 | F | TACCGCCGAAACATAACT | 154 |
| R | GACATACGCAGCCTACTG | ||
| ORF23 | F | GCCGAGACTGACCCTATGACA | 116 |
| R | CCCGAGATTCCCAAACTCAT | ||
| ORF29 | F | GGGTTACGTTTATGCGTGCCG | 245 |
| R | CTCCGTGAAAGACAAAGACAGATGG | ||
| ORF31 | F | GCCGTGGGATTATTGGTTT | 168 |
| R | AGTAGCGTTGGGTTTCTCG | ||
| ORF37 | F | GATTACACGAAACAAACCT | 181 |
| R | AACACTTCCGCAATACAA | ||
| ORF40 | F | CAACGGAACATACAACCTTACA | 249 |
| R | GCATCGCTTGAGCATAGTG | ||
| ORF50 | F | CCGTGTAGTAAGTAAATGCC | 137 |
| R | TTATGCTGCCGTAGTTGA | ||
| ORF60 | F | GGAAAGGGCATCCAACAA | 249 |
| R | GAACCGTGCGTGTCATCG | ||
| ORF67 | F | GTGGCATTATGGTAACTCAA | 128 |
| R | TGCTATGGTCTAACCGAAC | ||
| ORF68 | F | CCCGCAGTAACTCCTCAAC | 127 |
| R | ATGGCGCTTCATGTATCTT |
F, forward; R, reverse.
Electron microscopy.
ARPE-19 cells, NPCs, dNPCs, SY5Y cells, and dSY5Y cells were infected with rOka and 7D at an MOI of 0.001. rOka- and 7D-infected cells were harvested at 3 and 5 dpi, respectively, and fixed with 2.5% glutaraldehyde, followed by postfixation with 1% osmium tetroxide, dehydration in an ethanol series, and embedding with an Embed 812 kit (Electron Microscopy Sciences, Fort Washington, PA). The sections were sequentially stained with 3.5% aqueous uranyl acetate and 0.2% lead citrate. To analyze pORF7 and pORF40 localization in the virion, infected cell samples were fixed with fixative solution containing 3% PFA and 0.1% glutaraldehyde, dehydrated, and embedded with LR White. The sections were sequentially stained with anti-ORF7 (clone 8H3) or anti-ORF40 (clone 10F2) and 10-nm gold colloid-labeled goat anti-mouse IgG (GA1004; Boster). Images were obtained by using a Hitachi H-7000FA transmission electron microscope.
Statistical analysis.
For growth curve, virus entry, qPCR, and qRT-PCR analyses, each experiment was performed in triplicate, and the results are presented as means ± the standard deviations (SD) from three independent experiments. A Student t test was performed to analyze the statistical significance between different virus infections. Differences were considered to be significant when P < 0.05.
ACKNOWLEDGMENTS
We appreciate the technical support from Guo-Qiang Bi (University of Science and Technology of China) on microfluidic devices and the technical support with IVIS and electron microscopy from the Core Facility and Technical Support, Wuhan Institute of Virology.
This study was supported by National Natural Science Foundation of China (NSFC) projects 81601206 (W.-B.Z.) and 81601762 (W.W.) and by NSFC project 81427801 and Sino-Africa Joint Center project SAJC201605 (M.-H.L.). We have no conflicts of interest to declare.
REFERENCES
- 1.Tillieux SL, Halsey WS, Thomas ES, Voycik JJ, Sathe GM, Vassilev V. 2008. Complete DNA sequences of two Oka strain varicella-zoster virus genomes. J Virol 82:11023–11044. doi: 10.1128/JVI.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tseng HF, Schmid DS, Harpaz R, LaRussa P, Jensen NJ, Rivailler P, Radford K, Folster J, Jacobsen SJ. 2014. Herpes zoster caused by vaccine-strain varicella zoster virus in an immunocompetent recipient of zoster vaccine. Clin Infect Dis 58:1125–1128. doi: 10.1093/cid/ciu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiele S, Borschewski A, Kuchler J, Bieberbach M, Voigt S, Ehlers B. 2011. Molecular analysis of varicella vaccines and varicella-zoster virus from vaccine-related skin lesions. Clin Vaccine Immunol 18:1058–1066. doi: 10.1128/CVI.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller NH, Gilden DH, Cohrs RJ, Mahalingam R, Nagel MA. 2008. Varicella zoster virus infection: clinical features, molecular pathogenesis of disease, and latency. Neurol Clin 26:675–697. doi: 10.1016/j.ncl.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De SK, Hart JC, Breuer J. 2015. Herpes simplex virus and varicella zoster virus: recent advances in therapy. Curr Opin Infect Dis 28:589–595. doi: 10.1097/QCO.0000000000000211. [DOI] [PubMed] [Google Scholar]
- 6.Depledge DP, Yamanishi K, Gomi Y, Gershon AA, Breuer J. 2016. Deep sequencing of distinct preparations of the live attenuated varicella-zoster virus vaccine reveals a conserved core of attenuating single-nucleotide polymorphisms. J Virol 90:8698–8704. doi: 10.1128/JVI.00998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selariu A, Cheng T, Tang QY, Silver B, Yang LW, Liu C, Ye XZ, Markus A, Goldstein RS, Cruz-Cosme RS, Lin YZ, Wen LL, Qian HL, Han JL, Dulal K, Huang Y, Li YM, Xia NS, Zhu H. 2012. ORF7 of varicella-zoster virus is a neurotropic factor. J Virol 86:8614–8624. doi: 10.1128/JVI.00128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichelt M, Brady J, Arvin AM. 2009. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J Virol 83:3904–3918. doi: 10.1128/JVI.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerboni L, Sen N, Oliver SL, Arvin AM. 2014. Molecular mechanisms of varicella zoster virus pathogenesis. Nat Rev Microbiol 12:197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckingham EM, Jarosinski KW, Jackson W, Carpenter JE, Grose C. 2016. Exocytosis of varicella-zoster virus virions involves a convergence of endosomal and autophagy pathways. J Virol 90:8673–8685. doi: 10.1128/JVI.00915-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Cheng T, Zhu H, Xia NS. 2015. Insights into the function of tegument proteins from the varicella zoster virus. Sci China Life Sci 58:739–749. doi: 10.1007/s11427-015-4887-3. [DOI] [PubMed] [Google Scholar]
- 12.Zerboni L, Sobel RA, Ramachandran V, Rajamani J, Ruyechan W, Abendroth A, Arvin A. 2010. Expression of varicella-zoster virus immediate-early regulatory protein IE63 in neurons of latently infected human sensory ganglia. J Virol 84:3421–3430. doi: 10.1128/JVI.02416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Che X, Zerboni L, Sommer MH, Arvin AM. 2006. Varicella-zoster virus open reading frame 10 is a virulence determinant in skin cells but not in T cells in vivo. J Virol 80:3238–3248. doi: 10.1128/JVI.80.7.3238-3248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Che X, Oliver SL, Reichelt M, Sommer MH, Haas J, Rovis TL, Arvin AM. 2013. ORF11 protein interacts with the ORF9 essential tegument protein in varicella-zoster virus infection. J Virol 87:5106–5117. doi: 10.1128/JVI.00102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Cohen JI. 2013. Varicella-zoster virus ORF12 protein activates the phosphatidylinositol 3-kinase/Akt pathway to regulate cell cycle progression. J Virol 87:1842–1848. doi: 10.1128/JVI.02395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riva L, Thiry M, Bontems S, Joris A, Piette J, Lebrun M, Sadzot-Delvaux C. 2013. ORF9p phosphorylation by ORF47p is crucial for the formation and egress of varicella-zoster virus viral particles. J Virol 87:2868–2881. doi: 10.1128/JVI.02757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riva L, Thiry M, Lebrun M, L'Homme L, Piette J, Sadzot-Delvaux C. 2015. Deletion of the ORF9p acidic cluster impairs the nuclear egress of varicella-zoster virus capsids. J Virol 89:2436–2441. doi: 10.1128/JVI.03215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J Virol 79:6947–6956. doi: 10.1128/JVI.79.11.6947-6956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roller RJ, Haugo AC, Yang K, Baines JD. 2014. The herpes simplex virus 1 UL51 gene product has cell type-specific functions in cell-to-cell spread. J Virol 88:4058–4068. doi: 10.1128/JVI.03707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional analysis of the pseudorabies virus UL51 protein. J Virol 79:3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. 2006. Microfluidic culture platform for neuroscience research. Nat Protoc 1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 22.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. 2005. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX. 2000. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 24.Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedlackova L, Rice SA. 2008. Herpes simplex virus type 1 immediate-early protein ICP27 is required for efficient incorporation of ICP0 and ICP4 into virions. J Virol 82:268–277. doi: 10.1128/JVI.01588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb WW, Booy FP, Brown JC. 2007. Uncoating the herpes simplex virus genome. J Mol Biol 370:633–642. doi: 10.1016/j.jmb.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters MS, Kyratsous CA, Silverstein SJ. 2010. The RING finger domain of varicella-zoster virus ORF61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of Sp100-containing nuclear bodies. J Virol 84:6861–6865. doi: 10.1128/JVI.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White K, Peng H, Hay J, Ruyechan WT. 2010. Role of the IE62 consensus binding site in transactivation by the varicella-zoster virus IE62 protein. J Virol 84:3767–3779. doi: 10.1128/JVI.02522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller NH, Walters MS, Marcus RA, Graf LL, Prenni J, Gilden D, Silverstein SJ, Cohrs RJ. 2010. Identification of phosphorylated residues on varicella-zoster virus immediate-early protein ORF63. J Gen Virol 91:1133–1137. doi: 10.1099/vir.0.019067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stallings CL, Duigou GJ, Gershon AA, Gershon MD, Silverstein SJ. 2006. The cellular localization pattern of varicella-zoster virus ORF29p is influenced by proteasome-mediated degradation. J Virol 80:1497–1512. doi: 10.1128/JVI.80.3.1497-1512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri V, Sommer M, Rajamani J, Zerboni L, Arvin AM. 2008. Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection. J Virol 82:10231–10246. doi: 10.1128/JVI.01890-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver SL, Sommer M, Zerboni L, Rajamani J, Grose C, Arvin AM. 2009. Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo. J Virol 83:7495–7506. doi: 10.1128/JVI.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver SL, Sommer MH, Reichelt M, Rajamani J, Vlaycheva-Beisheim L, Stamatis S, Cheng J, Jones C, Zehnder J, Arvin AM. 2011. Mutagenesis of varicella-zoster virus glycoprotein I (gI) identifies a cysteine residue critical for gE/gI heterodimer formation, gI structure, and virulence in skin cells. J Virol 85:4095–4110. doi: 10.1128/JVI.02596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerboni L, Berarducci B, Rajamani J, Jones CD, Zehnder JL, Arvin A. 2011. Varicella-zoster virus glycoprotein E is a critical determinant of virulence in the SCID mouse-human model of neuropathogenesis. J Virol 85:98–111. doi: 10.1128/JVI.01902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oda S, Arii J, Koyanagi N, Kato A, Kawaguchi Y. 2016. The interaction between herpes simplex virus 1 tegument proteins UL51 and UL14 and its role in virion morphogenesis. J Virol 90:8754–8767. doi: 10.1128/JVI.01258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roller RJ, Fetters R. 2015. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol 89:3112–3122. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aleman N, Quiroga MI, Lopez-Pena M, Vazquez S, Guerrero FH, Nieto JA. 2003. L-particle production during primary replication of pseudorabies virus in the nasal mucosa of swine. J Virol 77:5657–5667. doi: 10.1128/JVI.77.10.5657-5667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichelt M, Joubert L, Perrino J, Koh AL, Phanwar I, Arvin AM. 2012. 3D Reconstruction of VZV-infected cell nuclei and PML nuclear cages by serial section array scanning electron microscopy and electron tomography. PLoS Pathog 8:e1002740. doi: 10.1371/journal.ppat.1002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol 82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandbaumhuter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol 15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 42.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan SD, Bhanot K, Ferrier A, De Repentigny Y, Chu A, Blais A, Kothary R. 2012. Microtubule stability, Golgi organization, and transport flux require dystonin-a2-MAP1B interaction. J Cell Biol 196:727–742. doi: 10.1083/jcb.201107096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.del Rio T, DeCoste CJ, Enquist LW. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J Virol 79:8614–8619. doi: 10.1128/JVI.79.13.8614-8619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael K, Klupp BG, Mettenleiter TC, Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J Virol 80:1332–1339. doi: 10.1128/JVI.80.3.1332-1339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanova L, Buch A, Dohner K, Pohlmann A, Binz A, Prank U, Sandbaumhuter M, Bauerfeind R, Sodeik B. 2016. Conserved tryptophan motifs in the large tegument protein pUL36 are required for efficient secondary envelopment of herpes simplex virus capsids. J Virol 90:5368–5383. doi: 10.1128/JVI.03167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. 2015. Herpes simplex virus capsid-organelle association in the absence of the large tegument protein UL36p. J Virol 89:11372–11382. doi: 10.1128/JVI.01893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan X, Li XJ, Liu XJ, Yuan H, Li JF, Duan YL, Ye HQ, Fu YR, Qiao GH, Wu CC, Yang B, Tian XH, Hu KH, Miao LF, Chen XL, Zheng J, Rayner S, Schwartz PH, Britt WJ, Xu J, Luo MH. 2013. Later passages of neural progenitor cells from neonatal brain are more permissive for human cytomegalovirus infection. J Virol 87:10968–10979. doi: 10.1128/JVI.01120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pugazhenthi S, Nair S, Velmurugan K, Liang Q, Mahalingam R, Cohrs RJ, Nagel MA, Gilden D. 2011. Varicella-zoster virus infection of differentiated human neural stem cells. J Virol 85:6678–6686. doi: 10.1128/JVI.00445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Rowe J, Wang W, Sommer M, Arvin A, Moffat J, Zhu H. 2007. Genetic analysis of varicella-zoster virus ORF0 to ORF4 by use of a novel luciferase bacterial artificial chromosome system. J Virol 81:9024–9033. doi: 10.1128/JVI.02666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markus A, Grigoryan S, Sloutskin A, Yee MB, Zhu H, Yang IH, Thakor NV, Sarid R, Kinchington PR, Goldstein RS. 2011. Varicella-zoster virus (VZV) infection of neurons derived from human embryonic stem cells: direct demonstration of axonal infection, transport of VZV, and productive neuronal infection. J Virol 85:6220–6233. doi: 10.1128/JVI.02396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao F, Shen ZZ, Liu ZY, Zeng WB, Cheng S, Ma YP, Rayner S, Yang B, Qiao GH, Jiang HF, Gao S, Zhu H, Xu FQ, Ruan Q, Luo MH. 2016. Identification and BAC construction of Han, the first characterized HCMV clinical strain in China. J Med Virol 88:859–870. doi: 10.1002/jmv.24396. [DOI] [PubMed] [Google Scholar]
- 54.Provost PJ, Krah DL, Friedman PA. November 1994. Process for attenuated varicella zoster virus vaccine production. US patent 5,360,736.
- 55.Kinchington PR, Hougland JK, Arvin AM, Ruyechan WT, Hay J. 1992. The varicella-zoster virus immediate-early protein Ie62 is a major component of virus particles. J Virol 66:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Wang X, Yang LW, Fu WK, Pan DQ, Liu J, Ye JH, Zhao QJ, Zhu H, Cheng T, Xia NS. 2016. Modulation of host CD59 expression by varicella-zoster virus in human xenografts in vivo. Virology 491:96–105. doi: 10.1016/j.virol.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Zhu R, Ye XZ, Yang LW, Wang YM, Huang YY, Wu J, Wang W, Ye JH, Li YM, Zhao QJ, Zhu H, Cheng T, Xia NS. 2015. A monoclonal antibody-based VZV glycoprotein E quantitative assay and its application on antigen quantitation in VZV vaccine. Appl Microbiol Biotechnol 99:4845–4853. doi: 10.1007/s00253-015-6602-5. [DOI] [PubMed] [Google Scholar]
- 58.Duan YL, Ye HQ, Zavala AG, Yang CQ, Miao LF, Fu BS, Seo KS, Davrinche C, Luo MH, Fortunato EA. 2014. Maintenance of large numbers of virus genomes in human cytomegalovirus-infected T98G glioblastoma cells. J Virol 88:3861–3873. doi: 10.1128/JVI.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu YR, Liu XJ, Li XJ, Shen ZZ, Yang B, Wu CC, Li JF, Miao LF, Ye HQ, Qiao GH, Rayner S, Chavanas S, Davrinche C, Britt WJ, Tang QY, Mcvoy M, Mocarski E, Luo MH. 2015. MicroRNA miR-21 attenuates human cytomegalovirus replication in neural cells by targeting Cdc25a. J Virol 89:1070–1082. doi: 10.1128/JVI.01740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]