Abstract
Background
Rodents represent the most diverse mammals on the planet and are important reservoirs of human pathogens. Coronaviruses infect various animals, but to date, relatively few coronaviruses have been identified in rodents worldwide. The evolution and ecology of coronaviruses in rodent have not been fully investigated.
Results
In this study, we collected 177 intestinal samples from thress species of rodents in Jianchuan County, Yunnan Province, China. Alphacoronavirus and betacoronavirus were detected in 23 rodent samples from three species, namely Apodemus chevrieri (21/98), Eothenomys fidelis (1/62), and Apodemus ilex (1/17). We further characterized the full-length genome of an alphacoronavirus from the A. chevrieri rat and named it as AcCoV-JC34. The AcCoV-JC34 genome was 27,649 nucleotides long and showed a structure similar to the HKU2 bat coronavirus. Comparing the normal transcription regulatory sequence (TRS), 3 variant TRS sequences upstream the spike (S), ORF3, and ORF8 genes were found in the genome of AcCoV-JC34. In the conserved replicase domains, AcCoV-JC34 was most closely related to Rattus norvegicus coronavirus LNRV but diverged from other alphacoronaviruses, indicating that AcCoV-JC34 and LNRV may represent a novel alphacoronavirus species. However, the S and nucleocapsid proteins showed low similarity to those of LRNV, with 66.5 and 77.4% identities, respectively. Phylogenetic analysis revealed that the S genes of AcCoV-JC34, LRNV, and HKU2 formed a distinct lineage with all known coronaviruses.
Conclusions
Both alphacoronaviruses and betacoronaviruses were detected in Apodemus chevrieri in the Yunnan Province of China, indicating that Apodemus chevrieri is an important host for coronavirus. Several new features were identified in the genome of an Apodemus chevrieri coronavirus. The phylogenetic distance to other coronaviruses suggests a variable origin and evolutionary route of the S genes of AcCoV-JC34, LRNV, and HKU2. These results indicate that the diversity of rodent coronaviruses is much higher than previously expected. Further surveillance and functional studies of these coronaviruses will help to better understand the importance of rodent as host for coronaviruses.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-017-0766-9) contains supplementary material, which is available to authorized users.
Keywords: Rodent, Coronavirus, Genetic diversity
Background
Coronaviruses (CoVs) are enveloped viruses in the Coronaviridae family that contain a positive-sense and single-stranded RNA genome of approximately 30 kilobases [1]. CoVs consist of 4 genera and have been identified in a wide range of animals and in humans. Members of the Alphacoronavirus (α-CoV) and Betacoronavirus (β-CoV) infect mammals, and members of the Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV) mainly infect avian species [2–4]. As important etiological agents, CoVs have been recognized in human and animals and cause upper respiratory diseases in most cases. To date, 6 human CoVs were discovered: 4 of them (HCoV-229E, NL63, OC43, and HKU1) mainly cause mild respiratory diseases, and the other 2, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) cause severe respiratory diseases [5, 6].
The SARS-CoV outbreak boosted the discovery of novel CoVs in various animals, particularly in bats. Over 140 novel bat coronaviruses (species or genotypes) have been discovered since the SARS outbreak [7, 8]. Furthermore, there is strong evidence to show that SARS-CoV, MERS-CoV, and HCoV229E may have evolved from bat CoVs [9–13].
Rodents are the most diverse mammals on the planet and have been documented as important carriers of human diseases [14]. Although murine hepatitis virus (MHV) has been used as a model to study CoV for a long time, limited information is available regarding the prevalence and diversity of rodent CoVs [15–18]. Recently, several novel α-CoVs and β-CoVs (LRNV, LAMV, LRLV, and HKU24) were identified in rodents in China and Europe [19–21]. These discoveries suggested that rodents may carry diverse, unrecognized CoVs [22]. In the present study, we describe the first discovery of CoVs in 3 different rodent species in the Yunnan Province of China and report a much higher (21.4%) detection rate of CoV nucleic acid in A. chevrieri than in other rodent species studied previously (<5%) [19, 20]. In addition, this is the first report of finding α-CoV and β-CoV in the same rodent species in China.
Methods
Sample collection
In October 2011, for pest control and routine pathogen surveillance, 177 rodents were captured in the bush and grass near the cropland ridge in Jianchuan County of the Yunnan Province (Additional file 1: Figure S1). Animal intestines were collected and transferred to liquid nitrogen for temporary preservation and transport. Following arrival at the laboratory, the samples were stored at –80 °C until they were used for virus detection. Animal species were first identified based on morphology and further by DNA sequencing of the mitochondrial cytochrome b (CytB) gene with ready-to-use methods [23].
RNA extraction
To extract viral RNA, 50 mg of intestinal tissue samples were homogenized using 1 ml PBS. Homogenates were centrifuged and RNA was extracted from 140 μL supernatant using the QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions. Extracted RNA was used as a template for amplifying the mitochondrial CytB gene with the primers CytBF (5′-ATGATATGAAAAACCATCGTTG-3′) and CytBR (5′-TTTCCNTTTCTGGTTTACAAGAC-3′). The 1.2-kb replicons were gel purified (Promega, Madison, USA) and directly sequenced using the forward and reverse primers with a 3100 Sequencer (Applied Biosystems, Waltham, MA, USA).
Reverse transcription PCR (RT-PCR) screening of CoVs
The 440-bp RNA-dependent RNA polymerase gene (RdRp) fragment of CoVs was amplified by previously described methods using a One-Step RT-PCR (Invitrogen, San Diego, USA) [24]. PCR target bands were gel purified and sequenced on a 3100 Sequencer (ABI, Waltham, MA, USA). Standard precautions were taken to avoid PCR contamination, and no false positives were observed in the negative controls. To determine the heterogeneity of the amplicons, we inspected the sequencing chromatograms. No overlapping multicolor peaks were found, indicating that no CoV co-infection occurred in the animals examined in this study. To confirm the PCR results, positive samples were verified by performing two independent PCRs. The CoV-positive samples were named using the rodent species name, the location (Jianchuan County), and the sample number. For example, a CoV detected in A. chevrieri (sample number 54) was named as A. chevrieri CoV JC54 (AcCoV-JC54).
Viral culture
Three positive rodent samples representing different CoVs (JC30, β-CoV; JC34 and JC54, α-CoV) were used to perform viral isolation in Vero E6 cells (African green monkey kidney cells, ATCC: CRL-1586).
Genome sequencing
To sequence the viral genome, 140 μL supernatant from a JC34 tissue homogenate was treated using viral metagenomics procedures and ready-to-use methods [25]. Synthesized DNA was used to construct the sequencing library, and next-generation sequencing (NGS) was performed using an HiSeq-PE150 instrument (Illumina/Solexa). BLAST searches were performed against the CoV database, and 413,599 reads homologous to CoV sequences were found. The reads were processed using Geneious (Version 5.5.9, Biomatters Limited, Auckland, New Zealand) to assemble a near full-length CoV genome contig. Subsequently, 5′ and 3′ RACE (Takara) were performed to confirm the ends of the genome sequence using two primers (5′-CAGGACGTCTAATGCAATACCT-3′ and 5′-AACACACTGAAATCAGACCTTG-3′), which were designed based on the obtained contig sequences and primers supplied in the kits. The replicons were both end sequenced. Finally, all sequences were assembled with the CoV contig to obtain a full-length CoV genome, designated as AcCoV-JC34.
Sequence analysis
The genome sequence of AcCoV-JC34 was compared to other representative CoVs with complete genomes available. The open reading frames (ORFs), deduced amino acid sequences, and potential cleavage sites in orf1ab were predicted by ORF Finder (NCBI) and ZCURVE_CoV 2.0 [26]. Sequence alignment and editing were performed using ClustalW (Version 2.0), BioEdit (Version 7.1.9), and Geneious (Version 5.5.9) [27, 28]. The spike (S) protein structure of AcCoV-JC34 was searched against sequences in the Protein Data Bank (PDB) and predicted using a web-based SWISS-MODEL server. The cleavage sites in the S protein were predicted by comparing amino acid sequences, combined with analysis using a web-based ProP server [29]. Phylogenetic trees were constructed using the maximum-likelihood (ML) algorithm, with bootstrap values determined by 1000 replicates in MEGA6 and PhyML software [30, 31].
Results
Detection of α-CoVs and β-CoVs in rodents
A total of 177 intestinal samples were obtained from rodents, including three different species. By RT-PCR, targeting a 440 base pairs (bp) partial RdRp gene sequence of CoVs, 23 rodents were identified as CoV positive, which included 21 of 98 (21.4%) A. chevrieri, 1 of 17 (5.9%) A. ilex, and 1 of 62 (1.6%) E. fidelis samples (Table 1). The obtained CytB gene sequences were deposited in GenBank under accession numbers KX964655–KX964657. The isolation of rodent CoV from VeroE6 cells was not successful.
Table 1.
Detection of coronavirus in rodents by RT-PCR
| Scientific name | No. of rodents tested | No. of positives for the following/total no. of animals tested (%): | Detected coronaviruses |
|---|---|---|---|
| Apodemus chevrieri | 98 | 21/98 (21.4%) | Alphacoronavirus and Betacoronavirus |
| Apodemus ilex | 17 | 1/17 (5.9%) | Betacoronavirus |
| Eothenomys fidelis | 62 | 1/62 (1.6%) | Alphacoronavirus |
| Total | 177 | 23/177 (13%) |
Partial RdRp sequences were searched against the CoV database, and the results indicated that 21 out of 23 sequences were β-CoVs, whereas the remaining two were α-CoVs. The β-CoV-related sequences had high nucleotide (nt) identities of 95–99% compared to the unclassified β-CoVs detected in rodents in China, Longquan-343 (A. agrarius) and HKU24 (R. norvegicus). The A. chevrieri CoV JC34 (AcCoV-JC34) and E. fidelis CoV JC54 (EfCoV-JC54) shared the highest nt identities of 84 and 98%, respectively, with the unclassified α-CoVs detected from R. norvegicus, LRNV (GenBank no: KF294380) in China or UKRn1 (GenBank no: KU739071) in Europe (Fig. 1). LRNV has a complete genome of 28,763 bp and UKRn1 (KU739071) has a partial RdRp sequence of 630 bp. Sequenced RdRp fragments in this study were deposited in GenBank under accession numbers: KX964650–KX964654.
Fig. 1.

Phylogenetic analysis of detected rodent CoVs with representative CoVs based on 440-bp partial RdRp sequences. The tree was constructed by the maximum-likelihood method with 1000 bootstrap replicates. Bootstrap values above 50% were shown. Rodent CoVs found in this study are shown in bold. CoV abbreviations: bat SL-CoV WIV1, bat SARS-like coronavirus WIV1; BCoV, bovine coronavirus; CCoV, canine coronavirus; CrCoV, canine respiratory coronavirus; ECoV, equine coronavirus; FCoV, feline coronavirus; HCoV 229E, human coronavirus 229E; HCoV HKU1, human coronavirus HKU1; HCoV NL63, human coronavirus NL63; HCoV OC43, human coronavirus OC43; IBV, infectious bronchitis virus; MERS-CoV, MERS coronavirus; MHV, murine hepatitis virus; PHEV, porcine hemagglutinating encephalomyelitis virus; SARS-CoV, SARS coronavirus; TGEV, transmissible gastroenteritis virus
Genome organization and ORFs of AcCoV-JC34
One positive sample (JC34) was chosen for further sequencing to obtain the full-length genome because it showed low sequence similarity to other CoVs and appeared to be a novel CoV. By random PCR and Illumina sequencing, a near full-length genome of CoV was assembled from 413,599 reads. After sequencing 5′- and 3′-rapid amplification of cDNA end replicons, a complete genome was characterized. This virus was named rodent AcCoV-JC34 and the complete genome sequence was deposited in GenBank under accession number KX964649.
The genome size of AcCoV-JC34 was 27,649 bp and the G + C content was 40%. Similar to other α-CoVs, AcCoV-JC34 has a concise genomic organization and genes characteristic of CoV, including (from 5′ to 3′) the ORF1ab, S, envelope (E), membrane (M), and nucleocapsid (N) genes (Table 2 and Fig. 2). In addition, ORFs likely coding for accessory proteins ORF3, ORF6, ORF8, and ORF9 were also found.
Table 2.
Gene similarities of AcCoV-JC34 and representative Alpha- and Beta-CoVs with full-length genome
| Viruses | Genome | Pairwise amino acid sequence identity between JC34 and other virus sequences (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Size (nt) | G + C content(%) | 3CLpro | RdRp | Hel | S | E | M | N | |
| Alphacoronavirus | |||||||||
| RatCoV-JC34 | 27,649 | 40.07 | - | - | - | - | - | - | - |
| BatCoV 1A | 28,326 | 38.14 | 57 | 76.6 | 74 | 17.6 | 36.5 | 50.2 | 23.3 |
| BatCoV HKU2 | 27,164 | 39.32 | 58.5 | 75.1 | 73.3 | 39.2 | 36 | 53.5 | 27.9 |
| BatCoV HKU8 | 28,773 | 41.79 | 61.2 | 76.5 | 73.5 | 18.6 | 33.8 | 51 | 23.5 |
| RatCoV LRNV# | 28,763 | 40.29 | 91.9 | 94.3 | 96 | 67.2 | 93.8 | 92.3 | 78 |
| BatCoV 512 | 28,203 | 40.12 | 60.9 | 73.3 | 71.8 | 18.3 | 35.5 | 53.1 | 24.6 |
| PEDV | 28,033 | 42.02 | 59.7 | 74.3 | 72 | 17 | 34.2 | 52.7 | 23.2 |
| HCoV-NL63 | 27,553 | 34.46 | 59.8 | 73.9 | 73 | 17.2 | 35.1 | 50.8 | 24 |
| HCoV-229E | 27,317 | 34.46 | 58.9 | 73.3 | 74.3 | 19 | 36.4 | 46.3 | 21.7 |
| MiCoV | 28,941 | 37.47 | 59.7 | 77.9 | 73 | 17.5 | 40.7 | 53.2 | 25.9 |
| FCoV | 29,277 | 38.34 | 57.8 | 75.3 | 71.4 | 16.8 | 42 | 55.6 | 24.7 |
| CCoV | 29,380 | 37.56 | 58.1 | 76.4 | 71.9 | 16.9 | 40.7 | 57.3 | 25.6 |
| TGEV | 28,586 | 37.6 | 58.5 | 76.5 | 71.9 | 17.3 | 37 | 56.9 | 26.1 |
| Betacoronavirus lineage A | |||||||||
| Betacoronavirus 1 | |||||||||
| HCoV-OC43 | 30,741 | 36.69 | 44.1 | 55.8 | 59.1 | 21.8 | 17.6 | 37.4 | 21 |
| PHEV | 30,480 | 37.25 | 44.1 | 55.9 | 59.1 | 21.4 | 18.8 | 37.4 | 20.9 |
| BCoV | 31,032 | 37.02 | 44.4 | 55.7 | 59.5 | 22.1 | 18.8 | 38.7 | 21.4 |
| ECoV | 30,992 | 37.23 | 44.4 | 56 | 59.5 | 21.5 | 17.6 | 37.4 | 21.7 |
| CrCoV | 31,028 | 37.01 | 44.4 | 55.8 | 59.5 | 21.9 | 18.8 | 38.3 | 21.2 |
| Murine coronavirus | |||||||||
| MHV | 31,386 | 41.88 | 42.9 | 55.5 | 57.5 | 21.1 | 18.1 | 36.8 | 21.3 |
| RCoV | 31,250 | 41.26 | 42.9 | 56 | 58.9 | 21 | 20.5 | 38 | 21.3 |
| RatCoV HKU24# | 31,249 | 40.07 | 43.3 | 56 | 60.5 | 21.6 | 20.5 | 39.6 | 21 |
| RatCoV-708#@ | 30,821 | 39.06 | 44.1 | 56 | 58.5 | 20.6 | 20.5 | 36.8 | 20.8 |
| HCoV-HKU1 | 29,926 | 32.06 | 44.8 | 55.8 | 58.3 | 21.2 | 30 | 37.9 | 21.8 |
| RabbitCoV HKU14 | 31,100 | 37.64 | 44.8 | 56 | 59.7 | 21.8 | 17.6 | 38.7 | 21.5 |
| Betacoronavirus lineage B | |||||||||
| SARS-CoV | 29,727 | 40.80 | 43 | 58.9 | 60.8 | 22.1 | 19 | 27 | 19.4 |
| Betacoronavirus lineage C | |||||||||
| MERS-CoV | 30,112 | 41.18 | 40.9 | 58.7 | 63.6 | 21.2 | 19.3 | 34.8 | 18.6 |
| Betacoronavirus lineage D | |||||||||
| BatCoV HKU9 | 29,114 | 41.05 | 39.8 | 57.2 | 62.3 | 22.2 | 17.7 | 29.6 | 16 |
#: novel detected unclassified rat coronaviruses; @: a near complete genome
Fig. 2.

Comparison of the genome organizations of AcCoV-JC34, LRNV, HKU2, FCoV, 229E, MHV, HKU24, and WIV1. Predicted ORFs and 5 conserved domains are indicted by the boxes. Abbreviations: 3CL, 3C-like protease; E, envelope; HE, hemagglutinin-esterase; Hel, helicase; M, membrane; N, nucleocapsid; PL1pro and PL2P, papain-like proteases 1 and 2; RdRp, RNA-dependent RNA polymerase; S, spike
A hexanucleotide transcriptional regulatory sequence (TRS) is in the 5′-leader sequence and is required for the transcription of subgenomic RNAs, which is a unique characteristic of CoVs. Similar to rat CoV LRNV, bat CoV HKU2, and human CoV NL63, a putative TRS motif, 5′-AACUAA-3′ was found upstream of each ORF except for S, ORF3, and ORF8 in the AcCoV-JC34 genome. Instead of the 5′-AACUAA-3′ motif, the S, ORF3, and ORF8 genes had a variant TRS, 5′-AACUUA-3′, 5′-UACUAA-3′, and 5′-CACUAA-3′, respectively (Table 3).
Table 3.
Locations of predicted ORFs in the genome of AcCoV-JC34
| ORF | Location (nt) | Length (nt/aa) | TRS location | TRS sequence(s) (distance to AUG) |
|---|---|---|---|---|
| 1ab | 437–20376 (shift at 12361) | 19940/6645 | 181 | CAACTCAACUAAACG(247)ATG |
| S | 20376–23756 | 3381/1126 | 20306 | TGGTAAACUUATTGTTAG(64)ATG |
| 3 | 23753–24397 | 645/214 | 23711 | TGTTGUACUAAACCT(36)ATG |
| E | 24397–24633 | 237/78 | 24392 | CCACTTAACUA A TG |
| M | 24643–25389 | 747/248 | 24636 | TTGATCAACUAAAATG |
| 6 | 25401–25901 | 501/166 | 25389 | GGTCTAAACUAAACCATTATG |
| N | 25903–27072 | 1170/389 | 25896 | AATTTCAACUAAAATG |
| 8 | 26530–27702 | 543/180 | 26484 | TAGATCGCACUAA(40)ATG |
| 9 | 27074/27391 | 318/105 | 27060 | TGATGAAACUAACGCCTAAAATG |
Start codons are underlined. The conserved (AACUAA) or variant (AACUUA, UACUAA, CACUAA) TRS core sequences are highlighted in bold
Sixteen putative nonstructural proteins (nsp1 to nsp16) coded by ORF1ab of the AcCoV-JC34 were predicted (Table 4). The overall amino acid (aa) identity between the ORF1a and ORF1b polyproteins of AcCoV-JC34 and those of LRNV were 76 and 93.5%, respectively, but <60% relative to those of the other α-CoVs. The most conserved proteins 3CLpro (nsp5), RdRp (nsp12), and Hel (nsp13) of AcCoV-JC34 possessed high aa identities, ranging from 91.9 to 96% compared to those of LRNV, but possessed low aa identities ranging from 57 to 77.9% compared to those of other known α-CoVs (Table 2). In addition, similar to the normal cleavage sites found in polyprotein of α-CoVs, 10 different cleavage sites were predicted between the nsps of AcCoV-JC34 (Table 4).
Table 4.
Prediction of nsp1 to nsp16 and the cleavage sites of polyproteins 1a and 1b of the AcCoV-JC34
| nsp | First-last amino acid residues | Protein size (aa) | Cleavage site | Putative functional domain(s) |
|---|---|---|---|---|
| nsp1 | M1-G218 | 218 | SCPCG|KSAFT | Unknown |
| nsp2 | K219-G820 | 602 | WVCKCG|AEVQLS | Unknown |
| nsp3 | A821-A2277 | 1457 | TRVGTA|DLAVFN | ADRP, PL1pro, PL2pro |
| nsp4 | D2278-Q2779 | 502 | LNAQ|SCAK | Hydrophobic domain |
| nsp5 | S2780-Q3042 | 263 | AKVQ|IEGA | 3CLpro |
| nsp6 | I3043-Q3319 | 277 | SGMQ|CSWA | Hydrophobic domain |
| nsp7 | C3320-Q3437 | 118 | STIQ|SKLT | Unknown |
| nsp8 | S3438-Q3715 | 278 | VKLQ|NNEI | Unknown |
| nsp9 | N3716-Q3820 | 105 | VRLQ|AGKP | Unknown |
| nsp10 | A3821-Q3955 | 135 | STVQ|SNII | Unknown |
| nsp11 | S3956-N3974 | 19 | - | Short peptide at the end of ORF1a |
| nsp12 | L3956-Q4881 | 926 | SVLQ|SAGL | RdRp |
| nsp13 | S4882-Q5478 | 597 | TDLQ|SVLS | Hel |
| nsp14 | S5479-Q5998 | 520 | PILQ|GLEN | ExoN |
| nsp15 | G5999-Q6334 | 346 | PQLQ|NSEW | NendoU |
| nsp16 | N6345-K6646 | 302 | - | O-MT |
Superscript numbers indicate positions in polyprotein pp1ab, with the supposition of a ribosomal frame shift resulting in a peptide bond between N3974/L3975 for the expression of ORF1ab
The S protein of AcCoV-JC34, consisting of 1126 amino acid residues, is predicted to be a type-I membrane glycoprotein with a signal peptide (residues 1 to 19), an extracellular region (residues 20 to 1070), a transmembrane domain (residues 1071 to 1093), and an intracellular region (residues 1094 to 1126) (Additional file 1: Figure S2). A fusion peptide (FP) and two heptad repeats (HR1 and HR2) important for membrane fusion and viral entry were located at residues 674 to 692 for FP, 753 to 840 for HR1, and 1029 to 1058 for HR2. The S protein of AcCoV-JC34 showed the highest aa similarity of 66.5% compared with rat CoV LRNV, followed by 39.2% compared with BtCoV HKU2. Proteolysis of the S protein plays a pivotal role in the activation of viral and cell membrane fusion. Two cleavage sites, one located at residue 508 at the S1/S2 interface (RRAR/AR), and the other located at residue 674 (R/S) at the S2′ position, were predicted by comparing aa sequences based on analysis with a web-based ProP program (Additional file 1: Figure S2). The S1 region of AcCoV-JC34 has an N-terminal domain (NTD) and C-terminal domain (CTD). Both the NTD and CTD showed low aa sequence identities of <25% with those of other very well characterized CoVs. One of them was responsible for receptor recognition and binding, but due to the high dissimilarity with known receptor-binding domains (RBDs), it was difficult to determine the precise location of the RBD of AcCoV-JC34.
The AcCoV-JC34 proteins ORF3, E, M, ORF6, N, ORF8, and ORF9 also had low aa identities with those of other known α-CoVs. The structural proteins E, M, and N of AcCoV-JC34 showed differences compared to homologues of known CoVs. The most conserved M protein had 46.3 to 92.3% sequence identity relative to those of α-CoVs. The N protein was most variable with only 21.3 to 77.4% sequence identity compared to those of α-CoVs at the aa-sequence level. Homologues of the ORF3, ORF6, ORF8, and ORF9 proteins could be found among some CoVs but with low similarity. Previous studies have shown that the ORF3, ORF6, and ORF9 proteins of CoVs may play different functions for the viral life cycle and pathogenesis, although more studies are needed to discern the functions of the NS proteins of AcCoV-JC34.
Phylogenetic features of rodent CoVs
The first phylogenetic tree was constructed based on 400-bp RdRp sequences. In this tree, JC54 and JC34 clustered in the α-CoVs, within rodent and shrew CoVs (Fig. 1). JC34 was distantly related to the branch formed by the closely related CoV strains JC54, UKRn1, and LNRV (Lucheng-19). The other 21 CoV sequences detected from A. chevrieri or A. ilex clustered in β-CoVs and formed an independent lineage together with HKU24 from R. norvegicus and Longquan-353 from A. agrarius, in China. The 20 sequences detected from A. chevrieri were further divided into two branches.
Using predicted protein sequences, we further analyzed the phylogenetic features of AcCoV-JC34. In the phylogenetic trees constructed based on polyprotein 1a and 1b, AcCoV-JC34 clustered in the same branch with a rat CoV LRNV. Interestingly, in the tree based on the S protein, AcCoV-JC34 clustered with a rat CoV LRNV and a bat CoV HKU2 and formed a branch that appeared distinct from α-CoVs, β-CoVs, γ-CoVs, and δ-CoVs (Fig. 3). In the trees based on other genes, AcCoV-JC34 and LRNV together formed independent branches. These results indicated that AcCoV-JC34 possessed a special evolutionary position and may have a common origin with LRNV and HKU2 for the S protein (Fig. 3).
Fig. 3.
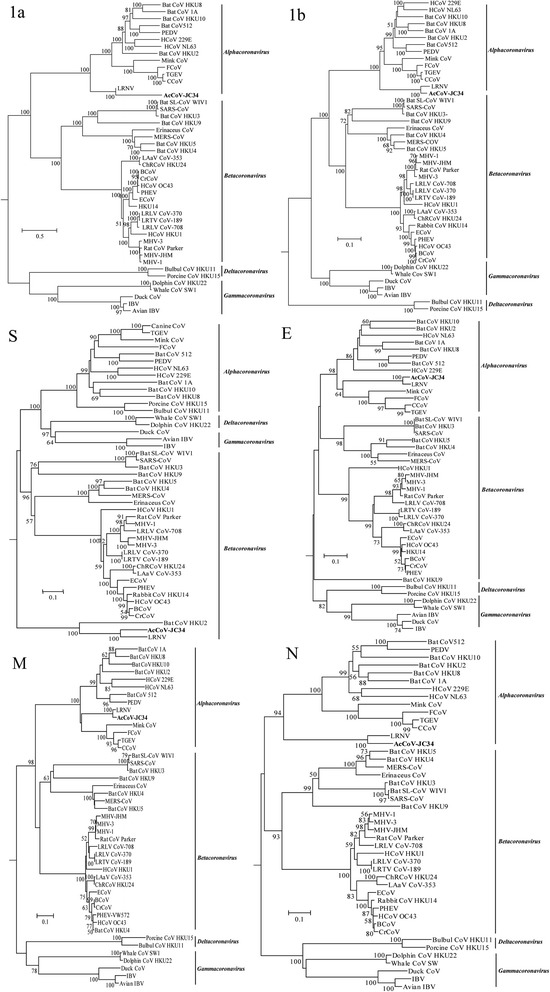
Phylogenetic analyses of AcCoV-JC34 based on amino acid sequences of ORF1a, 1b, S, E, M, and N. The trees were constructed by the maximum-likelihood method with 1000 bootstrap replicates. Bootstrap values above 50% are shown. AcCoV-JC34 identified in this study is shown in bold. The abbreviations and GenBank accession numbers are the same as those used in Fig. 1
Discussion
We detected CoVs in three different rodent species (A. chevrieri, A. ilex, and E. fidelis) from the Yunnan Province of China. In this study, we found a much higher (21.4%) detection rate of CoV nucleic acids in A. chevrieri than detected previously in other rodent species (<5%) [19, 20]. In addition, both α-CoV and β-CoV were found in A. chevrieri, suggesting that A. chevrieri may play an important role as a natural CoV host. A. chevrieri is known as Chevrier’s field mouse and is a dominant and endemic species in southwest China [32, 33]. In the Yunnan Province, A. chevrieri is an important pest in agriculture and human diseases that has been identified as a natural reservoir for plague bacilli and hantavirus [33]. The detection of both α-CoV and β-CoV in A. chevrieri with high infection rates highlighted the importance of viral surveillance in A. chevrieri in the Yunnan Province, which may be helpful for disease prevention and control.
We further characterized a full-length genome of a novel α-CoV, AcCoV-JC34, from A. chevrieri. In all five conserved replicase domains, AcCoV-JC34 was the most closely related to a R. norvegicus CoV LNRV, but diverged from other α-CoVs, indicating that AcCoV-JC34 and LNRV belong to a novel α-CoV species. To our knowledge, AcCoV-JC34 is one of the few rodent α-CoVs with a complete genome.
The genome of AcCoV-JC34 had some unique features compared to other CoVs, such as a shorter nsp5 (3CLpro) and three variant TRSs. These sequences containing the genes or elements were verified by PCR and NGS. Analysis of the aa sequence showed that the proteins encoded by AcCoV-JC34 had very low similarities to other α-CoVs. In particular, the S protein sequence had <20% sequence identity to other α-CoVs (except for LNRV and HKU2), but had a little higher identity (20.6 to 22.1%) compared to β-CoVs. In addition, the N proteins normally were conserved among each of CoV genera, but the N protein of AcCoV-JC34 only shared approximately 25% aa sequence identity with other α-CoVs (except for LNRV) (Table 2). The phylogenetic trees for ORF1a, 1b, and N showed that AcCoV-JC34 and LNRV formed a distinct branch within but at the root position of α-CoVs, suggesting that AcCoV-JC34 and LNRV may represent a special evolutionary position among α-CoVs. More interestingly, in the phylogenetic trees of S, AcCoV-JC34, LNRV, and HKU2 formed a root branch including all CoVs. These results suggested that other unknown CoVs exist in rodents in nature. Further studies should be continued to reveal the prevalence, diversity, and evolution of rodent CoVs.
All samples used in this study were from rodent intestines, suggesting a possible enteric tropism of rodent CoVs. During previous decades of research, different tissue tropisms of rodent CoVs have been observed. As the prototype of rodent CoVs, different MHV strains can infect variant tissues, and the A59 strain is primarily hepatotropic, but the JHM strain is neurotropic [15–18]. RCoV and a strain of sialodacryoadenitis virus (SDAV) both primarily infect the respiratory tract [34]. However, the tropisms of recently identified rodent CoVs from China and Europe have not been confirmed. A CoV in lineage A of β-CoV detected in the alimentary tract samples of Norway rats, HKU24, probably has enteric tropism [20]. Another cluster of α-CoVs (PLMg1, UKMa2, UKMa1, and UKRn1) were only detected in liver samples of Norway rats, the bank vole, the wood mouse, and the noncyclic field vole, suggesting that they are hepatotropic [21]. Additional research identified rodent α-CoV LRNV and β-CoVs LAMV and LRLV, which came from diverse tissue types that made it difficult to predict the tissue tropism of these viruses [19]. Nonetheless, the extensively studied rodent CoVs (MHV and RCoV) could lead to severe or mild diseases in their hosts. Further studies are needed to determine the potential pathogenicity of AcCoV-JC34 along with other recently detected rodent CoVs.
In the AcCoV-JC34 genome, a predicted ORF3 protein (214 aa) was located between the S and E genes. The ORF3 protein of AcCoV-JC34 possessed 30 to 78% aa sequence identity with the homologous proteins encoded by other α-CoVs. This protein has variant names in different CoVs and was named ORF4 protein in human coronavirus 229E [35], non-structural protein 3 in human coronavirus NL63 [36], 3c-like protein or non-structural protein 3c in ferret coronavirus, 3c protein in feline coronavirus [37], 3b protein in transmissible gastroenteritis virus (TGEV) [38], and ORF3 protein in porcine epidemic diarrhea virus (PEDV) [39]. Normally, the ORF3 protein was considered as an accessory non-structural protein, but several studies showed that the ORF3 protein was a membrane protein related to virulence [35, 37, 38, 40]. However, with low similarities between the ORF3 of AcCoV-JC34 and previously studied proteins, more experiments are needed to understand its function.
The S protein of CoVs is responsible for receptor recognition, binding, and membrane fusion, and serves as the first key factor of host restriction by meditating viral entry. In different CoVs, the RBD can be located at the NTD or CTD in S1. For example, among the α-CoVs, CTD was characterized as RBD in HCoV NL63 (aa 476–616), 229E (aa 417–547), and PEDV [41–44], but the NTD was characterized as an RBD in the TGEV [45]. Here, the S1 of AcCoV-JC34 shared <20% aa sequence identity with those of very well characterized α-CoVs, which made it difficult to predict whether the RBD was located in NTD or CTD and which host molecule could be the possible receptor for AcCoV-JC34. The S2 of AcCoV-JC34 showed 40 to 50% identity to that of β-CoVs. By sequence alignment and SWISS-MODEL analysis (data not shown), we deduced the precise positions of FH, HR1, and HR2. The higher similarities between S2 of AcCoV-JC34 (HKU2) and β-CoVs than that to α-CoVs suggested that the structure and functional mechanism of S2 of AcCoV-JC34 may more homologous to β-CoVs.
Emerging infectious diseases caused by CoV are mostly due to interspecies transmission from animals to humans. Previous data indicated that bats are natural reservoirs for α- and β-CoVs [46]. A number of human CoVs, including SARS-CoV, MERS-CoV, HCoV229E, and NL63 might have originated from bats [47, 48]. Among the rodent CoVs, the receptor usage, tissue tropism, and pathogenesis of MHV have been studied in detail [49]. However, novel CoVs, like AcCoV-JC34, HKU24, LRNV, LAMV, and LRLV are not fully understood. Identification of the receptor for these viruses could help in evaluating the potential host range and ability for interspecies transmission from rodents to other mammals. Although most of these novel rodent CoVs have been characterized with full-length or near full-length genome sequences, the lack of successfully isolating those viruses thoroughly restricts future studies. More positive samples and cell lines will facilitate viral culture in the future. In addition, more attention should be paid to the diversity of CoVs in rodent, which could help to better understand the role of rodents in the evolution and ecology of CoVs.
Conclusions
The results of this study revealed that diverse α-CoVs and β-CoVs are circulating in rodents in the Yunnan Province of China and highlighted the importance of rodents as a natural reservoir for CoVs. The complete genome of Ac-JC34 with new characteristics and a special S gene provided new insights into the genetics and evolution of CoVs. These findings should be useful for future genomic studies of CoVs and for further functional studies of S proteins.
Acknowledgements
Not applicable.
Funding
This work was jointly funded by the National Natural Science Foundation of China (81260437, 81660558), the Scientific and Technological Basis Special Project from the Minister of Science and Technology of China (2013FY113500) (to Y-ZZ), the Yunnan Centre for Collaborative Innovation in Public Health and Disease Prevention and Control (2015YNPHXT05) (to Y-ZZ), and the China Mega-Project for Infectious Disease (2014ZX10004001-003) (to Z-LS).
Availability of data and materials
Nucleotide sequences are available in GenBank under accession numbers KX964649–KX964657. Other relevant data are included in the manuscript.
Authors’ contributions
YZ-Z, WH-Y, and JH-Z collected the samples. XY-G, B-L, YZ-Z, and W-Z performed the experiments. XY-G and ZL-S analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Yunnan Institute of Endemic Diseases Control and Prevention. Sampling procedures were performed following the guidelines and protocols for the Laboratory Animal Use and Care from the Chinese Centers for Disease Control and the Rules for the Implementation of Laboratory Animal Medicine (1998) from the Ministry of Health, China.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- A. chevrieri
Apodemus chevrieri
- A. ilex
Apodemus ilex
- aa
Amino acid
- CoVs
Coronaviruses
- CTD
C-terminal domain
- CytB,
Cytochrome b
- E
Envelope
- E. fidelis
Eothenomys fidelis
- FP
Fusion peptide
- HR
Heptad repeats
- M
Membrane
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MHV
Mouse hepatitis virus
- N
Nucleocapsid
- NGS
Next-generation sequencing
- nt
Nucleotide
- NTD
N-terminal domain
- ORFs
Open reading frames
- PDB
Protein Data Bank
- PEDV
Porcine epidemic diarrhea virus
- RBD
Receptor-binding domain
- RdRp
RNA-dependent RNA polymerase gene
- RT-PCR
Reverse transcription PCR
- S
Spike
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- SDAV
Sialodacryoadenitis virus
- TGEV
Transmissible gastroenteritis virus
- TRS
Transcription regulatory sequence
- α-CoV
Alphacoronavirus
- β-CoV
Betacoronavirus
- γ-CoV
Gammacoronavirus
- δ-CoV
Deltacoronavirus
Additional file
Geographical map of Jianchuan country and the sampling areas. Figure S2. Alignment and predicted domains and cleavage sites of AcCoV-JC34 spike protein. NTD, N-terminal domain; RBD, receptor-binding domain; HR, heptad repeat; TM, transmembrane anchor. The signal peptide corresponds to residues 1 to 19. The cleavage sites were indicated by arrows. (PDF 223 kb)
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-017-0766-9) contains supplementary material, which is available to authorized users.
Contributor Information
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Yun-Zhi Zhang, Email: zhangyunzhi1818@163.com.
References
- 1.Masters PS, Perlman S. Chapter 28: Coronaviridae. In: Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 825–79.
- 2.Cavanagh D, Britton P. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Family Coronaviridae. In: Andrew MQ, King MJA, Carstens EB, Lefkowitz EJ, editors. International Committee on Taxonomy of Viruses. Leiden: Academic Press Elsevier; 2012. p. 770–92.
- 3.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Ge XY, Hu B, Shi ZL. Bat coronaviruses. In: Lin-Fa Wang CC, editor. Bats and viruses: from pathogen discovery to host genomics. NY: Wiley; 2015. p. 127–55.
- 8.Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Shi Z, Yu M, Ren W, Smith C, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 11.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luis AD, Hayman DTS, O'Shea TJ, Cryan PM, Gilbert AT, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc R Soc B-Biol Sci. 2013;280. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3574368/. [DOI] [PMC free article] [PubMed]
- 15.Lane TE, Hosking MP. The pathogenesis of murine coronavirus infection of the central nervous system. Crit Rev Immunol. 2010;30:119–130. doi: 10.1615/CritRevImmunol.v30.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner LP. Pathogenesis of demyelination induced by a mouse hepatitis. Arch Neurol. 1973;28:298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]
- 17.De Albuquerque N, Baig E, Ma X, Zhang J, He W, et al. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J Virol. 2006;80:10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funk CJ, Manzer R, Miura TA, Groshong SD, Ito Y, et al. Rat respiratory coronavirus infection: replication in airway and alveolar epithelial cells and the innate immune response. J Gen Virol. 2009;90:2956–2964. doi: 10.1099/vir.0.014282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Lin XD, Guo WP, Zhou RH, Wang MR, et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau SK, Woo PC, Li KS, Tsang AK, Fan RY, et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsoleridis T, Onianwa O, Horncastle E, Dayman E, Zhu M, et al. Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses. 2016;8:84. doi: 10.3390/v8030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabre PH, Hautier L, Dimitrov D, Douzery EJ. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol Biol. 2012;12:88. doi: 10.1186/1471-2148-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin DM, Kocher TD, Wilson AC. Evolution of the Cytochrome-B Gene of Mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 24.de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, et al. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge X, Li Y, Yang X, Zhang H, Zhou P, et al. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Ou HY, Chen LL, Zheng WX, Zhang CT. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2–3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 28.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Criscuolo A. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol. 2011;61:944–948. doi: 10.1016/j.ympev.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 32.Xing-Yuan M, Xian-Guo G, Wen-Ge D, Ai-Qin N, Ti-Jun Q, et al. Ectoparasites of Chevrier's field mouse, Apodemus chevrieri, in a focus of plague in southwest China. Med Vet Entomol. 2007;21:297–300. doi: 10.1111/j.1365-2915.2007.00679.x. [DOI] [PubMed] [Google Scholar]
- 33.Yue H, Fan Z, Liu S, Liu Y, Song Z, et al. A mitogenome of the Chevrier's field mouse (Apodemus chevrieri) and genetic variations inferred from the cytochrome b gene. DNA Cell Biol. 2012;31:460–469. doi: 10.1089/dna.2011.1301. [DOI] [PubMed] [Google Scholar]
- 34.Liang SC, Schoeb TR, Davis JK, Simecka JW, Cassell GH, et al. Comparative severity of respiratory lesions of sialodacryoadenitis virus and Sendai virus infections in LEW and F344 rats. Vet Pathol. 1995;32:661–667. doi: 10.1177/030098589503200607. [DOI] [PubMed] [Google Scholar]
- 35.Dijkman R, Jebbink MF, Wilbrink B, Pyrc K, Zaaijer HL, et al. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol J. 2006;3:106. doi: 10.1186/1743-422X-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez SR, Sims GE, Wentworth DE, Halpin RA, Robinson CC, et al. Genomic analysis of 16 Colorado human NL63 coronaviruses identifies a new genotype, high sequence diversity in the N-terminal domain of the spike gene and evidence of recombination. J Gen Virol. 2012;93:2387–2398. doi: 10.1099/vir.0.044628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bank-Wolf BR, Stallkamp I, Wiese S, Moritz A, Tekes G, et al. Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis. Vet Microbiol. 2014;173:177–188. doi: 10.1016/j.vetmic.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Hasoksuz M, Spiro D, Halpin R, Wang S, et al. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim YK, Cho YY, An BH, Lim SI, Lim JA, et al. Molecular characterization of the spike and ORF3 genes of porcine epidemic diarrhea virus in the Philippines. Arch Virol. 2016;161:1323–1328. doi: 10.1007/s00705-016-2758-2. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor JB, Brian DA. The major product of porcine transmissible gastroenteritis coronavirus gene 3b is an integral membrane glycoprotein of 31 kDa. Virology. 1999;256:152–161. doi: 10.1006/viro.1999.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin HX, Feng Y, Wong G, Wang L, Li B, et al. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J Gen Virol. 2008;89:1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- 42.Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng F, Ye G, Liu Q, Navid MT, Zhong X, et al. Identification and Comparison of Receptor Binding Characteristics of the Spike Protein of Two Porcine Epidemic Diarrhea Virus Strains. Viruses. 2016;8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reguera J, Ordono D, Santiago C, Enjuanes L, Casasnovas JM. Antigenic modules in the N-terminal S1 region of the transmissible gastroenteritis virus spike protein. J Gen Virol. 2011;92:1117–1126. doi: 10.1099/vir.0.027607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo PC, Lau SK, Lam CS, Tsang AK, Hui SW, et al. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in gammacoronavirus. J Virol. 2014;88:1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Huynh J, Li S, Yount B, Smith A, Sturges L, et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng G, Sun D, Rajashankar KR, Qian Z, Holmes KV, et al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci U S A. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Nucleotide sequences are available in GenBank under accession numbers KX964649–KX964657. Other relevant data are included in the manuscript.


