Abstract
Ten-eleven-translocation (TET) proteins oxidize 5-methylcytosine (5mC) to form stable or transient modifications (oxi-mCs) in the mammalian genome. Genome-wide mapping and protein interaction studies have shown that 5mC and oxi-mCs have unique distribution patterns and alternative roles in gene expression. In addition, oxi-mCs may interact with specific chromatin regulators, transcription factors and DNA repair proteins to maintain genomic integrity or alter DNA replication and transcriptional elongation rates. In this review we will discuss recent advances in our understanding of how TETs and 5hmC exert their epigenetic function as tumor suppressors by playing alternative roles in transcriptional regulation and genomic stability.
Introduction
Cytosine modifications have been implicated in a wide variety of biological processes such as maintenance of pluripotency, development and differentiation of mammalian cells [1]. 5-methylcytosine (5mC) is the most abundant CpG modification in the mammalian genome and is typically associated with gene silencing [2]. The discovery of the Ten-Eleven-Translocation (TET) family of enzymes and their ability to generate oxidative products of 5mC (oxi-mCs) have led to intense investigation into the role these modifications play in reversing DNA methylation signatures and the regulation of transcriptional activity. Recent evidence points towards additional roles for TETs and oxi-mCs in protecting the genome from the accumulation of mutations and chromosomal lesions that may predispose cells to malignancy.
TET-mediated DNA oxidation
The TET proteins (TET1-3) are a family a α-ketoglutarate (α-KG) and Fe2+-dependent dioxygenases that catalyze the hydroxylation of 5-methylcytosine (5mC) in the mammalian genome to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5CaC) through iterative oxidation reactions [3–6] (Figure 1). The oxidative products of 5mC (oxi-mCs) can exist as stable modifications in the genome or as transient modifications that provide a trigger for DNA demethylation [3,5,7,8]. Passive dilution of 5mC can occur during DNA replication if DNA methyltransferase 1 (DNMT1), which normally targets hemimethylated DNA, is unable to recognize 5hmC [9]. The TET proteins are also able to promote DNA demethylation by triggering base excision repair (BER) of oxi-mCs (5fC or 5caC) [10] or 5hmU generated by deamination of 5hmC [11]. Research into how these modifications shape the epigenetic landscape has been fuelled by the discovery that their dysregulated abundance or loss of normal patterning is a hallmark of cancer.
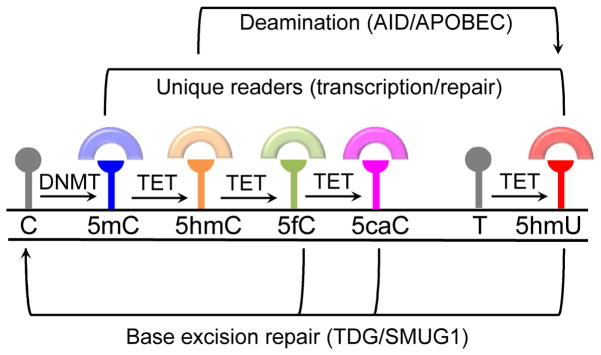
Figure 1. TET-mediated DNA oxidation products.
DNA methyltransferases (DNMT) and a methyl group to cytosine (C) forming 5-methylcytosine (5mC) that can be converted by TET proteins to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5CaC) through iterative oxidation reactions. The oxidative products of 5mC (oxi-mCs) can exist as stable modifications in the genome, or as transient modifications that provide a trigger for DNA demethylation. The TET proteins are able to promote DNA demethylation by triggering base excision repair (BER) via DNA glycosylases (TDG/SMUG1) that target oxi-mCs (5fC or 5caC). 5hmU generated by deamination of 5hmC (via AID/APOBEC deaminases) or by TET-mediated oxidation of thymidine (T) can also trigger BER. Each oxidation product of TET can recruit unique readers (colored semi-circles) that may exert different biological functions in response to the presence of the DNA modification in the genome.
5hmC and TETs are tumor suppressors
Decreased expression of TET proteins and loss of 5hmC has been documented in multiple tumor tissues [12–14] and mutations or deletions in TET genes are prevalent in human cancer genomes. Of the three family members, TET1 and TET3 are most frequently mutated in solid tumors such as cutaneous squamous cell carcinoma, melanoma and colorectal cancer [15–17]. TET2 is primarily mutated in hematopoietic malignancies and is one of the most frequently mutated genes in patients with myelodysplastic syndrome and acute myeloid leukemia (AML) [18–22].
Functional and structural studies using TET2 mutant proteins such as those found in AML patients have revealed that truncation or mutation of the catalytic domain affects the ability of the enzyme to bind Fe2+ or α-KG, leading to impaired oxidation of 5mC and DNA hypermethylation [10,23–25]. Isocitrate dehydrogenase (IDH1/2) mutations are also widespread in patients with AML [26] or glioma [27] and generate 2-hydroxyglutarate (2-HG) instead of α-KG that inhibits TET function [18,26,28]. In addition, mutation in the Wilms’ tumor 1 gene (WT1), a DNA-binding partner of TET2, also blocks 5hmC generation due to impaired DNA recruitment [29,30]. Mutations or deletions in IDH1/2, WT1 and TET1-3 tend to be mutually exclusive in the majority of tumors [15–17,26] and in AML have been shown to drive overlapping aberrant DNA hypermethylation signatures [18,29–31]. Given that decreased expression or mutation of TET genes confers a poor prognosis in multiple cancers [22,32,33], understanding how loss of 5hmC and aberrant DNA methylation in the genome affects tumor biology is an important research objective.
TETs and 5hmC in the regulation of gene expression
A great deal of focus has been placed on studying the role of 5hmC and TETs in transcriptional regulation given the historical association of DNA methylation with gene silencing [2]. All TET proteins bind preferentially at transcriptional start sites (TSSs) and promoters with affinity that positively correlates with CpG density [34–39]. TET1 plays a dual role, activating or repressing its direct target genes [34–36] whereas TET2 binding at promoters positively correlates with gene expression [38]. However, recent studies have shown that TET2 can also act as a negative regulator of lineage specific genes [40]. These findings are consistent with genome-wide mapping studies that show 5hmC localization within gene bodies and promoter regions of both active and repressed genes [35,36,41,42]. 5hmC enrichment has also been observed at active enhancers and is depleted upon loss of TET function [43,44]. 5fC and 5caC are even more abundant than 5hmC at poised and active enhancers and promoter TSSs compared to gene bodies [45–47]. Overall these studies would suggest that TETs, by modulating the balance between inter-genic and intra-genic oxi-mCs, play an important role in dictating the transcriptional outcome of bivalent genes.
Although not experimentally proven, TET proteins may also regulate gene expression by altering chromosomal architecture through its ability to modulate 5mC. The insulator protein, CCCTC-binding factor (CTCF) is inhibited by 5mC from binding at gene bodies [48] and has been shown to require TET-mediated oxidation of 5mC to regulate alternative splicing [49]. TETs could also regulate chromosomal architecture by protecting large undermethylated genomic regions known as DNA methylation “canyons” from hypermethylation [50]. 5hmC enrichment at canyon edges and the aberrant expression of canyon-associated genes in cancer implicates an important role for TETs and 5hmC in their maintenance [51].
TETs and oxidized mC in genomic integrity
The most common single nucleotide polymorphisms (SNPs) and somatic mutations found in cancer genomes is the transition of C to T in the context of CpG dinucleotides [52,53]. This is attributed to an increased tendency for spontaneous deamination of 5mC compared to C [54,55] and the differential recruitment of error-prone repair enzymes. Deamination of an unmethylated C or 5hmC generates U:G or 5hmU:G mismatches that are efficiently repaired by thymine and uracil DNA glyosylases, TDG and SMUG1 [11,56]. 5mC deamination generates T:G mismatches that can recruit TDG but also the error-prone mismatch repair (MMR) complex, MutSα [57,58]. Cytosine deaminases (AID and APOBEC1-3) also have increased activity on 5mC than 5hmC [59,60]. Consequently, the less mutagenic variant of cytosine, 5hmC, may provide protection from deamination-mediated C-T transitions in the mammalian genome. Comparing base-resolution maps of 5mC and 5hmC in normal human brain, kidney and myeloid cells with cytosine mutations in tumors from these tissues, it was indeed shown that 5hmC sites exhibit a 50% decrease in C-T mutation frequency than 5mC and a mutation load similar to that of unmodified cytosines [61]. Another study also reported a minor reduction in C-T transition SNP rates at 5hmC compared to 5mC in mammalian genomes [54].
Interestingly, a higher rate of C-G transversion at 5hmC sites has also been reported in human and mouse ESCs [54]. These transversions may be a consequence of breakdown in repair of the less stable 5fC or 5caC modifications. Unlike 5hmC:G base pairs, TDG recognizes and excises 5fC/5caC:G mismatches [6,62,63] and 5caC:G pairs are bound as strongly as T:G mismatches by the MMR complex [62]. In addition, these oxi-mCs could be recognized by different error-prone DNA polymerases, recruit unique DNA damage response protein proteins or depending on the nucleotide context may differ in their efficiency of repair. Ultimately, in cancer genomes, C-G transversions are orders of magnitude less frequent than C-T transitions [53,55] and 5hmC is vastly more abundant than 5fC and 5caC [4,6,63]. In protecting the genome from DNA hypermethylation, TET proteins and 5hmC may play a dual role in maintaining genomic integrity by limiting the rate of cytosine loss through deamination of 5mC.
DNA repair and damage sensing by oxi-mCs
The placement of 5hmC at the precipice between a stable epigenetic modification and the formation of 5fC and 5caC makes it an ideal intermediate for the recruitment of DNA damage sensing and repair enzymes. In HeLa cells, 5hmC but not 5mC was recently found to co-localize by immunostaining with γH2AX, 53BP1 and RAD51 foci in what appeared to be G1-specific 53BP1 nuclear bodies [64] (Figure 2). In this study, the co-localization at sites of DNA damage was dependent on TET2 and exacerbated by genototoxic insults such as aphidicolin treatment to induce double strand breaks (DSBs). 5hmC foci were never seen without 53BP1 staining, suggesting that damage-induced 5hmC is removed along with 53BP1 during S phase [64,65]. Interestingly, 5mC, 5fC or 5caC staining was absent at DNA damage foci providing further evidence that 5hmC at sites of DNA damage is a stable epigenetic mark [64]. TET proteins and 5hmC are however not essential for the recruitment of DNA repair proteins given studies performed in Tet-triple knockout (Tet-TKO) mESCs, showed that γH2AX foci still formed and could be resolved in response to aphidicolin treatment [64]. Instead, at low dose aphidicolin treatment, which mimics replication stress [65], a greater frequency of chromosome segregation defects were found to occur in Tet-TKO ESCs [64]. Tet-TKO mESCs have also been reported to exhibit increased telomere–sister chromatid exchange [43], a mechanism of homologous recombination (HR) and telomere elongation independent of telomerase [66]. A failure to adequately repair DNA damage sustained during S phase can lead to under-replicated regions that interfere with the separation of chromosomes in mitosis [67]. Accumulation of γH2AX also occurs in the absence of DSBs to regulate cell cycle progression by inhibiting DNA replication [68,69]. In this context the overexpression of TET proteins in NIH-3T3 cells has been shown to induce 5hmC, accumulate γH2AX and delay DNA replication [70]. Given that genomic stability relies on DNA replication fidelity during S phase and correct segregation of sister chromatids during mitosis, the role of oxi-mCs in these highly coordinated cellular processes should be investigated further.
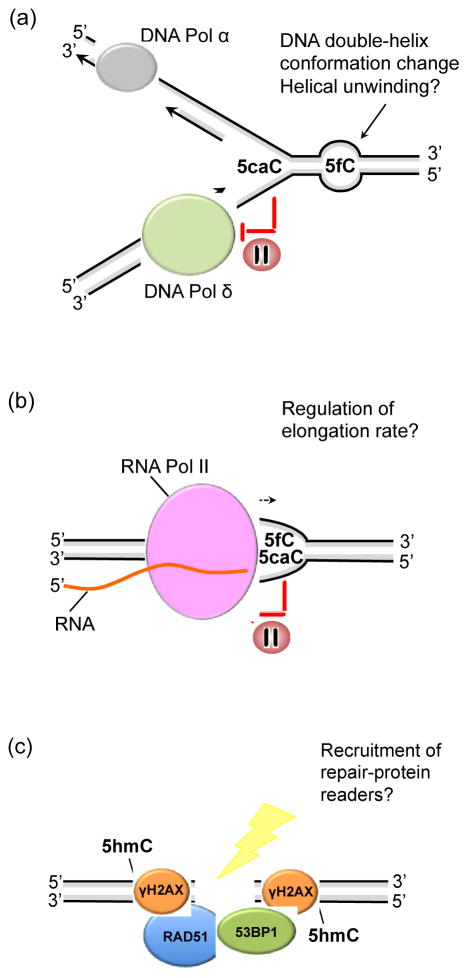
Figure 2. Alternative roles for oxi-mCs in replication, transcription and repair.
A) 5caC is recognized by the proofreading DNA polymerase δ causing a brief stalling at this modified base that may delay DNA synthesis. 5fC alters the structure of the DNA double-helix, characterized by helical unwinding, suggesting that recognition of the altered DNA conformation rather than the modified base itself might trigger biological events. B) 5fC and 5caC have been reported to induce a transient pause in the RNA polymerase II elongation complex that may regulate the rate of transcription as well as promote alternative splicing. C) 5hmC co-localizes with γH2AX, 53BP1 and RAD51 at sites of DNA damage and double strand breaks.
Loss of TET function and genomic instability in cancer
Reducing 5hmC levels below a critical threshold may lead to overt genomic instability in adult cells. Recent studies in mouse models have shown that TET-deficiency plays a causal role in genomic instability and disease progression. Our laboratory discovered that Tet1 deficiency causes decreased expression of DNA repair genes and a spontaneous increase in γ-H2AX foci in developing B cells [71]. B cell progenitors also exhibit aberrant self-renewal and mice spontaneously develop B cell lymphoma upon advanced age [71].
Combined Tet2/Tet3 loss in mouse hematopoietic cells drives an aggressive leukemia that correlates with the spontaneous accumulation of γH2AX and impaired DNA damage repair (DDR) in myeloid progenitors [72]. Gene expression differences in both HR and non-homologous end-joining (NHEJ) repair pathways were also observed in Tet2/3 DKO immature myeloid cells [72]. It is not clear however whether these expression changes are the cause or consequence of defects in DNA damage responses. These studies suggest that Tet2 and Tet3 functionally overlap to maintain a threshold of 5hmC that protects developing myeloid cells from genomic instability as neither KO alone causes spontaneous γH2AX accumulation [72]. Interestingly, TET-deficient hematopoietic stem cells (HSCs) do not show defects in DDR [72,73]. This may be because loss of TET function and the subsequent dilution of 5hmC in the genome require several rounds of DNA replication and the replicative history of HSCs is much younger than progenitors. If this is the case, long-lived HSCs with increased rounds of replication may exhibit greater accumulation of DNA damage in TET-deficient mice.
Readers of oxi-mCs
5hmC could serve as a stable intermediate not only for DNA demethylation but also for the recruitment of chromatin or transcriptional regulators and repair proteins in times of DNA damage or replication stress. 5hmC and 5mC are indistinguishable to DNA polymerases [74] however in a screen to find interacting proteins, these marks exhibit unique reader profiles, and overlap in their recruitment of only a small number of transcriptional regulators, such as MeCP2 and Uhrf1 [75]. DNA glycosylases (Mpg and Neil3) and helicases (Recq1) are uniquely recruited to 5hmC and not 5mC in mESCs whereas 5fC and 5caC recruit a large number of DNA repair proteins, including BER (Neil1, Neil3, and Mpg) and MMR (Msh3 and Exo1) factors [75]. Of all the oxi-mCs, 5fC is the only modification that alters the structure of the DNA double-helix, characterized by helical unwinding, suggesting that recognition of the altered DNA conformation rather than the modified base itself might trigger biological events [76]. This finding could explain why the enriched binding of DNA repair-associated proteins is most pronounced for 5fC [75].
Oxidation of 5hmC may also control the rate of DNA replication and RNA transcription (Figure 2). 5caC is recognized by the proofreading DNA polymerase δ causing a brief stalling at this modified base that may delay DNA synthesis [62]. 5fC and 5caC have also been reported to induce a transient pause in the RNA polymerase II elongation complex that may regulate the rate of transcription as well as promote alternative exon usage [77,78]. TET enzymes also target thymine for oxidation, generating 5hmU, which when paired with adenine is selectively repaired by SMUG1 [79] and attracts its own complement of specific protein readers [75,80]. Given the large number of proteins that recognize oxi-mCs, a combination of diverse biological processes have the potential to regulate gene expression and DNA fidelity by these modifications.
5hmC and asymmetric epigenetic inheritance
5mC sites are highly symmetric, where 99% of methylated CpGs are found to be methylated on both strands [81]. 5mC is copied onto newly synthesized DNA by DNMT1 however 5hmC maintenance, through the action of TET enzymes on newly synthesized DNA strands, has not been reported. Recent sequencing efforts have discovered that 5hmC is distributed asymmetrically in the genome.
Initial studies of whole-genome 5hmC mapping in bulk cell populations led to conflicting reports on the existence of 5hmC strand bias in mouse ESCs [82,83]. A newly described method for genome-wide, strand-specific, single-cell 5hmC sequencing has now confirmed that 5hmC varies in abundance on the two DNA strands of a given chromosome up to 10-fold [84]. Based on the observation that 5hmC marks are passively lost during replication [73] Mooijman et al (ref [84]) hypothesized that differences in strand age between the plus and minus strands of a chromosome could be a potential mechanism for generating the observed 5hmC strand bias [84]. Assuming that cells divide symmetrically, with random chromosome segregation, each of the two daughter cells would inherit one strand of an original mother chromosome. Comparing the distribution of the strand bias between oocytes and single cells from haploid two-cell embryos, the oocytes were shown to have no significant strand bias, whereas single cells from two-cell embryos displayed strong strand bias, with each sister cell receiving an old strand. This heritable 5hmC bias allowed the authors to develop a model to infer sister-cell relationships and cell lineage reconstruction in sets of 4-cell embryos [84]. Asymmetric inheritance of 5hmC has also been observed by immunofluorescence in an adult stem cell model of non-random chromosome segregation [85]. However, the observations from single-cell sequencing of 5hmC in embryos suggests that asymmetric 5hmC inheritance is a general phenomenon and not exclusive to asymmetric cell divisions. The asymmetric inheritance of 5hmC upon DNA replication may serve as a source of chromosome-wide epigenetic memory and has also been observed with histones [86]. Strand-bias of 5hmC may dictate asymmetric distribution of gene regulation or DNA fidelity of template strands upon cell division. Whether this is mediated via the strand-specific recruitment of oxi-mC readers or DNA repair machinery is not yet known.
Conclusions and Future Directions
The tumor suppressive role of TETs and oxi-mCs and their association with the prevention of aberrant DNA hypermethylation extends beyond our conventional view of gene regulation. Unmodified and modified cytosines exhibit different rates of mutation and elicit unique DNA repair responses. The absence of TETs or 5hmC may limit the recruitment of many of the readers of oxi-mCs that are intimately involved in DNA damage repair independently of their role in transcriptional regulation. Steric effects of oxi-mCs in genomic topology could also influence the rate of DNA replication or gene transcription. 5hmC strand bias may be another mechanism by which TETs regulate asymmetric gene expression or DNA repair to instruct cell fate, drive differentiation and protect cells from replication induced DNA damage. DNA hypermethylation and deficient DNA repair may therefore predispose cells to the acquisition of oncogenic mutations independently of the transcriptional role for TETs and 5hmC. Future studies should continue to explore, at single-base resolution, the correlation between mutation frequency, oxi-mC and asymmetric distribution of these modifications in genome-wide sequencing studies to shed light on how TETs and oxi-mCs protect cells from malignant transformation.
Acknowledgments
This research was supported by the US National Institutes of Health 5RO1CA194923, 1R01CA169784, 1R01CA133379, R01CA149655 and 5R01CA173636), the Leukemia & Lymphoma Society (TRP#6340-11 and LLS#6373-13), The Chemotherapy Foundation, The V Foundation for Cancer Research, Alex’s Lemonade Stand Foundation for Childhood Cancer, St. Baldrick’s Cancer Research Foundation and the Howard Hughes Medical Institute (I.A.). The work was also supported by the New York State Department of Health (#CO030132).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 3.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell research. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 8.Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 10.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer science. 2012;103:670–676. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21:1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 21.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nature genetics. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 22.Kosmider O, Gelsi-Boyer V, Ciudad M, Racoeur C, Jooste V, Vey N, Quesnel B, Fenaux P, Bastie JN, Beyne-Rauzy O, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57:662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, Lee MC, Ko BS, Tang JL, Yao M, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Yu SJ, Hong Q, Yang Y, Shao ZM. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated with Poor Prognosis of Patients with Early Breast Cancer. PLoS One. 2015;10:e0133896. doi: 10.1371/journal.pone.0133896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell cycle. 2011:10. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Molecular cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, Guzman A, Lei Y, Huang YH, Rao A, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48:1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 42.Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neri F, Incarnato D, Krepelova A, Rapelli S, Anselmi F, Parlato C, Medana C, Dal Bello F, Oliviero S. Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marina RJ, Sturgill D, Bailly MA, Thenoz M, Varma G, Prigge MF, Nanan KK, Shukla S, Haque N, Oberdoerffer S. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2016;35:335–355. doi: 10.15252/embj.201593235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiehle L, Raddatz G, Musch T, Dawlaty MM, Jaenisch R, Lyko F, Breiling A. Tet1 and Tet2 Protect DNA Methylation Canyons against Hypermethylation. Mol Cell Biol. 2016;36:452–461. doi: 10.1128/MCB.00587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia J, Han L, Zhao Z. Investigating the relationship of DNA methylation with mutation rate and allele frequency in the human genome. BMC Genomics. 2012;13(Suppl 8):S7. doi: 10.1186/1471-2164-13-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Supek F, Lehner B, Hajkova P, Warnecke T. Hydroxymethylated cytosines are associated with elevated C to G transversion rates. PLoS Genet. 2014;10:e1004585. doi: 10.1371/journal.pgen.1004585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. Cytosine to thymidine (C-T) transitions are the most frequent somatic point mutation to accumulate with age. The majority of somatic mutations in the small intestine and colon adult stem cells in this study correlate to a signature consistent with spontaneous deamination of methylated cytosine residues into thymidine at CpG sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kemmerich K, Dingler FA, Rada C, Neuberger MS. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung−/− Msh2−/− mice. Nucleic Acids Res. 2012;40:6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U:G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- 59.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rangam G, Schmitz KM, Cobb AJ, Petersen-Mahrt SK. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One. 2012;7:e43279. doi: 10.1371/journal.pone.0043279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Tomkova M, McClellan M, Kriaucionis S, Schuster-Boeckler B. 5-hydroxymethylcytosine marks regions with reduced mutation frequency in human DNA. Elife. 2016:5. doi: 10.7554/eLife.17082. Tissue-specific 5hmC patterns in brain, kidney and myeloid cells in the blood correlate with lower CpG>T mutation frequency in cancers originating in these respective tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibutani T, Ito S, Toda M, Kanao R, Collins LB, Shibata M, Urabe M, Koseki H, Masuda Y, Swenberg JA, et al. Guanine- 5-carboxylcytosine base pairs mimic mismatches during DNA replication. Sci Rep. 2014;4:5220. doi: 10.1038/srep05220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Kafer GR, Li X, Horii T, Suetake I, Tajima S, Hatada I, Carlton PM. 5-Hydroxymethylcytosine Marks Sites of DNA Damage and Promotes Genome Stability. Cell Rep. 2016;14:1283–1292. doi: 10.1016/j.celrep.2016.01.035. 5hmC is found to co-localize with γH2AX at sites of DNA damage induced by aphidicolin or microirradiation in human cancer cells lines that is dependent on TET activity. In TET triple KO ESCs, chromosome segregation defects accumulate in response to replication stress. [DOI] [PubMed] [Google Scholar]
- 65.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 66.Lazzerini-Denchi E, Sfeir A. Stop pulling my strings - what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol. 2016;17:364–378. doi: 10.1038/nrm.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 68.Fernando RN, Eleuteri B, Abdelhady S, Nussenzweig A, Andang M, Ernfors P. Cell cycle restriction by histone H2AX limits proliferation of adult neural stem cells. Proc Natl Acad Sci U S A. 2011;108:5837–5842. doi: 10.1073/pnas.1014993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turinetto V, Orlando L, Sanchez-Ripoll Y, Kumpfmueller B, Storm MP, Porcedda P, Minieri V, Saviozzi S, Accomasso L, Cibrario Rocchietti E, et al. High basal gammaH2AX levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells. 2012;30:1414–1423. doi: 10.1002/stem.1133. [DOI] [PubMed] [Google Scholar]
- 70.Nakatani T, Yamagata K, Kimura T, Oda M, Nakashima H, Hori M, Sekita Y, Arakawa T, Nakamura T, Nakano T. Stella preserves maternal chromosome integrity by inhibiting 5hmC-induced gammaH2AX accumulation. EMBO Rep. 2015;16:582–589. doi: 10.15252/embr.201439427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H, Lobry C, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, Rao A. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071. doi: 10.1038/ncomms10071. Double-deletion of Tet2 and Tet3 in mice causes the spontaneous accumulation of DNA damge in myeloid progenitors, and an inability to repair DNA damage in reponse to whole body irradiation. Double KO mice exhibit a dramatic decrease in disease latency normally seen with Tet2 loss alone provding causal evidence that a threshold level of TET expression and 5hmC may be required to maintain genomic integrity in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Renciuk D, Blacque O, Vorlickova M, Spingler B. Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine. Nucleic Acids Res. 2013;41:9891–9900. doi: 10.1093/nar/gkt738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 76••.Raiber EA, Murat P, Chirgadze DY, Beraldi D, Luisi BF, Balasubramanian S. 5-Formylcytosine alters the structure of the DNA double helix. Nat Struct Mol Biol. 2015;22:44–49. doi: 10.1038/nsmb.2936. Structural studies reveal that 5fC is the only oxi-mC that causes a bending in the DNA double helix reminiscent of helical unwinding that may trigger the recruitment of DNA repair enzymes through recognition of steric changes in DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, Chong J, Li H, He C, Fu XD, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523:621–625. doi: 10.1038/nature14482. By determining the X-ray crystal structure of yeast elongating RNA pol II in complex with oxi-mCs the authors find that interaction with 5caC causes a positional shift for incoming nucleoside 5′-triphosphate (NTP), thus compromising nucleotide addition. Additional elongation studies in vivo reveal that 5fC and 5caC delay RNA Pol II elongation on gene bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 80.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 81.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, Wang H, Sun D, Cheng X, Zhu Z, et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 2013;3:567–576. doi: 10.1016/j.celrep.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84••.Mooijman D, Dey SS, Boisset JC, Crosetto N, van Oudenaarden A. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol. 2016;34:852–856. doi: 10.1038/nbt.3598. 5hmC strand bias on DNA had been previously observed in bulk cell populations. By using a single-cell 5hmC sequencing technique the strand bias of 5hmC was not only confirmed, but could also be used to reconstruct mother to daughter cell asymmetric inheritance of 5hmC. [DOI] [PubMed] [Google Scholar]
- 85.Huh YH, Cohen J, Sherley JL. Higher 5-hydroxymethylcytosine identifies immortal DNA strand chromosomes in asymmetrically self-renewing distributed stem cells. Proc Natl Acad Sci U S A. 2013;110:16862–16867. doi: 10.1073/pnas.1310323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]