Abstract
The advent of single-cell sequencing has been revolutionary to the field of cancer genomics. Perfectly suited to capture cancer’s heterogeneous nature, single-cell analyses provide information bulk sequencing could never hope to uncover. Many mechanisms of cancer have yet to be fully understood, and single-cell approaches are showing promise in their abilities to uncover these mysteries. Here we focus on the most recent single-cell methods for cancer genomics, and how they are not only providing insights into the inner workings of cancer, but are also transforming individualized therapy and non-invasive monitoring and diagnosis.
Introduction
Genomic analysis has been widely applied in cancer studies. The identification of genomic, epigenomic, and transcriptomic changes in cancer has led to precise classification, biomarker discovery, and mechanical understanding of cancer, and has played an essential part in cancer diagnosis, monitoring, and treatment [1]. However, until recently, bulk sequencing has been the only viable option for cancer genomic analysis. One major limitation is that bulk sequencing cannot detect the heterogeneity within a tumor. This limitation has important clinical consequences. For example, cancer is often composed of multiple clones, and the most aggressive clone is difficult to identify and target since it may not be the one that metastasizes.
Throughout every stage of cancer, cells accumulate distinct mutations, which define the further evolution and progression of the disease. It is commonly viewed that cancer originates from an accumulation of mutations in oncogenes and tumor suppressors such that cell growth becomes unregulated and invasive [2]. The progeny of these cells in turn accumulate further mutations and selective pressures drive clonal evolution. The cancer will eventually metastasize, spreading to other parts of the body through the circulatory or lymphatic systems to form further distinct subpopulations. In addition, targeted cancer therapy may drive further evolution and eventually lead to drug resistance.
The recent advent of single-cell sequencing has revolutionized the field of cancer genomics, opening the door to a vast number of possibilities (Table 1). From the ability to resolve intra-tumoral heterogeneity [10•,17••,27•,34••], map clonal evolution [50,51], and track the development of therapy resistance [10•,58•], to the capacity to analyze rare tumor cell populations such as tumor stem cells and circulating tumor cells [47,48], single-cell techniques have opened new avenues for cancer research. A better understanding of the mechanisms of cancer can in turn inform more effective and personalized treatments.
Table 1.
A summary of relevant single-cell methods and their applications to cancer.
| Method Type | Specific Methods | Application to Cancer Genomics | Refs Experimental Methods |
|---|---|---|---|
| Single-cell whole genome amplification | DOP-PCR, MDA, MALBAC | Used in conjunction with next-generation sequencing to detect intra-tumor CNVs and SNPs. | [7,8,9] |
| Single-cell spatial genomics | STAR-FISH | Detects the spatial distribution of intra-tumor CNVs and SNPs. Can be combined with longitudinal analysis to reveal migratory cells. | [10•] |
| Single-cell transcriptome amplification | Smart-seq, Tang et al. method, single-cell qPCR | Identifies cancer-specific gene expression signatures, cancer cell types, alternative-splicing events. | [11,12,13] |
| Single-cell spatial transcriptomics | smFISH, SeqFISH, MERFISH, FISSEQ, TIVA | Can provide spatially-resolved gene expression signatures in tumors. Has potential applications in tracing cell migratory paths and locating tumor-like stem cells. | [16,17••,18•,19,20•,21,22] |
| Single-cell DNA methylomics | scRRBS, PBAT | Enables the discovery of differential methylation in cancer cells. Potential for broadening understanding of phenotypic plasticity of cancer cells. | [25,26,27•] |
| Single-cell chromatin accessibility | ATAC-seq, Pico-Seq | Can give insight into the differential binding of transcription factors in cancer cells. | [29•,30•,31] |
| Chromosome conformation capture | Hi-C, ChIP-seq | Potential for understanding the mechanisms of cancer heterogeneity through mapping transcription factor-regulatory element interactions. | [32,33] |
| Simultaneous multiple single-cellomics | G&T-seq, scTrio-seq, Darmanis et al. method | Provides an integrated view of intra-tumoral heterogeneity through measuring direct interactions between genomic, transcriptomic, epigenetic, and proteomic variation. | [34••,27•,36•] |
|
| |||
| Computational Methods | |||
|
| |||
| Single-cell spatial transcriptomic inference | Seurat, Achimetal. method | Infers cell location through scRNA-seq data and an in situ RNA reference map of several landmark genes, enabling mapping of intra-tumor spatial heterogeneity. | [37•,38] |
| Pseudo-time ordering | Monocle, TSCAN, Waterfall, SCUBA, Wanderlust, Wishbone | Projects gene expression values from a single time-point to a continuous trajectory over cell differentation. Potential use in understanding differentiation from stem-like cancer cell to matured cancer cell. | [39,40,41,42,43,44•,45] |
| Rare cell-type detection | RaceID, StemID, GiniClust | Potential use in the detection of circulating tumors cells and stem-like cancer cells. | [46,47,48] |
| Clonal evolution inference | SCITE, OncoNEM | Builds lineage trees for understanding evolutionary events such as the development of therapy resistance. | [50,51] |
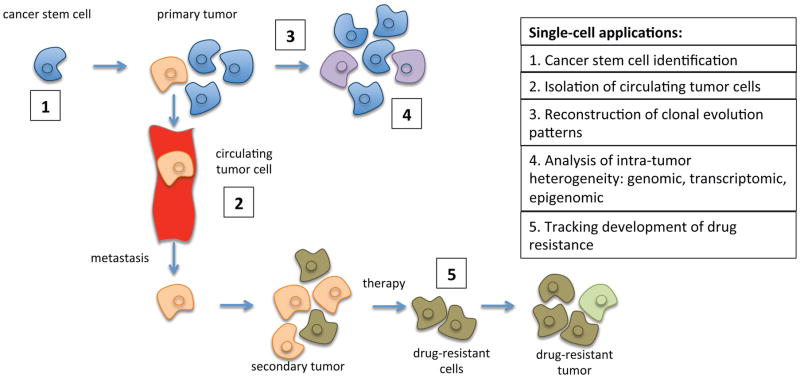
In this paper, we review recent progress in single-cell analysis techniques and their applications in cancer genomics (Figure 1), focusing on topics that have not been covered by previous reviews [3–6].
Figure 1.
Single-cell methods provide novel insights into every stage of cancer progression, from primary tumor development to metastasis, to the development of drug resistance.
Intra-tumor genome sequence heterogeneity
Understanding the genomic heterogeneity of cancer cells first and foremost necessitates methods for single-cell DNA sequencing. The earliest developments for single-cell genomics involve whole genome amplification, providing ample amounts of DNA for subsequent sequencing. Degenerate oligonucleotide primed PCR (DOP-PCR) is appropriate for CNV detection, with low coverage but uniform amplification [7]. Multiple displacement amplification (MDA) is a linear amplification method capable of higher coverage through the use of Phi-29 polymerase, making it suitable for SNP detection [8]. MALBAC (multiple annealing and looping-based amplification cycles) combines MDA and PCR for a high coverage, uniform amplification method suitable for either CNV or SNP detection [9]. These methods have been extensively applied to the characterization of intra-tumor CNVs and SNPs in various cancer types.
However, one major limitation of the aforementioned methods is that spatial information is lost as soon as single cells are isolated. Such information is integral to understanding the interaction of the cell with its micro-environment and may prove valuable for evaluating drug responsiveness. Recently, a new technology, STAR-FISH (specific-to-allele PCR-FISH) [10•], has been developed which can detect the spatial distribution of both SNVs and CNVs using a combination of in situ PCR and FISH. PCR primers are built to target mutant and wild type mRNAs, one gene at a time. Amplification is followed by hybridization of fluorophores to a 5′ overhang built into each probe. Janiszewska et al. use their method to study the commonly reported His1047Arg mutation in PIK3CA and ERBB2 (commonly known as HER2) amplification in HER2+ breast cancer, before and after chemotherapy. They were able to identify changes in mutational frequency of mutated cells, which help gain an understanding of the development of drug resistance in HER2+ breast cancer [10•]. When combined with longitudinal analysis, this method was used to pinpoint migratory cells [10•]. Currently, the technology can only be used to detect the location of known mutations.
The introduction of spatial methods to single-cell cancer genomics allows genomic heterogeneity to be mapped in space. This presents new opportunities in studying cell-to-cell interactions, and in identifying migratory cancer cells and their roles in metastasis.
Intra-tumor transcriptomic heterogeneity
Like single-cell genome analysis, the first efforts in single-cell transcriptomics were in the amplification of the transcriptome to allow for quantification and sequencing of the transcriptome. Whole transcriptome amplification methods include poly-A tailing methods [11] and template-switching methods like Smart-seq [12]. Targeted gene expression profiles can also be quantified by multiplexing qPCR with high sensitivity [13].
In conjunction with single-cell RNA sequencing and qPCR, these methods have been used in various cancer studies. Cancer-specific gene expression signatures and alternative-splicing events have been identified for melanoma [12]. Gene expression signatures have led to the identification of cancer cell types, such as cancer stem cells [14]. The relative contributions of clonal evolution and multi-lineage differentiation in transcriptomic heterogeneity have been studied in the context of colon cancer [15].
Recent technologies have been developed to quantify gene expression levels in situ, thereby preserving spatial information. Here we review recent single-cell spatial transcriptomic methods and their potential for future use in cancer studies. These methods share the same fundamental principle as single-molecule fluorescence in situ hybridization (smFISH), whereby fluorescently-labeled DNA oligonucleotide probes are hybridized to their complementary target mRNA, and are then identified via fluorescence microscopy [16,17••]. The newer techniques described below have greatly enhanced detection efficiency and throughput.
SeqFISH (sequential FISH) is an adaptation of smFISH that uses sequential hybridization to allow for multiplexing [18•]. Each mRNA is assigned a unique sequence of fluorophores that create a barcode through which each mRNA can be decoded. In the first round of this process, probes that target the same mRNA are labeled with the same fluorophore. These probes are hybridized, imaged, and then purged. In the next round, the same probes are labeled with a different fluorophore, and the same sequence of steps is followed. Several rounds of this create a unique barcode of colors for the particular mRNA. Each probe set targeting a particular mRNA is labeled with a unique barcode in this way. For F fluorophores and N hybridization rounds, this means FN mRNAs can be visualized. As this number scales up rapidly with an increasing number of fluorophores and hybridization rounds, this technique can potentially be used to sequence all known genes with limited numbers of fluorophores and hybridization cycles. The authors initially applied this method to immobilized yeast cells and mouse embryonic stem cells [18], but have since extended the method so that it is now applicable to deep tissues such as the brain [19].
MERFISH (multiplexed error-robust FISH) is a similar approach which also allows for error correction by using a smart choice of barcodes [20•]. Specifically, barcode sequences are chosen to include only those that are separated by a certain Hamming distance (Hamming distance=number of changes in a barcode sequence required to transform one sequence into another). Since not all possible barcodes encode a particular mRNA, this encoding scheme provides a means to error detection and correction. The authors use this approach to simultaneously measure 1001 genes in human fibroblast cells. Two fluorophores and 14 hybridization rounds allow all encoding sequences to be separated by a Hamming distance of 2 [20•]. Of note, these authors show that their barcode design helps reduce the error rate significantly.
FISSEQ is another in situ technique which is based on sequencing. RNA is first reverse-transcribed and amplified [21]. The amplicons are crosslinked to the cellular matrix and sequenced by using the SOLiD SBL (sequencing-by-ligation) technique. The method has been applied to a simulation of the wound healing response in primary fibroblasts where the authors found differentially expressed genes between migrating cells and contact-inhibited cells [21]. Such a method could similarly be applied to find differentially expressed genes in migratory vs. non-migratory tumor cells.
In addition, transcriptomic profiles can also be measured in vivo by using a technology called TIVA (transcriptome in vivo analysis). In this approach, a photoactivatable biotin-labeled TIVA-tag is inserted into live cells, attached to mRNA upon selective photoactivation, and recaptured via streptadavin beads. The captured mRNA is subsequently sequenced [22]. TIVA was used on live mouse and human brain tissue, as well as mouse brain cells in culture. A comparison of live and culture mouse brain cells shows significant differences in gene expression levels, emphasizing that cells removed from their natural environment may not be representative of the same cells in vivo [22].
The aforementioned methods give increasingly multiplexed ways of spatially resolving gene expression patterns. While most of the applications to date have been limited to cell culture, we expect that soon they will be applicable to tissue samples. If they can be adapted to tumor cross-sections, these methods will have great impact on investigating the cancer progression path. For example, the location of tumor-like stem cells could be mapped within the tumor. If longitudinal measurements are taken, cell migratory paths may be traced.
Intra-tumor epigenetic heterogeneity
Epigenetics plays an important role in regulating gene expression in cancer, and exploring the heterogeneity of epigenetic patterns may aid in understanding underlying transcriptomic heterogeneity. As a dynamic process, epigenetics may contribute to the phenotypic plasticity of cancer cells, for example aiding in the differentiation of cancer stem cells [23]. Studies have shown abnormally low levels of global DNA methylation along with hyper-methylation in specific regions, such as tumor suppressor gene promoter regions, giving strong evidence for the role of epigenetic aberrations in cancer proliferation [24].
The characterization of intra-tumor epigenetic heterogeneity has been less extensively studied due to its technical difficulty. Nonetheless, multiple epigenetic methods have recently been adapted for single-cell purposes. Determining DNA methylation patterns has traditionally been performed by bisulfite sequencing methods, but bulk techniques have performed poorly in the single-cell setting due to DNA degradation during bisulfite conversion. Methods have adapted bisulfite sequencing for single-cell, including scRRBS (reduced representation bisulfite sequencing) [25] and PBAT (post-bisulfite adapter-tagging) [26]. In each, a modified version of bisulfite sequencing is applied to each cell individually. ScRRBS mitigates the issue of high DNA loss by replacing the multiple purification steps prior to bisulfite sequencing with a single-tube reaction. A restriction enzyme that recognizes CpG islands is used to cut the genome, selecting CpG island regions for subsequent conversion and sequencing. By sequencing only these regions, this method provides low-cost but low-coverage sequencing [25]. ScRRBS has been applied to human hepatocellular carcinoma tissue in conjunction with simultaneous transcriptome sequencing (discussed in greater detail in the next section) [27•]. Methylation levels at all CpG sites were measured and subsequently used to cluster the tissue into two subpopulations via unsupervised hierarchical clustering. A large amount of heterogeneity was found between and within these subpopulations. Interestingly, when the same clustering method was applied using CNV patterns, an identical clustering was found [27•].
PBAT is a more unbiased whole-genome approach that addresses the issue of bisulfite-conversion-induced DNA degradation by performing suitable library preparation after bisulfite sequencing. Traditionally, adapter-tagging is performed before bisulfite conversion and sequencing templates become degraded, but switching the order of these events alleviates this problem [26,28]. In an application of PBAT, differential methylation of distal regulatory elements was discovered in mouse embryonic stem cells [28]. These elements cannot commonly be captured by scRRBS, making it promising for higher-coverage cancer methylation studies.
Chromatin structure also plays an important role in gene regulation. Most transcription factors can only bind to open chromatin regions, whereas a small number of pioneer factors may bind to closed chromatin, opening it up so that other factors can bind. The genome-wide landscape of chromatin accessibility can be measured by using either ATAC-seq (assay for transposase-accessible chromatin) [29•,30•] or DNase-seq [31]. The difference between these two methods is the DNA-cutting enzymes, corresponding to Tn5 and DNase I, respectively. Both methods have been adapted to single-cell analysis. Two single-cell methods have modified ATAC-seq. A combinatorial indexing approach [29•] tags nuclei with unique barcodes so they can then be grouped and processed together. Groups of nuclei are placed in wells, barcoded, and then passed through a second set of wells and barcoded again. Given that each nuclei is highly likely to pass through a unique combination of wells, the barcoding is overwhelmingly cell-specific [29•]. In a microfluidic approach [30•], cells are captured and assayed separately. The microfluidic technique has been used to find a high variability of transcription factor motif accessibility in cancer cell lines [30•]. For DNase-seq, a single-cell method called Pico-Seq [31] sorts cells using FACS before DNaseI treatment. To prevent a large loss of digested DNA during subsequent library preparation, circular carrier DNA is added after digestion. This DNA will not be amplified in the PCR that follows due to its incompatibility with the adaptor ligation process. Of note, the authors applied their method to formalin-fixed paraffin-embedded follicular thyroid cancer patient tissue and, in one patient, found a SNV that prevents the binding of tumor suppressor protein p53 [31].
The aforementioned methods have started to provide new mechanistic insights into cancer heterogeneity. In addition, two additional single-cell methods, Hi-C and ChIP-seq, have been recently developed and show potential for use in future cancer epigenetic studies. A type of chromosome conformation capture that quantifies interactions between genomic loci, Hi-C can be used to find trans-regulatory elements and their targets [32]. ChIP-seq, which characterizes interactions between DNA and DNA-binding proteins, can determine transcription factor-regulatory element interactions [33].
Simultaneous multiple omic analysis
Ideally, the different omic approaches should be applied to study a particular tumor so that the information can be integrated. However, this multiple-omic approach is much more technologically challenging. We review some recent studies in this direction.
Simultaneous transcriptomic and genomic sequencing for single cells has recently been achieved by the G&T-seq method [34••]. Cells are first isolated and lysed to release mRNA and genomic DNA. Poly-A mRNA is then separated from genomic DNA through the use of biotinylated oligo-dT primers coupled with streptavidin-coated magnetic beads. The primers are hybridized directly to the poly-A tail, and subsequently recruited by streptavidin-coated magnetic beads through a strong biotin-streptavidin interaction. Standard single-cell techniques can then be used to separately sequence the isolated mRNA and genomic DNA [34••].
The ability to measure transcriptomic and genomic landscapes in the same cells opens a window into understanding the direct effect of genomic variation on transcriptomic variation. Macaulayet al. use their method on HCC38 breast cancer cells to discover the chromosomal rearrangement responsible for the fusion transcript MTAP-PCDH7, found in a majority of HCC38 cells [34••]. They also conclude that a trisomy found in a subset of HCC38_BL (B lymphoblastoid) cells results in proportionally increased mRNA expression in these cells [34••]. To date, the application of G&T has been limited to cell lines; however, it provides hope to analyze the direct effect of copy number variants on transcript levels in tumor samples in the near future.
An extension of this idea of concurrent sequencing has been implemented via the scTrio-seq method [27•]. This technique simultaneously sequences not only the genome and transcriptome, but the DNA methylome as well. In this method, separation of genomic DNA and mRNA is performed through centrifugation of lysed single cells, where a special centrifugation technique allows for the separation of cytoplasm from intact nuclei. The mRNA found in the cytoplasm is sequenced separately from the genomic DNA, which is subjected to scRRBS, providing methylomic and genomic data. The ability to simultaneously quantify genomic, transcriptomic, and epigenomic changes in the same cells has provided new insights into the gene expression regulatory mechanisms. The authors use their method in the analysis of the heterogeneity of human hepatocellular carcinoma. Their results corroborate those of Macaulay et al. in that CNV gene dosage is found to have a proportional effect on transcript levels. DNA methylome results, however, show that CNVs have no similar effect on methylation levels [27•].
The transcriptome is often used as a proxy for protein levels, as single-cell proteomic analyses have not reached the degree of multiplexing that single-cell transcriptomic analyses have. However, mRNA molecules have shorter half-lives than proteins, and previous studies have shown that the mRNA and protein levels may not correspond well [35]. However, their relationship remains unclear at the single-cell level. Recently, Darmanis et al. have developed a new technique to simultaneously measure the transcriptomes and proteomes of single cells [36•]. This is achieved by the splitting of cell lysate and independent processing of each fraction, much like the methods above. The mRNA fraction is subjected to qPCR, and the protein fraction to proximity extension assay (PEA). During PEA, pairs of oligo-labeled antibodies bind to target proteins, where each pair’s oligos are complementary to one another and bind upon being brought in proximity, creating a PCR amplicon, which is then quantified with PCR. The authors apply this technique to quantify cancer pathway proteins that were determined a priori to be of relevance in BMP4-treated glioblastoma cells, and find poor correlation between mRNA and protein levels in these cells. They conclude that protein levels are better predictors of treatment response, leading to the conclusion that perhaps single-cell transcriptomic methods are not sufficient in determining treatment response [36•].
Computational methods for analyzing single-cell genomic and transcriptomic data
With the advent of single-cell technologies comes the necessity for new computational methods to process the data collected. These methods fall into two categories. First are methods that modify bulk sequencing methods to adjust for nuances unique to single-cell data: sparse, noisy data that lacks technical replicates. The second set of methods implement new applications possible only with single cell data. Here we mention methods of the second variety which are of special relevance to cancer genomics. Other methods are extensively covered in previous reviews [5,6].
Inference of spatial patterns
As described above, exciting technologies have been developed to profile single-cell gene expression patterns in situ. Computational methods are still lacking to systematically detect the spatial patterns and classify samples using such patterns.
In some cases, spatial patterns can be inferred by integrating single-cell RNA-seq data collected from isolated cells with in situ expression patterns of a small number of landmark genes [37•,38]. Location of the cells is inferred through correlation between their expression levels and those of the in situ data landmark genes. This approach has been used in developmental biology for the analysis of embryos, where cells are predictably distributed across the dorsal-ventral and animal-vegetal axes [37•]. An analogous method has been used to map cells back to annelid brain regions [38]. However, there is a possibility for difficulties in measuring spatial heterogeneity in tumors due to their typical lack of spatial patterning [37•].
Pseudo-time ordering with bifurcation
Single-cell RNA-seq data is only capable of producing a static view of gene expression levels within cells. Pseudo-time ordering computational methods now allow for a window into continuous changes in gene expression levels, which have thus far given insights into the transcriptional kinetics of cell differentiation. Making the assumption that cells at various stages of differentiation can be found in one scRNA-seq dataset, a time series of transcriptional changes is produced, onto which each cell is mapped. Applying these methods to cancer data can be used to track genes activated at various stages of differentiation from cancer stem cell to matured cancer cell.
Monocle was the first of a series of pseudo-time-ordering algorithms, and uses a combination of dimensionality reduction and a minimal spanning tree (MST) algorithm to build a differentiation trajectory [39]. Monocle2 has since been released, which uses reverse graph embedding and is capable of handling data from much larger scRNA-seq experiments than before [40]. TSCAN (pseudo-Time reconstruction in Single-Cell RNA-seq Analysis) was built as an improvement upon the original Monocle method, reporting more robust results. Instead of creating an MST on all cells, cells are first clustered via hierarchical clustering, and these clusters are used as the MST inputs [41]. A reduced space from which to build a trajectory allows for more stable inference, hence more robust final results. Waterfall is a similar method that also conducts clustering before MST creation [42]. An alternative approach to reconstruct pseudo-time is by fitting the data by a principal curve [43]. This method has been applied to analyzing CyTOF data.
Cell differentiation often involves bifurcation, where two or more distinct cell-types may emerge from a common stem/progenitor cell population. If the temporal information is known, SCUBA can be used to detect bifurcation events [43]. However, in most cases, the temporal information is unavailable. Some pseudo-time methods also build bifurcation events into their models. Instead of assuming one trajectory for all cells, these methods allow for a branching trajectory to account for differentiation into multiple cell types. One method, Wishbone [44•], is an updated version of Wanderlust [45] with the added ability to account for bifurcations. The initial Wanderlust algorithm represents cells as nodes in a graph, where the shortest path between two nodes represents their phenotypic distance. An early cell is chosen and distances are calculated between each cell and the early cell. To adjust for the fact that longer paths are noisier than shorter paths, random waypoint cells are introduced, and each cell’s position is iteratively refined with respect to these waypoint cells. Repeating the graph-building process several times and averaging cell positions from all these graphs mitigates “short circuits,” or edges that occur erroneously between developmentally distant cells [45]. Wishbone updates this algorithm by introducing a step to identify branch points through discrepancies in waypoint distances. Additionally, “short circuits” are avoided via a different approach, where the initial graph is rebuilt in a reduced space to remove noise [44•].
The ability to order cells of complex lineage relationship may have important applications in development. Already, these methods have been used to study the development of cells such as human B lymphocytes [45] and human neural cells [42]. In the future, pseudo-time ordering may be used in mapping the altered mechanisms of cell development in cancer.
Rare cell-type detection
The detection of rare cell types is pertinent to cancer, where the ability to identify circulating tumor cells (CTCs), cancer stem cells, or drug resistant cells will have important clinical implications. Most clustering methods to date are only able to identify major cell groups.
RaceID [46] is a method aimed at detecting rare cell types from scRNA-seq data. Cells are first clustered into major groups by k-means. Outliers of each cluster, which are determined not to be a cause of technical or biological noise, are then grouped into rare cell clusters based on transcriptome correlation [46]. RaceID was recently updated for more robust clustering, where the newer RaceID2 [47] replaces k-means with k-medoid clustering. Grun et al. have integrated RaceID2 into a stem-cell detection algorithm named StemID, which uses the identified cell clusters to guide inference of a lineage tree. Stem cells are then defined by this differentiation trajectory. In this manner, the authors were able to classify stem cells from mouse bone marrow cells, and predict novel pancreatic pluripotent cells [47].
GiniClust [48] is an alternative approach for detecting rare cell-types, by using an innovative approach to choose genes that are likely to be associated with rare cells types, using a statistic called the Gini index. The high Gini genes are identified and subsequently used as input into DBSCAN (density-based spatial clustering of applications with noise) [49]. The authors used this approach on both scRNA-seq and qPCR data. Among other findings, they were able to discover a novel stem cell type characterized by a high expression of ZSCAN4 in mouse embryonic stem cells, and were able to identify rare normal cells in glioblastoma primary tumor samples [48].
Clonal evolution inference
Cancer undergoes a process of clonal expansion and selection that can be inferred through single-cell sequencing data using computational tools. Two such methods are OncoNEM (oncogenetic nested effects model) [50] and SCITE (single cell inference of tumor evolution) [51], which create tumor lineage trees from the single-cell sequencing data. Building lineage trees can guide understanding of the development of therapy resistance; if a sample is taken post-treatment, we can infer a timeline of mutational events that take place before, during and after treatment. Furthermore, these methods can identify mutations that occur early on in tumor development and are propagated throughout each subsequent clone, and guide treatment targeted towards these mutations. These two methods differ in their algorithms—SCITE uses Markov chain Monte Carlo and OncoNEM uses a heuristic search—but importantly, both implement a probabilistic model instead of the traditional maximum parsimony model. Single-cell sequencing data suffers from a large amount of technical error as compared to bulk data that can easily be propagated through subsequent tree-building methods. Using maximum likelihood principles, SCITE and OncoNEM build sequencing error estimation into their models to account for this [50,51].
Biological insights obtained through single-cell analyses
Cancer stem cells
The cancer stem cell hypothesis postulates that there exists a sub-population of self-renewing cells with differentiation potential that serves to initiate and maintain the larger tumor cell population. These cells are estimated to make up less than 1% of the total tumor cell population [52]. Single-cell techniques have provided a powerful tool for identifying and molecularly characterizing cancer stem cells.
As a starting point, Patel et al. [14] use scRNA-seq to analyze the transcriptomes of cells from 5 human glioblastomas in search of glioblastoma stem-like cells (GSC). The authors derive a transcriptome signature that corresponds with “stemness” by comparing the transcriptomes of GSCs and DGCs (differentiated glioblastoma cells) modeled in culture. They then use this signature to identify GSCs in vivo, and find a continuous gradient of stemness-indicating gene expression [14]. Lawson et al. similarly identifies stem-like cells in metastatic breast cancer tumors by a stem-cell-like gene expression signature [53••]. Early stage metastases contain these stem-like cells, while later stage metastases contain cells closer to primary tumor cells in gene expression, supporting the theory that as cancer progresses, tumor cells with stem-like properties initiate and propagate metastatic tumors [53••].
Circulating tumor cells
Single-cell analysis has also provided a powerful tool for the detection and characterization of circulating tumor cells, which are cells that are shed from the tumor into the vasculature or lymphatics and circulate through the bloodstream. Monitoring the presence of CTCs may be used to track the evolution of tumors over time with a simple series of blood tests. However, at an estimated frequency of as little as 1 in 109 of all blood cells [54], it is extremely challenging to capture and analyze these cells. Because of the large amount of heterogeneity in these cells, which may derive from the original tumor or any metastases, single cell methods are necessary. The rarity of these cells requires tools for isolation from hematological cells. A common method involves identifying circulating tumor cells (CTCs) through the presence of EpCAM (epithelial cell adhesion molecule)—found in epithelial cells but not blood cells—on the surface of the cell. Separation of these cells from the blood is then performed using magnetic beads coated with anti-EpCAM antibody. Other recent methods have been developed to overcome a major limitation of this method: the expression of EpCAM is variable from tumor cell to tumor cell, especially those in the epithelial-mesenchymal transition. These alternative methods include isolation of CTCs by microscopic imaging, cell size, and passive capture through removal of all other blood cells.
Genomic and transcriptomic profiling of CTCs have been applied to studying cancer progression. Ni et al. elucidate the pathway of metastasis in lung cancer through the whole-genome sequencing of CTCs from lung cancer patients [55]. As these circulating tumor cells reproducibly share similar CNV patterns to the same patient’s metastatic tumors, the CNV patterns of these CTCs can be used as proxies for the metastatic tumors. These CNV patterns are different from those of the primary tumors, suggesting that metastasis may occur through a set of copy number changes [55]. Several papers point to the sequencing of CTCs as a tool for noninvasively tracking the development therapy resistance. Miyamoto et al. and Dago et al. study the progression of prostate cancer over the course of androgen receptor inhibitor treatment, discussed in the next section [56,57].
Development of therapy resistance
The ability to detect mutations at a single-cell level has lead to yet another possibility: tracking the development of cancer therapy resistance. The main approach towards this goal is longitudinal single-cell measurements before and after various therapies. A common method for treating cancer is chemotherapy before a round of targeted therapy; longitudinal data therefore may consist of measurements before and after each of these events. Noting differences in mutational frequencies over time gives insight into how tumor cells respond to therapy and the mechanisms by which they develop resistance. These studies may in addition be used to validate two prevalent theories of therapy resistance: adaptive resistance, in which a mutation present at low frequency in the original population is selected for during therapy and rises in frequency, or acquired resistance, whereby resistance-conferring mutations arise as a consequence of therapy.
One study evaluates the response of BRAFV600E melanoma to treatment with RAF or combined RAF/MEK inhibitors in both cell culture and tissue [58•]. A comparison of scRNA-seq data from biopsies taken from patients before and after treatment with either RAF or RAF/MEK inhibitors finds that post-treatment tissues contain a higher proportion of cells overexpressing AXL, a known marker of resistance. A follow-up experiment in melanoma cell lines, in which cells are treated to increasing doses of RAF/MEK inhibitors, also reveals an increase in AXL-positive cells. These AXL-positive cells preexisted in the treatment-naive sample and were selected for by treatment, a demonstration of the adaptive resistance mechanism [58•].
The Dago et al. and Miyamoto et al. studies mentioned above use CTC tracking to analyze the development of resistance to androgen receptor (AR)- targeted therapy in prostate cancer patients [56,57]. Through whole-genome sequencing of CTCs before and after treatment, the former find the emergence of two distinct resistant subpopulations with AR amplification. One of these subpopulations is found to be a descendant of a clone found in the therapy-naive population, indicating support for the adaptive resistance hypothesis [56]. Miyamoto et al. use scRNA-seq of CTCs to show the acquisition of heterogeneous resistance-conferring changes in the AR-independent Wnt signaling pathway [57]. Both studies demonstrate the relevance of CTCs in the non-invasive monitoring of therapy resistance.
Authors of the aforementioned Janiszewska et al. paper [10•] use their STAR-FISH technique to study the implications of chemotherapy in the development of resistance to subsequent HER2-targeted trastuzumab therapy in HER2+ breast cancer patients. HER2 amplification and frequency of the His1047Arg mutation in PIK3CA were observed before and after chemotherapy in HER2+ breast tumor samples. Chemotherapy is found to result in an increased frequency of PIK3CA mutants (known to be a determinant of resistance to trastuzumab) and a decreased frequency of HER2 amplification (giving trastuzumab less target sites). These results suggest that trastuzumab may be ineffective for patients who have already received chemotherapy. The spatial information provided by the STAR-FISH method may also be informative in studying resistance, as the authors found that chemotherapy increases the dispersion of cancer cells with the PIK3CA mutation. This increased dispersion may be an indicator of poor prognosis [10•].
A new study extends this type of study to single-cell proteomic data [59]. Wei et al. collect proteomic data on 12 proteins and phosphoproteins in cells of a patient-derived in vivo brain cancer glioblastoma model before and after treatment with an mTOR kinase inhibitor. Correlations between protein expression levels are then used to build signaling networks, and these networks are compared pre- and post- targeted therapy. The drug decreases mTORC1/C2 signaling (the intended target), but upon reaching a state of resistance, the signaling is reactivated, once again an example of adaptive resistance. In addition, upon reaching a state of resistance, new correlations can be seen in the ERK/Src pathways. This is an indication that increased signaling in these pathways may promote downstream mTOR signaling, and consequently that an effective targeted therapy must simultaneously target both pathways [59].
Conclusion
Single-cell biology is a fast-evolving field. As discussed in the paper, a lot of the technical and computational development has been made in just a few years. These methods have greatly empowered researchers to systematically interrogate the cellular heterogeneity within a tumor especially in terms of spatial heterogeneity and multi-omics integration. All the methods reviewed here share a common goal: improving our understanding of tumor cell heterogeneity for the purpose of informing personalized cancer treatment.
Studying intra-tumoral heterogeneity and the spatial orientation of subclones in the primary tumor via new spatial transcriptome methods and simultaneous multiple omic sequencing will allow for the proper drug targeting of the subclones. Examining the nature of stem-like tumor cells and the transcriptomic mechanisms required to give rise to new tumor populations will give clarity to the origination of metastases. Targeting these stem-like cells could hamper the spread of cancer throughout the body. Being able to isolate and longitudinally sample CTCs will permit non-invasive diagnosis and monitoring over the course of treatment. Treatment approaches can be constantly updated upon tracking the response and evolution of CTCs throughout treatment. Finally, treatment resistance can be prevented with a more accurate modeling of the development of resistance to current drugs.
Much work remains to make these possibilities realities. But as single-cell sequencing methods continue to become cheaper, capable of higher coverage, and able to process a greater number of cells faster, no doubt these goals will become more and more attainable.
Acknowledgments
This work was supported by a Claudia Barr Award and NIH grant R13CA124365 to GCY. DT’s research was supported by an NIH training grant T32GM074897.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Van Loo P, Voet T. Single cell analysis of cancer genomes. Curr Opin Genet Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 6.Saadatpour A, Lai S, Guo G, Yuan GC. Single-Cell Analysis in Cancer Genomics. Trends Genet. 2015;31:576–586. doi: 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA, et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet. 2015;47:1212–1219. doi: 10.1038/ng.3391. This paper introduces a method, STAR-FISH, which identifies SNVs and CNVs in single cells in situ, and applies it to study the development of drug resistance in HER2+ breast cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 12.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 17••.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. This paper introduces smFISH, a method for spatially analyzing transcriptomic heterogeneity. Various newer, more advanced methods are developed based on the same principle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. This paper introduces seqFISH, a multiplexing in situ method that detects different mRNAs in single cells by multiple rounds of hybridization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. This paper introduces MERFISH, a method for spatially analyzing transcriptomic heterogeneity capable of multiplexing using an error-robust barcoding scheme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notaro S, Reimer D, Fiegl H, Schmid G, Wiedemair A, Rossler J, Marth C, Zeimet AG. Evaluation of folate receptor 1 (FOLR1) mRNA expression, its specific promoter methylation and global DNA hypomethylation in type I and type II ovarian cancers. BMC Cancer. 2016;16:589. doi: 10.1186/s12885-016-2637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40:e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. This paper introduces scTrio-seq, a method for simultaneous profiling of the genome, transcriptome, and DNA methylome in single cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. These two papers adapt ATAC-seq to single cells for study of heterogeneity in chromatin accessibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. This paper introduces G&T-seq, a method that simultaneously sequences genomes and transcriptomes in single cells, and applies to breast cancer cell lines where they identify the genomic origin of a fusion transcript. [DOI] [PubMed] [Google Scholar]
- 35.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 36•.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, et al. Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. This study introduces a method for simultaneous quantification of mRNA and proteins in single cells and applies it to glioblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. This paper introduces a computational method for inference of spatial patterns through the integration of scRNA-seq data with in situ data for a small number of landmark genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Achim K, Pettit JB, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33:503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 39.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C. Monocle: Differential expression and time-series analysis for single-cell RNA-Seq. 2016. [Google Scholar]
- 41.Ji Z, Ji H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 2016;44:e117. doi: 10.1093/nar/gkw430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci U S A. 2014;111:E5643–5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe’er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. This paper introduces Wishbone, a pseudo-time ordering method capable of accounting for bifurcation, with applications in the modeling of tissue development and cell differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bendall SC, Davis KL, Amir e-A, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 47.Grun D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang L, Chen H, Pinello L, Yuan GC. GiniClust: detecting rare cell types from single-cell gene expression data with Gini index. Genome Biol. 2016;17:144. doi: 10.1186/s13059-016-1010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ester M, Kriegel H-P, Sander J, Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. 2nd International Conference on Knowledge Discovery and Data Mining; Portland, OR: AAAI; 1996. pp. 226–231. [Google Scholar]
- 50.Jahn K, Kuipers J, Beerenwinkel N. Tree inference for single-cell data. Genome Biol. 2016;17:86. doi: 10.1186/s13059-016-0936-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shipitsin M, Polyak K. The cancer stem cell hypothesis: in search of definitions, markers, and relevance. Lab Invest. 2008;88:459–463. doi: 10.1038/labinvest.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. Using a stem-like gene expression signature to identify stem-like cells in metastatic breast cancer tumors, the study finds a decline in stem-like cells from early to late stage metastases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dago AE, Stepansky A, Carlsson A, Luttgen M, Kendall J, Baslan T, Kolatkar A, Wigler M, Bethel K, Gross ME, et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One. 2014;9:e101777. doi: 10.1371/journal.pone.0101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. This study shows evidence for the adaptive resistance hypothesis; through scRNA-seq of prostate cancer cells before and after AR-targeted therapy, the authors show selection for a preexisting AXL-positive cell subpopulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D, et al. Single-Cell Phosphoproteomics Resolves Adaptive Signaling Dynamics and Informs Targeted Combination Therapy in Glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]