Abstract
An endophytic species of Micrococcus was isolated from Aloe vera leaf (syn. Aloe barbadensis) and screened for protease production with five other species of Micrococcus. Data indicated that endophytic Micrococcus aloeverae AE-6 MCC 2184T and Micrococcus yunnanensis DSM 21948T showed efficient protease production potential and secreted active protease at high salt (10%), temperature (40 °C) and in wide range of pH 8–10. Unlike M. yunnanensis DSM 21948T, protease production by M. aloeverae AE-6 MCC 2184T was stringently controlled by pH. Protease induction study using different group of peptides, peptide carbohydrates and peptide macronutrient combinations showed variable response with both the organisms. Result indicated that the amount of protease was not directly related to cell biomass but it depends on nature of inducible peptides. In this study we also developed a modified agar-well assay for semi-quantitative data from large number of replicates.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-017-0638-4) contains supplementary material, which is available to authorized users.
Keywords: Protease, Endophytes, Peptides, Growth yield
Introduction
Isolation of extremophiles and screening them for industrially valuable enzymes are current research interests of microbiologists [1, 2]. Due to stability at high pH and use in laundry industry as detergent additive, alkaline protease or serine protease is one of the most industrially important and extensively studied enzymes and represent about 60% of all the industrial enzyme’s sales in the world [3–5]. Owing to high cost of substrates and media, the overall cost of enzyme production is high. Use of new microorganisms with high protease production efficiency, development of novel processes to increase the yield using cheap substrates may lower the production cost which is highly appreciable for commercial production.
The important protease producing bacteria are Bacillus, Pseudomonas, Halomonas, Arthrobacter and Serratia [6] and most of the alkaline proteases have been reported from genus Bacillus. Members of other groups are less explored and understudied [3, 4, 7]. It has been realized that microbes are less expensive natural sources of new biomolecules. Therefore, to discover the novel group of protease with better efficiency, specificity and activity for various applications, exploration of protease from other groups of microbes is essential.
Survival potential in wide range of salt, pH and temperature and growth at high salt and alkaline pH makes Micrococcus an ideal candidate for study of industrial enzymes, especially alkaline protease. Although, a few studies related to alkaline protease production have already been done using some species of Micrococcus [8–12] but extensive data on screening, characterization and optimization of medium conditions and protease production processes using different strains of Micrococcus from diverse niches are still lacking. Our study focuses on isolation of halo-tolerant, efficient alkaline protease producing endophytic species of Micrococcus in comparison to reported species of the said genus.
In current study two different species of the genus Micrococcus: Micrococcus aloeverae AE-6 MCC 2184T and Micrococcus yunnanensis DSM 21948T were selected for comparative study based on their efficient protease production ability among the selected species, screened for protease production. M. aloeverae AE-6 MCC 2184T was isolated from inner fleshy leaf tissues of Aloe vera and published as a novel species of the genus Micrococcus based on the polyphasic characterization [13], while M. yunnanensis DSM 21948T was isolated from surface sterilized root of Polyspora axillaris.
Materials and Methods
Isolation and Characterization of Bacteria
Fresh leaves from healthy A. vera (syn. Aloe barbadensis) were aseptically collected, and processed according to Prakash et al. [13]. Endophytic isolates were cultured as described by Gayathri et al. [12]. Leaves were washed with distilled water and sterilized with 70% alcohol and mercurous chloride (HgCl2) for 0.5 and 3.0 min respectively and washed three times with sterile distilled water. Inner tissues were crushed in normal saline and 100 µl suspension plated on tryptic soy agar (TSA) and Luria–Bertani (LB) agar plates and incubated at 30 °C. Morphologically distinct colonies were purified and maintained in deep freezer for future study. Micrococcus aloeverae AE-6 was characterized and deposited in Microbial Culture Collection (MCC) and in DSMZ Germany with culture collection number MCC 2184T and DSM 2747 respectively. Isolation of genomic DNA, 16S rRNA gene sequencing and phylogenetic characterization was done as discussed in Prakash et al. [13]. Small subunit rRNA gene sequence accession number of M. aloeverae AE-6 is KF524364.
Screening for Protease Production
Micrococcus aloeverae was selected for protease production due to its growth ability in wide range of salt (up to 12%), pH (6–12) and temperature (10–43 °C). In order to conduct a comparative study other species of Micrococcus viz. M. yunnanensis M. endophyticus, M. luteus were also tested simultaneously. Selected strains were screened for protease production on skimmed milk agar plate. Cultures were spot inoculated and incubated at 30 °C and colonies were observed for the zone of clearance and reconfirmed by coomassie blue method [12, 14, 15].
Agar-Well Assay
Modified skimmed milk agar plate method was followed [1, 16].
Equal amount of skimmed milk agar was poured and wells were created in the centre of plates using 5 mm sterile borer. Each well was inoculated with equal volume (200 µl) of overnight grown cultures in triplicate in order to provide the almost equal amount of cellular biomass for comparison purpose and plates were incubated at 30 °C. After growth, diameters of zone of clearance were measured and mean and standard deviation were calculated.
Determination of Range of Salt, pH and Temperature for Protease Production
Micrococcus yunnanensis and M. aloeverae were selected for comparative study. Protease production potentials of both the strains were studied within wide range of pH, at high temperature (40 °C) and salt (10%). pH range and optima for protease production was studied on skimmed milk agar without any buffer as both the organisms did not alter the pH of the medium. To ensure protease production in alkaline condition, medium at pH-10 was buffered with bicarbonate. Skimmed milk agar with different pH and NaCl concentrations was prepared as earlier and well assay was employed to screen the protease production. pH and salts plates were incubated at optimum temperature (30 °C). To assess the temperature sensitivity of protease and its production at higher temperatures plates were incubated at 40 °C. After visible growth, diameter of zones was measured. Mean and standard deviation were calculated.
Effects of Various Peptide Sources and Their Combination with Different Sugars and Macronutrients on Protease Production
A total of 125 different combinations of different sources of peptides, peptide plus carbohydrates and peptide plus macronutrients (65 sets for each organism) were tested in order to formulate a low cost medium and to enhance the protease production efficiency of Micrococcus.
Six different peptides (peptone, tryptone, beef extract, yeast extract, soy meal and casein) were selected for assessment. The test medium comprised 0.5% peptides (each separately) and 0.5% NaCl. pH of the medium was adjusted with 1N-NaOH and 1N-HCl and kept at 8.0 (optimum for protease production). Medium was inoculated in replicate of three with 1% overnight grown culture and incubated at 30 °C. Five ml culture was harvested at different time intervals and amount of protease was assessed in culture supernatant using enzyme assay for protease as discussed below. For growth yield whole contents of flask was harvested at 6000 rpm for 30 min. Cell pellet obtained was dried at 70 °C for 48 h and weighed.
Total 30-different combinations of above mentioned six different peptides (peptone, tryptone, beef extract, yeast extract, soy meal and casein) with five different carbohydrates (maltose, sucrose, inositol, starch and dextrose) were designed. Similarly, total 30 different combinations of peptides plus five major macronutrients (KCl, FeCl3, MgCl2·6H2O, CaCl2·2H2O and KH2PO4) were also developed. In case of peptide carbohydrate combination media contains 0.5% peptide, 0.5% skimmed milk and 0.2% carbohydrate while peptide and macronutrient combinations contain 0.5% peptide, 0.5% skimmed milk and 0.1% macronutrients. pH was kept at 8.0 with 1N-NaOH and 1N-HCl and solidified with 1.8% molecular grade agar (Sigma–Aldrich). Wells were cut and inoculated. Plates were incubated at 30 °C and zone of clearance was measured at different time intervals.
Assay for Protease Activity
Assay for protease activity was conducted as described by Anson [14] and using the modified and optimized protocol of Sigma-Aldrich (www.sigmaaldrich.com/img/assets/18160/Protease_casein_substrate.pdf). Culture supernatant obtained after centrifugation of grown culture at 4000 rpm for 20 min. Casein was used as substrate for the enzyme while culture supernatant was used as crude enzyme. Five ml 0.65% (w/v) casein solution was added to 1 ml culture supernatant and incubated at 37 °C for 10 min. The reaction was stopped by adding trichloroacetic acid. Tyrosine was measured at 600 nm using UV–visible spectrophotometer (Cary 300 UV–Vis, Agilent Technologies) applying the method of Folin and Ciocalteau [14, 15]. Enzyme units were calculated using the standard curve generated during this study with pure tyrosine. Unit was defined as amount of enzyme that release 1 µM tyrosine min−1 from casein.
Results
Isolation and Characterization of Endophytic Isolates
Six morphologically distinct endophytic isolates were obtained from inner tissues of A. vera leaf. Small subunit rRNA gene sequencing and phylogenetic analysis indicated that isolated endophytic bacteria belonged to three different genera including Micrococcus, Staphylococcus and Kocuria (Supplementary Figure 1). Screening result for protease production indicated that M. aloeverae produced wider zone of clearance on skimmed milk agar among the selected endophytes. Due to efficient protease production and survival in wide range of physiological conditions M. aloeverae AE-6 was selected for protease study along with other procured species of genus Micrococcus (M. yunnanensis, M. endophyticus, M. luteus, M. lylae and M. flavus). Comparative data on protease production with other spp. of Micrococcus indicated that only M. yunnanensis and M. aloeverae AE-6 produced good amount of protease and selected for comparative study (Table 1). Agar well assay as well as coomassie staining method confirmed the protease production ability of both the organisms.
Table 1.
Comparative view of growth range, site of isolation and protease production on Micrococcus spp. isolated from different niches
| Type strains of Micrococcus spp. | Site of isolation | Protease production potential | 30 °C/48 h/pH-10* | 40 °C/72 h/pH-8 | pH for optimum production | Growth range in | References | ||
|---|---|---|---|---|---|---|---|---|---|
| pH* | Salt %* | Temperature* | |||||||
| M. aloeverae AE-6 MCC 2184T | Aloe vera leaf | +++ | 17 mm | 19 mm | 8 | 5–12 (9) | 0–11 | 15–41 | [13] |
| M. yunnanensis DSM 21948T | Polyspora axillaris roots | ++++ | 20 mm | 20 mm | 8 | 6–8 (7–8) | 0–15 | 4–45 | [23] |
| M. endophyticus DSM 17945T | Aquilaria sinensis roots | ++ | ND | ND | ND | 6–9 (7–8) | 0–10 | 15–37 | [24] |
| M. luteus DSM 20030T | Human Skin | + | ND | ND | ND | 5–10 (7) | 0–10 | 20–40 | [25] |
| M. lylae DSM 20315T | Human Skin | – | ND | ND | ND | 6–9 (7) | 0–10 | 20–45 | [26] |
| M. flavus JCM 14000T | Activated sludge | – | ND | ND | ND | 5–9 (6) | 0–10 | 26–34 | [27] |
ND Not detected, +, positive for protease, mm, diameter of halo formation in millimetre
* Data taken from Prakash et al. [13]
Agar-Well Assay
We observed that bacterium grew as thick biofilm inside the wells and secreted extracellular enzyme distributed homogenously through the pores of agar gel and gave reproducible zone of clearance (Supplementary Figure 2).
Effect of pH, Temperature and Salt on Protease Production by Active Cells
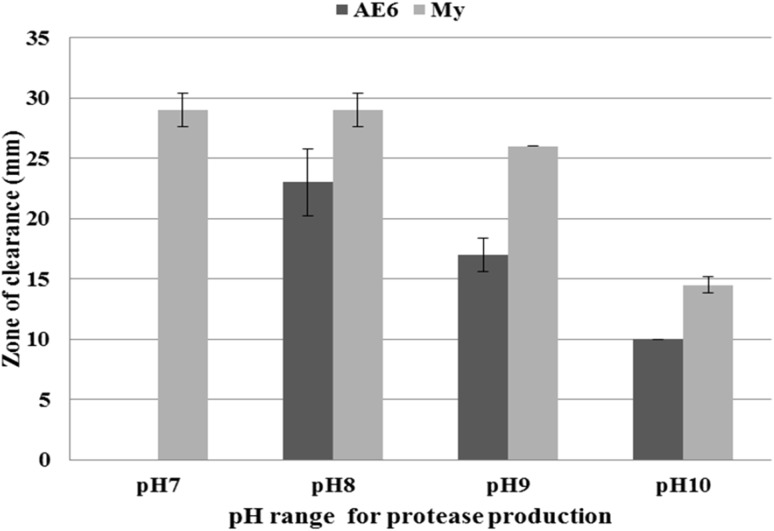
A comparative study of protease production of both organisms in variable conditions of pH, temperature and NaCl is presented in Table 1. Both the strains showed growth between pH 6–12 with pH optima between 8 and 9. Both strains produced highest protease at pH-8 which decreased with increasing pH (Fig. 1). Quantity of protease produced at pH 7 and 8 was almost similar in case of M. yunnanensis while M. aloeverae AE-6 did not produce any protease at pH-7 despite its good growth (Fig. 1). Both the selected strains also secreted protease at 10% NaCl, pH-10 and temperature 40 °C.
Fig. 1.
pH range and optimum pH for protease production in M. aloeverae AE-6 MCC 2184T and M. yunnanensis DSM 21948T. Secretion of protease started only after pH-8 in M. aloeverae AE-6. Data are replicate of three readings. Bar showing the standard deviation of the data
Effect of Different Peptide and Peptide Carbohydrate Combination on Protease Production
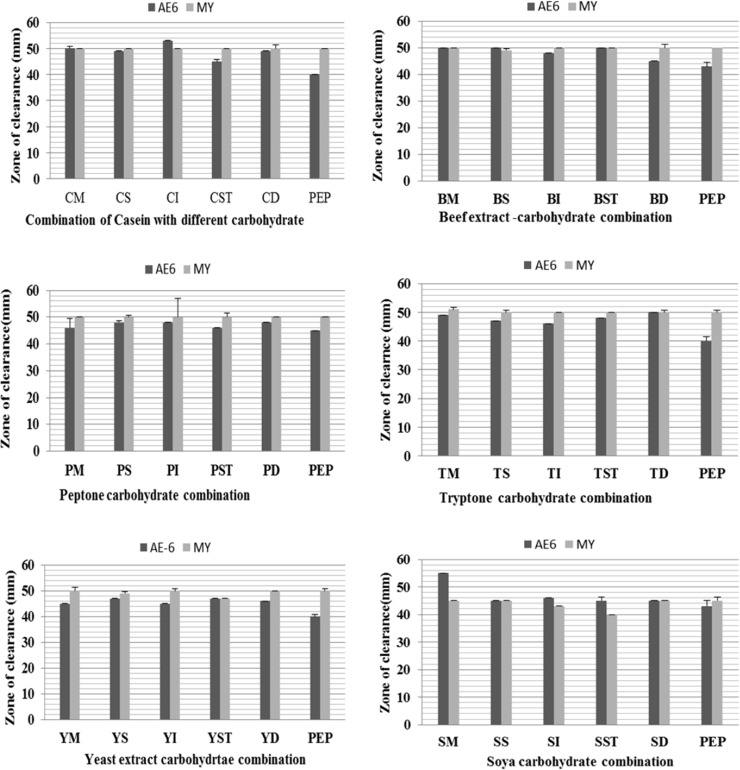
In M. aloeverae AE-6 casein induced maximum protease followed by tryptone and peptone. Beef and yeast extract induced similar amount of protease secretion. Trend of protease induction by peptides was slightly different in case of M. yunnanensis, casein induced maximum levels of protease followed by yeast extract, while almost similar levels of protease was induced by tryptone, peptone and beef extract (Table 2; Fig. 2). Soya-meal induced least amount of protease in both the organisms (Table 2).
Table 2.
Comparison of growth response, growth yield and protease production potential of Micrococcus aloeverae AE-6 MCC2184T and M. yunnanensis DSM 21948T on different peptides
| Peptides | M. aloeverae AE-6 MCC 2184T | M. yunnanensis DSM 21948T | ||||||
|---|---|---|---|---|---|---|---|---|
| Zone (mm) | Growth response | Growth yield* | Enzyme unit (ml−1) | Zone (mm) | Growth response | Growth yield* | Enzyme unit (ml−1) | |
| Yeast-extract | 45.3 ± 1.5 | ++ | 108.0 ± 2.6 | 17.3 ± 0.7 | 55.0 ± 0.5 | +++ | 150.3 ± 1.5 | 16.4 ± 0.8 |
| Beef-extract | 43.3 ± 1.5 | ++ | 84.3 ± 4.0 | 15.8 ± 0.7 | 52.8 ± 0.7 | +++ | 90.1 ± 1.5 | 14.4 ± 0.4 |
| Peptone | 46 ± 1.0 | ++ | 65.0 ± 2.0 | 17.6 ± 2.0 | 54.2 ± 0.4 | +++ | 100.3 ± 0.5 | 15.1 ± 0.3 |
| Tryptone | 47.3 ± 1.5 | ++ | 53.3 ± 4.1 | 18.6 ± 0.6 | 53.3 ± 0.5 | +++ | 99.6 ± 1.5 | 15.4 ± 0.6 |
| Casein | 48.3 ± 0.5 | ++ | 83.6 ± 3.5 | 20.2 ± 0.8 | 56.5 ± 0.5 | +++ | 100.6 ± 1.1 | 20.4 ± 0.6 |
| Soya-meal | 40.3 ± 1.5 | +++ | 110.6 ± 4.0 | 10.6 ± 0.3 | 40.0 ± 1 | ++++ | 161.0 ± 1.3 | 9.1 ± 0.3 |
* Growth yield was reported in mg after incubation of cell biomass for 48 h at 70 °C. Data are mean of three replicates. ±, indicates the standard deviation from mean
Fig. 2.
Comparative study of effect of peptide carbohydrate combination on protease production efficiency of Micrococcus sp. starin AE-6 MCC 2184T and Micrococcus yunnanensis DSM 21948T. Letters P, C, T, Y and B indicates about peptone, casein, tryptone, yeast extract and beef extract respectively while M, S, I, ST, and D represent for maltose, sucrose, inositol, starch and dextrose respectively. Two or three letter in combination indicates the combinations of nutrients
Addition of macronutrient with peptide had no significant effect on enzyme production. In contrast, carbohydrate and peptide combinations showed inhibitory as well as stimulatory effects on protease production. Micrococcus aloeverae AE-6 gave wider zone on casein-inositol, beef-inositol, tryptone-dextrose and soya dextrose combinations and showed stimulatory effect on protease production (Fig. 2). In contrast beef-dextrose, tryptone-maltose and peptone-maltose combinations did not work well and showed inhibitory effect. In case of M. yunnanensis beef-starch, tryptone-sucrose, tryptone-starch, soya-maltose, soya-inositol and soya dextrose combinations showed good protease production while, casein-maltose, beef-dextrose, tryptone-dextrose and soya-sucrose did not work well. In addition almost all the combinations of carbohydrate with yeast and peptone showed similar results with both the strains (Fig. 2). Micrococcus aloeverae AE-6 and M. yunnanensis showed highest growth yield (cell biomass) on yeast extract and least on tryptone and beef extract respectively but unit of enzyme production was more on peptone (Table 2). Our well assay report also indicated that growth in soya meal corresponded to highest cell density in comparison to other peptides but amount of enzyme secretion was least on soymeal (Table 2).
Effect of Incubation Time
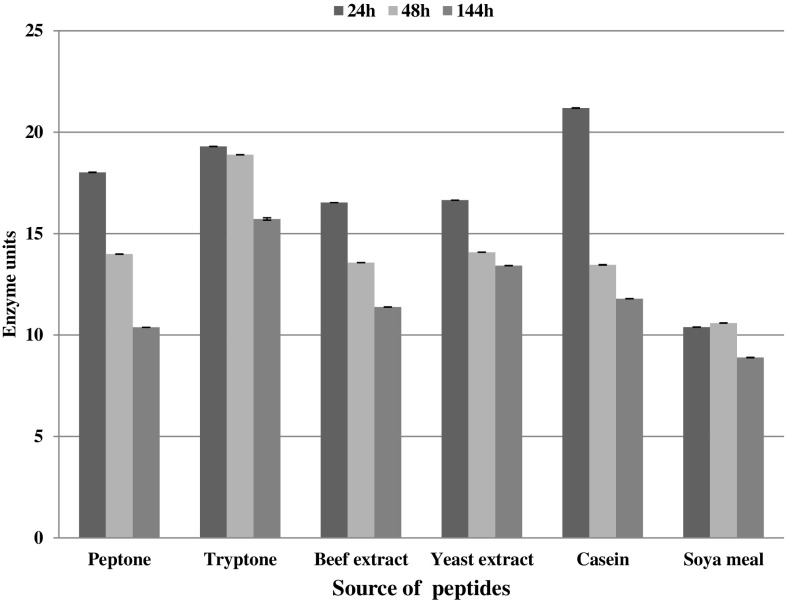
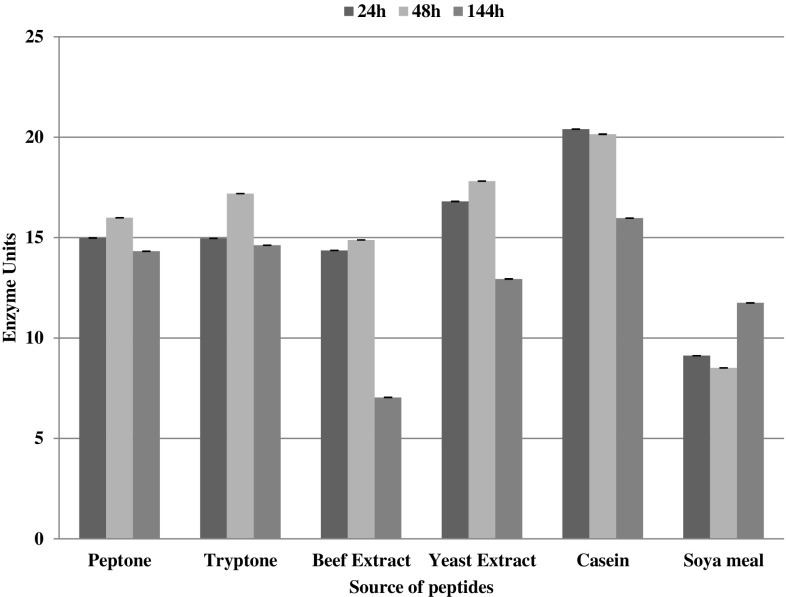
Strain AE-6 secreted maximum amount of protease at 24 h of incubation and after that the levels of protease declined gradually at 48 and 144 h (Fig. 3). In contrast protease data for M. yunnanensis showed more agreement with Bacillus spp. and protease production was maximum at 48 h (Fig. 4).
Fig. 3.
Amount of enzyme secreted by M. aloeverae AE-6 MCC 2184T ml−1 of the culture supernatant during the course of incubation. Data are replicate of three readings and error—bar is showing the standard deviation of the data. Unit was defined as amount of enzyme that release 1 µM tyrosine min−1 from casein
Fig. 4.
Amount of enzyme secreted by M. yunnanensis DSM 21948T ml−1 of the culture supernatant during the course of incubation. Data are replicate of three readings and error—bar is showing the standard deviation of the data. Unit was defined as amount of enzyme that release 1 µM tyrosine min−1 from casein
Discussion
Micrococcus species are widely distributed in nature and their potential to grow and secrete active protease at high salt and alkaline pH makes them an ideal agent for treatment of high alkaline and saline effluents coming from leather and other industries using the concept of bacterial biofilm.
Growth of Strain AE-6 and M. yunnanensis at 0–10% salt and 8–9 pH (optimum for growth as well as protease secretion) indicated that both the organisms are halo-alkali-tolerant in nature and protease production in both the organisms is directly related to pH conditions of the medium. Proteolytic activities of proteases from both the organisms at pH-8 and above indicated that proteases secreted by Strain AE-6 and M. yunnanensis are active at high temperature, pH and salt conditions indicating that these factors are not inhibitory for production of protease in Strain AE-6 and M. yunnanensis and protease was active and stable in extreme growth conditions Induction effect of pH on protease production has also been reported by other researchers in different species of bacteria including Micrococcus, Bacillus, Pseudomonas, Burkholderia, and Geobacillus etc. [12, 17–19] but pH required for induction of protease production varied from genus to genus. Similar to our observations, Eftekhar et al. [20] and Mahendran et al. [21] also reported protease production in alkaline medium at pH 10 and 10.5 respectively by the Bacillus species and showed optimum production of protease at pH-8 with continuous decrease in alkalinity.
The effects of peptides and carbohydrates in the growth media vary with species to species of the same genus or even with strains of the same species. Although few other studies also supplied peptides as an additional source of organic nitrogen in growth medium and observed the effects on protease production on other group of bacteria but the studies were not too extensive, included only few bacterial genera like Pseudomonas fluorescence, Bacillus sp. and Prevotella ruminicola and only two or three different peptides [2, 17–19]. In contrast to our observations Tambekar and Tambekar [2] and Sinha and Satyanyranan [22] reported high levels of protease on soya tone and soybean meal using Bacillus pseudofirmus, Cohnella sp. In conclusion same kind of peptide exerts different effect on different group of bacteria and it can induce or repress the secretion of extracellular protease depending on the group and nature of organisms.
In this study we also reported that despite maximum growth yield on soya-meal amount of enzyme secreted was minimum in comparison to other used peptides. Similar to our findings, Tambekar and Tambekar [2] also reported that despite the best growth on beef extract, peptone and tryptone B. pseudofirmus, Cohnella thermotolerans and Bacillus odyssey secreted lower amount of extracellular protease than other peptides produced less cell growth. Thus, it is clear that protease production is not directly related to cell biomass but it is related with nature of peptide used. The period of optimum production of protease vary from late log phase to stationary phase but it depends on nature of medium used as well as type of organism selected for study [11, 16, 19]. Our observations support the idea that late log phase or early stationary phase of the culture is the best stage for the secretion of the extracellular protease.
Our data indicated that, it is possible to induce the bacterium for better protease secretion in same or even low cost by using right combination of peptide and carbohydrates and selection of right pH conditions. Casein inositol combination is stimulatory for M. aloeverae AE-6 MCC 2184T but combinations of casein with other carbohydrates are inhibitory or not stimulatory for same bacterium. Similarly M. yunnanensis DSM 21948T started secretion of protease from pH-6 MCC 2184T while protease production was tightly regulated by pH in the case of M. aloeverae AE-6 MCC 2184T. The results indicate that optimization and selection of right combination is must for induction of right amount of enzymes.
Unlike previous studies conducted on Bacillus, Pseudomonas and Serratia here we concluded that casein and alkaline pH (pH-8) induced maximum protease in both the species of Micrococcus. We also concluded that induction of secretion of extra cellular protease is multifactorial and depends on diverse set of physicochemical conditions. Furthermore, we also observed that amount of enzyme secreted is not proportional to cell biomass. Even properly induced lower cell biomass can produce higher amount of enzyme. Therefore, optimization of substrate, medium conditions and physicochemical conditions for every strain is mandatory before its exploitation on industrial scale.
In conclusion, we report two new species of endophytic Micrococcus (M. aloeverae and M. yunnanensis) as efficient protease producers, surviving in wide range of ecological conditions and secrete active and stable protease at high salt, temperature and alkaline pH. Due to survival and protease secretion in harsh conditions Micrococcus can be used for treatment of protein contamination from industrial waste water. In addition, we also observed that endophytic micrococci produced better quantity of protease in comparison to other species of micrococcus isolated from diverse habitats and this the interesting question for future investigation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1: Small subunit rRNA gene sequence based phylogenetic tree shows relationship of the strains isolated from inner tissues of Aloe vera leaf (syn. Aloe barbadensis). (DOCX 52 kb)
Supplementary Figure 2: Coomassie stained skimmed milk agar plate showing the bacterial growth inside the wells of agar-gel and formation of zone of clearance of skimmed milk by secreted extracellular protease. (DOCX 2031 kb)
Acknowledgements
This work was supported by the Department of Biotechnology (DBT; Grant No. BT/PR/0054/NDB/52/94/2007) Government of India, The authors thank University Grants Commission (UGC), New Delhi, India for providing the financial support in form of Dr. D.S. Kothari Postdoctoral Fellowship to NB.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-017-0638-4) contains supplementary material, which is available to authorized users.
References
- 1.Vazquez SC, Hernandez E, Cormack WPM. Extracellular proteases from the Antarctic marine Pseudoalteromonas sp. P96-47 strain. Rev Argent Microbiol. 2008;40:63–71. [PubMed] [Google Scholar]
- 2.Tambekar SD, Tambekar DH. Optimization of production and partial characterization of alkaline protease from thermo-halo-alkaliphilic Lonar-Lake bacteria. Biosci Discov. 2013;4:30–38. [Google Scholar]
- 3.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Res. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- 5.Kirk O, Borchert TV, Fuglsan CC. Industrial enzyme applications. Curr Opin Biotechnol. 2002;13:345–351. doi: 10.1016/S0958-1669(02)00328-2. [DOI] [PubMed] [Google Scholar]
- 6.Sarethy IP, Saxena Y, Kapoor A, Sharma M. Alkaliphilic bacteria: applications in industrial biotechnology. J Ind Microbiol Biotechnol. 2011;38:769–790. doi: 10.1007/s10295-011-0968-x. [DOI] [PubMed] [Google Scholar]
- 7.Wieser M, Denner EBM, Kämpfer P, Schumann P, Tindall BJ. Emended descriptions of the genus Micrococcus, Micrococcus luteus (Cohn 1872) and Micrococcus lylae (Kloos et al. 1974) Int J Syst Evol Microbiol. 2002;52:629–637. doi: 10.1099/00207713-52-2-629. [DOI] [PubMed] [Google Scholar]
- 8.Bhowmik T, Marth EH. Protease and Peptidase Activity of Micrococcus species. J Dairy Sci. 1988;71:2358–2365. doi: 10.3168/jds.S0022-0302(88)79819-7. [DOI] [Google Scholar]
- 9.Clark DJ, Hawrylik SJ, Kavanagh E, Opheim DJ. Purification and characterization of a unique alkaline elastase from Micrococcus luteus. Prot Expr Purif. 2000;18:46–55. doi: 10.1006/prep.1999.1166. [DOI] [PubMed] [Google Scholar]
- 10.Manikandan M, Kannan V, Pasić L. Extraction, purification and characterization of a protease from Micrococcus sp. VKMM 037. Environ Technol. 2011;32:1487–1495. doi: 10.1080/09593330.2010.540718. [DOI] [PubMed] [Google Scholar]
- 11.Odu NN, Akujobi CO. Protease production capabilities of Micrococcus luteus and Bacillus species isolated from abattoir environment. J Microbiol Res. 2012;2:127–132. doi: 10.5923/j.microbiology.20120205.03. [DOI] [Google Scholar]
- 12.Gayathri S, Saravanan D, Radhakrishnan M, Balagurunathan R, Kathiresan K. Bioprospecting potential of fast growing endophytic bacteria from leaves of mangrove and salt-marsh plant species. Ind J Biotechnol. 2010;9:397–402. [Google Scholar]
- 13.Prakash O, Nimonkar Y, Munot H, Sharma A, Vemuluri VR, Chavadar MS, Shouche YS. Description of Micrococcus aloeverae sp. nov., an endophytic actinobacterium isolated from Aloe vera. Int J Syst Evol Microbiol. 2014;64:3427–3433. doi: 10.1099/ijs.0.063339-0. [DOI] [PubMed] [Google Scholar]
- 14.Anson ML. The estimation of pepsin, trypsin, papain and cathepsin with hemoglobin. J Gen Physiol. 1938;22:79–89. doi: 10.1085/jgp.22.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folin O, Ciocalteau VJ. On tyrosine and tryptophan determinations in proteins. J Biol Chem. 1929;73:62–650. [Google Scholar]
- 16.Kumar A, Sachdev A, Balasubramanyam SD, Saxena AK. Optimization of conditions for production of neutral and alkaline protease from species of Bacillus and Pseudomonas. J Microbiol. 2002;42:233–236. [Google Scholar]
- 17.Wang HT, Hsu J. Optimal protease production condition for Prevotella ruminicola 23 and characterization of its extra cellular crude protease. Anaerobe. 2005;11:155–162. doi: 10.1016/j.anaerobe.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Kalaiarasi K, Sunitha PU. Optimization of alkaline protease production from Psuedomonas fluorescens isolated from meat waste contaminated soil. Afr J Biotechnol. 2009;8:7035–7041. [Google Scholar]
- 19.Palsaniya P, Mishra R, Beejawat N, Sethi S, Gupta L. Optimization of alkaline protease production from bacteria isolated from soil. J Microbiol Biotech Res. 2012;2:858–865. [Google Scholar]
- 20.Eftekhar F, Fouladi J, Faghihi M. Isolation and identification of an alkaline protease producing Bacillus from soil. Iran J Biotechnol. 2003;1:183–185. [Google Scholar]
- 21.Mahendran S, Sankaralingam S, Shankar T, Vijayabaskar P. Alkalophilic protease enzyme production from estuarine Bacillus aquimaris. World J Fish Mar Sci. 2010;2:436–443. [Google Scholar]
- 22.Sinha N, Satyanarayana T. Alkaline protease production by thermophilic Bacillus licheniformis. Enz Microbiol Technol. 1999;8:370–372. [Google Scholar]
- 23.Zhao GZ, Li J, Qin S, Zhang YQ, Zhu WY, Jiang CL, Xu LH, Li WJ. Micrococcus yunnanensis sp. nov., a novel actinobacterium isolated from surface-sterilized Polyspora axillaris roots. Int J Syst Evol Microbiol. 2009;59:2383–2387. doi: 10.1099/ijs.0.010256-0. [DOI] [PubMed] [Google Scholar]
- 24.Chen HH, Zhao GZ, Park DJ, Zhang YQ, Xu LH, Lee JC, Kim CJ, Li WJ. Micrococcus endophyticus sp. nov., isolated from surface-sterilized Aquilaria sinensis roots. Int J Syst Evol Microbiol. 2009;59:1070–1075. doi: 10.1099/ijs.0.006296-0. [DOI] [PubMed] [Google Scholar]
- 25.Cohn F. Untersuchungen u¨ber Bakterien. Beitr Biol Pflanz. 1872;1:127–244. [Google Scholar]
- 26.Kloos WE, Tornabene TG, Schleifer KH. Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. Int J Syst Bacteriol. 1974;24:79–101. doi: 10.1099/00207713-24-1-79. [DOI] [Google Scholar]
- 27.Liu XY, Wang BJ, Jiang CY, Liu SJ. Micrococcus flavus sp. nov., isolated from activated sludge in a bioreactor. Int J Syst Evol Microbiol. 2007;57:66–69. doi: 10.1099/ijs.0.64489-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Small subunit rRNA gene sequence based phylogenetic tree shows relationship of the strains isolated from inner tissues of Aloe vera leaf (syn. Aloe barbadensis). (DOCX 52 kb)
Supplementary Figure 2: Coomassie stained skimmed milk agar plate showing the bacterial growth inside the wells of agar-gel and formation of zone of clearance of skimmed milk by secreted extracellular protease. (DOCX 2031 kb)