Abstract
In the present study, gold nanoparticles (AuNPs) synthesis was carried out by using a rare bacteriophage which is morphologically similar to 7–11 phages of the C3 morphotype of tailed phage belonging to Podoviridae family as green route. Effect of various physiological parameters like pH, temperature and concentration of gold chloride salt on AuNPs synthesis was studied. The reaction mixtures have shown vivid colours at various physiological parameters. Phage inspired AuNPs were further characterized by using different techniques such as UV–Vis spectrophotometry, scanning electron microscopy (SEM), energy dispersive spectroscopy (EDS), X-ray diffraction (XRD) and dynamic light scattering (DLS). DLS study revealed synthesis of various sizes of AuNPs in the range of 20–100 nm. SEM studies revealed synthesis of varied shaped AuNPs, viz., spheres, hexagons, triangles, rhomboids and rectangular etc. The presence of Au in the nanostructures was confirmed by EDS. The XRD pattern reflects the crystalline nature and nano size of AuNPs. These phage inspired AuNPs showed anti-bacterial activity against different bacterial pathogens. Anti-biofilm activity of AuNPs was evaluated on a glass slide. It was noticed that at 0.2 mM concentration of these AuNPs about 80% of biofilm formation by Pseudomonas aeruginosa, a human pathogen was inhibited. Thus, the phage inspired AuNPs synthesis could be potential therapeutic agents against human pathogens.
Keywords: Bacteriophages, Biofilms, Biopharmaceuticals, Biotechnology, Viruses
Introduction
Bacteriophages are a class of viruses with ability to multiply and survive in bacterial host cells [1]. Phages are found to be abundant with ability to persist in nature without their host. They have potential of a high resistance to various environmental conditions as compared to the host cells [2]. They have high specificity for their host and are explored extensively with respect to their applications in different fields. Lytic phages are considered as therapeutic agents for the treatment of human, animal and plant diseases. Phages have been used in food industries as biocontrol agents to control human and plant pathogens [3].
Recently, viruses are considered as valuable biological resources in the field of nanotechnology. Virus mediated nanoparticles synthesis is an attractive and could be a promising ‘green technology’. M13 viruses are used in preparation of cobalt oxide nanowires and Co–Pt crystals [4]. Hybrid gold–cobalt oxide nano-wires are also synthesized by using gold binding peptides and incorporated into the filament coat of virus. Such nano-wires are being used in battery industries to improve the power of battery [4, 5]. Phagemid vectors are also used for expression of metal binding peptides on the major coat proteins and synthesis of semiconductor metallic nanowires [6, 7]. FCC iron nanoparticles are synthesized by using M13 virus for uranium reduction and uranium dioxide nanocrystals synthesis [8]. Some specific binding peptide motifs with high affinity for various semiconductor materials like ZnS, CdS and PbS etc. are identified. Thus, they have a key role in mineralization of the inorganic crystals [9]. Modified pII minor coat protein of phages was used as a biological agent for ZnS and CdS nanoparticles synthesis [10]. ZnS nanoparticles were self-assembled into ordered film structures. Similarly, anti-streptavidin viruses were used to align different nanoparticles such as gold, organic dyes and biological molecules [11].
Biological molecular assemblies are known as an excellent source for the development of nano-engineered systems with several biomedical applications. Therefore, biologically active molecular networks in which bacteriophages were directly assembled with AuNPs [12]. Genetically and chemically manipulated bacteriophages were used in anti-cancer therapy [13]. A covalent coupling of AuNPs to retargeted adenoviral vectors were used as selective agents for delivery of the nanoparticles to tumor cells [14]. In this study, a rare 7–11 phage belonging to Podoviridae family is used as a novel green route for AuNPs synthesis. This is the first report on green synthesis of AuNPs by using a virus as reducing agent. Such bio-inspired AuNPs were also tested for their potential anti-bacterial and anti-biofilm activity against, P. aeruginosa (PAO1).
Materials and Methods
Preparation of Gold Solution
The stock of gold chloride solution was prepared (1 mM) by adding gold chloride salt (HAuCl4) in distilled water and used further for all experiments as per the requirement.
Phage Isolation, Purification and Enrichment
Lytic phages specific to Salmonella serovar Paratyphi B were isolated from the surface water of the Pavana river, Pune, India [15]. Water sample was collected in a sterile screw capped bottles of 250 ml capacity. Water was used as a source of phages. The phages were isolated from the filtrate on nutrient agar medium by the double agar layer plaque assay (DAL) method [16] in which the mid-log phase culture (O.D650 0.80) of Salmonella serovar Paratyphi B (0.5 ml) and the filtrate (0.2 ml) were mixed together in 3 ml sterile soft agar (0.6% w/v) and then poured onto sterile nutrient agar medium. Plates were incubated at 37 °C for 24 h and checked for the formation of plaques after every 3 h. After incubation, plate showed varied sized clear and turbid plaques. Clear plaque formation is a characteristic of the lytic phages whereas the turbid plaques are formed by the lysogenic phages. A single clear plaque (6 mm diameter) was picked up from the plate using sterile cork borer and suspended in 500 ml flask containing 200 ml phage broth in 500 ml conical flask (gram/litre, peptone; 10, meat extract; 3, sodium chloride; 5, glucose; 1, CaCl2·H2O; 0.2, MgSO4·7H2O;0.5 and pH 7.4). Thereafter, 0.5 ml of Salmonella serovar Paratyphi B culture was inoculated into the same flask and incubated at 37 °C for 24 h. After incubation, broth was centrifuged at 10,000 rpm for 20 min at 4 °C. Supernatant was filtered through membrane filter (0.2 μm) and the filtrate was used as phage source. Plaque characters of the purified phages were determined in triplicate by double agar layer plaque assay technique. After incubation at 37 °C, plaques were observed and then a single plaque was picked up from the plate, enriched in the phage broth using host, Salmonella serovar Paratyphi B.
Determination of Phage Titre
Bacteriophage titre of the lysate was determined by using double agar layer plaque assay technique as described earlier [15, 16]. Briefly, 100 µl of diluted phage lysate, 500 µl of S. Serovar paratyphi B suspension and 4 ml of molten agar were mixed properly and poured on sterile nutrient agar plates. All plates were incubated at 37 °C and plaque forming units (PFU) were counted after 12–18 h. Thus, phage titre was determined.
Transmission Electron Microscopy (TEM) of Phages
Phage morphology was studied by using TEM. Bacteriophage samples were prepared by centrifugation of purified phages at 25,000 rpm for 1 h at 4 °C by using a Beckman J2-21 centrifuge (Beckman Instruments, Palo Alto, CA). Further, phages were washed twice with 0.1 M ammonium acetate buffer (pH 7.0) and stained with 2% phosphotungstate (pH 7.2) solution and deposited on carbon-coated Farmvar films and examined under Philip EM 300 electron microscope.
Preparation of Phage Lysate in SM Buffer for Synthesis of AuNPs
Nutrient agar plates with plaques were flooded with SM buffer (g/l, Nacl; 5.8, MgSO4·7H2O; 2, 1 M Tris; 50 ml, pH 7.5) and kept at 4 °C for 12–18 h. Further, SM buffer with phages was centrifuged at 10,000 rpm at 4 °C for 10 min for sedimentation of bacteria. Supernatant was filtered through membrane filter (0.20 µm). Phage titre of the filtrate was determined as described earlier and these phages were used for AuNPs synthesis.
Synthesis of AuNPs by Using Phage Lysate
The reaction mixture contained 0.5 ml of phage lysate (6 × 1011 PFU/ml) and 2 ml of gold chloride solution. The reaction mixture was incubated at room temperature for 74 h. After completion of reaction, a change in colour was observed. The AuNPs were synthesized by using phages at various environmental conditions, viz., pH, temperature and gold chloride salt concentration.
Characterization of AuNPs
Characterization of AuNPs was carried out by using UV–Vis spectroscopy, SEM–EDX and XRD. UV–Vis spectrum measurements were performed by using a Jasco V-530 UV–Vis spectrophotometer. The SEM attached with EDS of AuNPs was recorded. The XRD pattern of AuNPs was measured by using thin films of AuNPs in the transmission mode on a D 8 Advanced Brucker instrument with Cu Kα radiation using λ = 1.54 Å. The particle size was analyzed by using DLS equipment (25 °C at a fixed angle of 90°) with Brookhaven 90 plus particle sizing software.
Anti-bacterial Activity of AuNPs
Antibacterial activity of phage inspired AuNPs synthesis was tested by well agar diffusion technique [17]. Different aliquots of AuNPs (50 and 100 µl) were added into wells prepared on sterile nutrient agar plates seeded with bacterial culture. Plates were kept for 10–15 min at refrigeration temperature to allow diffusion of AuNPs into the medium. All Plates were incubated at 37 °C for 24 h. Then plates were observed for zone of growth inhibition by phage inspired AuNPs.
Anti-biofilm Activity of AuNPs on the Glass Surface
Anti-biofilm activity of phage inspired AuNPs was studied against the bacterial biofilm of P. aeruginosa (PAO1) on a glass slide surface. The test culture was grown in nutrient broth for 24 h and inoculated into sterile petriplates containing 20 ml of sterile nutrient broth medium. The phage inspired AuNPs of different concentrations (0.05, 0.1 and 0.2 mM) were transferred into each individual plate. The plates without AuNPs were considered as control. The glass slides were dipped into sterile nutrient broth medium containing bacteria and incubated at 37 °C for 24 h. All experiments were performed in triplicates. Biofilm formation on glass slides in presence and absence of phage inspired AuNPs was observed and evaluated by fluorescence microscopy. For fluorescence microscopy, biofilms were stained with 0.01% acridine orange stain. Biofilm formation by bacterial pathogen was quantified by crystal violet assay. A wild-type strain of P. aeruginosa, was inoculated into sterile nutrient broth and incubated at 37 °C for 24 h on rotary shaker (120 rpm). The absorbance (A 600 nm) of the freshly grown culture was adjusted to 0.8. Further, 1 ml of each culture was added into 15 ml of nutrient broth and adjusted with different concentrations of AuNPs (0.05, 0.1 and 0.2 mM), separately and mixed thoroughly. About, 200 μl of mixture was added into wells of flat bottomed polysterene microtiter plates. The plates were incubated at 30 °C for 10, 15, 20 and 25 h. The wells without AuNPs were considered as control [18].
Statistical Analysis
All the experiments were performed in triplicate. The arithmetic mean was used for data analysis. The unpaired ‘t’ test was used for comparison between control (without AuNPs) and test biofilm formation (with AuNPs). A probability level of p (0.05) was used throughout the study. All statistical analysis of data was carried out by using the GraphPad InStat [DATASET1.ISD] software.
Results and Discussion
Effect of pH on Phage Inspired AuNPs Synthesis
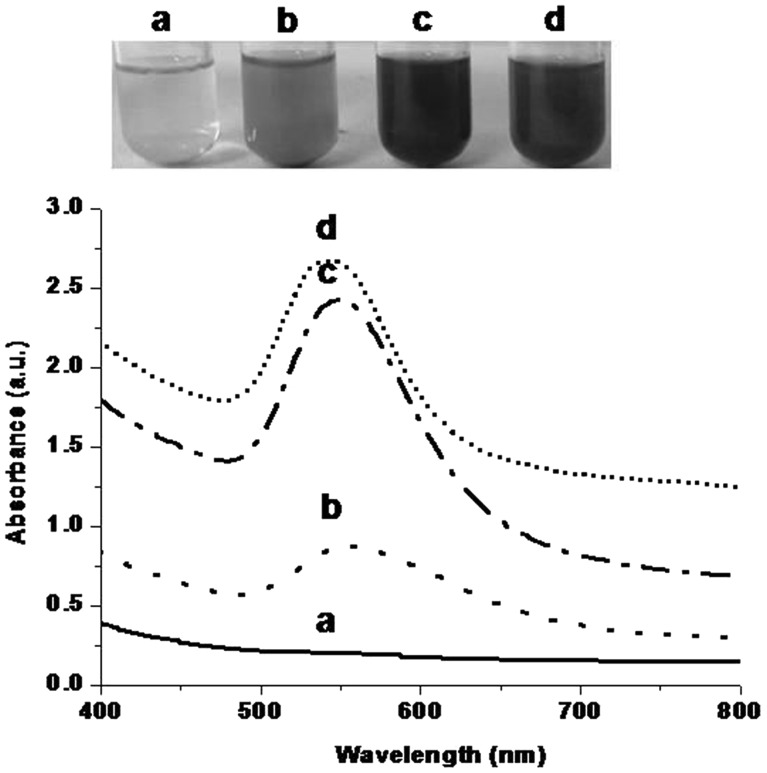
Electron micrographic study revealed that the phage belonged to Podoviridae family which is morphologically similar to the 7–11 phages of the C3 morphotype of tailed phages, characterized by a very long, cigar-shaped head (162 × 58 nm) and short tail (12 × 7 nm). These phages are very rare in their occurrence [15]. The phage inspired AuNPs were synthesized at different pH values (2, 3, 4 and 5) of the reaction mixtures. Figure 1 showed that the yellow color of gold chloride solution did not distinctly turn into any other color after reaction carried out at pH 2. The UV–Visible spectrum of the reaction mixture did not show a characteristic peak of AuNPs. Absence of Au characteristics peak suggested that AuNPs were not synthesized at pH 2. Yellow color of gold chloride solution was turned into light pink color with a broad peak in the range of 500–650 nm at pH 3. When the reaction took place at pH 4, ruby red color was formed with a sharp narrow peak in the range of 530–570 nm. Dark reddish color appeared at pH 5 with a strong peak in the range of 520–580 nm. The pale yellow color of the reaction mixture turned into different colors with characteristic peaks at pH 3, 4 and 5. Thus, visual observations and UV–Vis spectra studies indicate that the phage inspired AuNPs was synthesized. Intensity of a peak was varying at different pH values of reaction mixtures. Similar, results were obtained with biological reducing agents such as bacteria and banana peel extract when used for bio-inspired AuNPs synthesis. Thus, results are in corroboration with our previous studies [17, 19].
Fig. 1.
Visual observations and UV–visible absorption of phage inspired AuNPs synthesized at pH a 2 b 3 c 4 d 5
Effect of Temperature on Phage Inspired AuNPs Synthesis
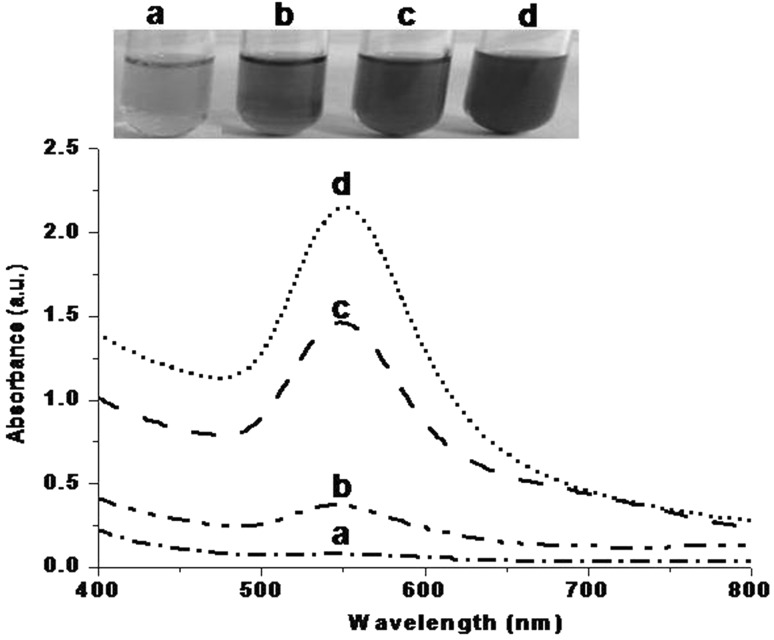
As indicated in Fig. 2, AuNPs were synthesized at different temperatures (20, 25, 30 and 40 °C). At 20 °C, light pinkish color was formed with a small broad peak in the range of 500–600 nm. Dark pink color appeared when reaction was performed at 25 °C. A broad peak was observed in the range of 500–600 nm with increasing absorbance. Red color was developed at 35 °C and a broad peak was seen in the range of 500–600 nm. Intensity of red color was increased at 40 °C with sharp peaks in between 540 and 560 nm. Intensity of color was increased with increasing temperatures suggested that AuNPs formation was superior at higher temperature. Results obtained substantiate our previous reports [17, 19].
Fig. 2.
Visual observations and UV–visible absorption of phage inspired AuNPs synthesized at temperature (°C) a 20 b 25 c 30 d 40
Effect of Metal Salt Concentrations on Phage Inspired AuNPs Synthesis
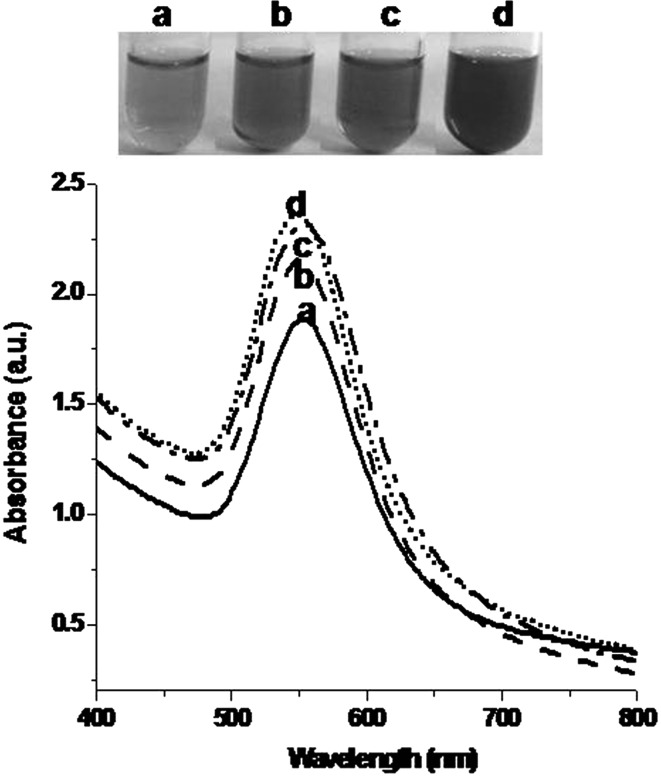
Figure 3 shows that phage inspired AuNPs synthesis was studied at different concentrations of gold chloride (0.5, 1, 1.5 and 2 mM). Pink color was observed when reaction was performed with 0.5 mM of gold chloride solution. A sharp narrow peak was seen in the range of 530–570 nm. Reactions with 1 and 1.5 mM of gold chloride solutions showed dark pink color with characteristic sharp peaks in the range of 510–580 nm. Ruby reddish color was observed with 2 mM of gold chloride solution with typical sharp peak in the range of 510–590 nm. The colour change of reaction mixture was due to the collective coherent oscillation of conduction electrons at the surface of AuNPs. Thus, colour change in reaction mixture under different experimental conditions is an indication of AuNPs synthesis [19–21].
Fig. 3.
Visual observations and UV–visible absorption of phage inspired AuNPs synthesized at different concentration of gold chloride (mM) a 0.5 b 1.0 c 1.5 d 2.0
SEM Attached with EDS and XRD Studies of Phage Inspired AuNPs
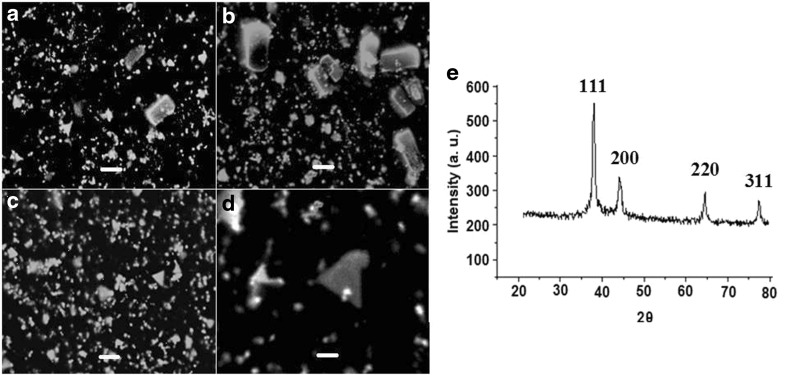
Figure 4a–d shows the SEM micrographs of AuNPs in the form of different shapes such as spheres, hexagons, triangles, rhomboids and rectangular. Nanoparticles with varied morphologies have been reported in previous study [22]. The spot EDS of AuNPs showed peaks of gold in the selected area (figure not shown). SEM demonstrated a polymorphic distribution in size and shapes of AuNPs. Maximum nanoparticles were in the range of 20–50 nm. Few nanoparticles were in the range of 50–150 and 150–500 nm. Size of phage inspired AuNPs was further determined by using DLS (figure not shown). The size of AuNPs was determined by DLS and is in conformity with size obtained by SEM images. Figure 4e is a representative XRD profile of the AuNPs that gives the structural information and its crystalline nature. A thin film of gold chloride before reaction did not show the characteristic peaks. The reaction of Au with phages showed the diffraction peaks at 2θ = 38.1°, 44.5°, 64.21° and 77.78°. The XRD analysis revealed that AuNPs has predominant (111) and (200) peaks, that are expected for nanoplates and microstructures with (111) dominant facets. The XRD pattern of AuNPs has similarity with the pattern reported earlier [17, 19].
Fig. 4.
Scanning electron micrographs of phage inspired AuNPs a–c are magnified ×20,000. Inset bar represent 2 μm and d is magnified ×60,000. Inset bar represent 0.1 μm. e Representative XRD profile of thin film phage inspired AuNPs
Anti-bacterial and Anti-biofilm Activity of Phage Inspired AuNPs
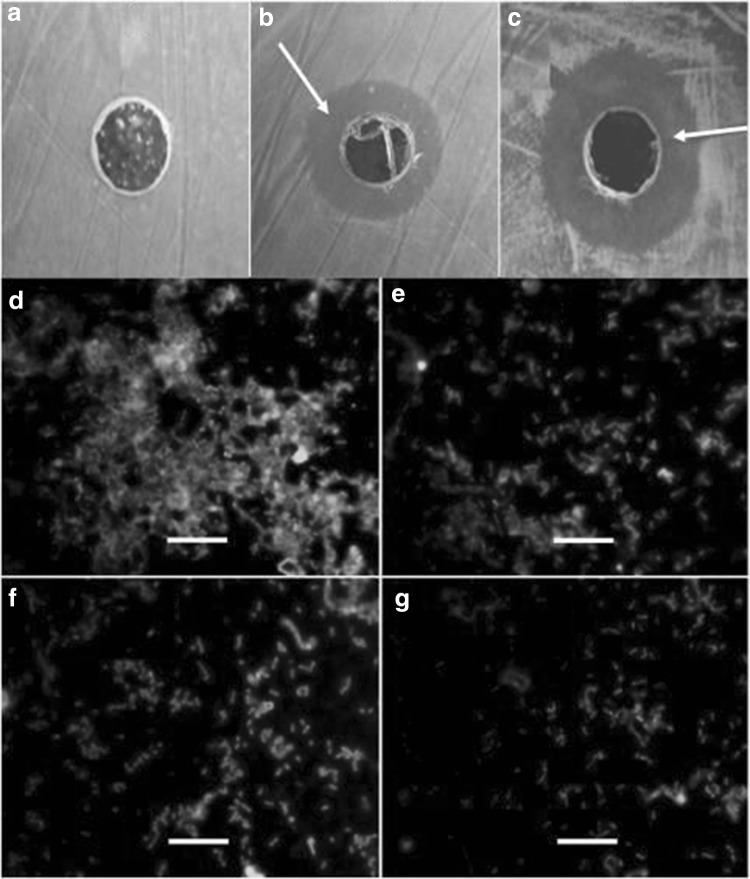
Anti-bacterial activity of phage inspired AuNPs was tested against several bacterial cultures, viz., Klebsiella pneumoniae, P. aeruginosa (MTCC 728), Escherichia coli (MTCC 728), Proteus vulgaris (MTCC 426) and S. serovar Paratyphi B. Figure 5a–c shows the representative zone of growth inhibition of P. aeroginosa (MTCC 728) and Salmonella serovar Paratyphi B by AuNPs. AuNPs exhibited highest antibacterial activity against Klebsiella pneumoniae, P. aeruginosa (MTCC 728) and Salmonella serovar Paratyphi B etc. Antibacterial activity of bacterial cell mediated AuNPs has been reported against P. aeruginosa. A inhibition zone diameter was 1.4 ± 0.2 cm at 2 mM of AuNPs [17, 23]. In this study, zone diameter by phage inspired AuNPs synthesis was 2.1 ± 0.3 cm reported. Thus, phage inspired AuNPs exhibited higher antimicrobial activity against P. aeruginosa as compared to bacterial cell mediated AuNPs. The inhibitory mechanism of AuNPs is not yet fully understood. AuNPs are reported to interfere with DNA, RNA and protein synthesis [23]. Anti-biofilm activity of phage inspired AuNPs was observed against P. aeruginosa (MTCC 728). Biofilm formation by P. aeruginosa on the glass surfaces was inhibited by AuNPs. Biofilm inhibition (%) was increased in presence of different AuNPs concentration when compared with control (p < 0.05) as shown in Fig. 5d–g. About 70–80% of biofilm was inhibited by phage inspired AuNPs in case of pathogenic strain of P. aeruginosa (MTCC 728). Similarly, anti-biofilm activity of AuNPs was observed by bio-inspired nanoparticles as reported earlier [19, 23].
Fig. 5.
a Control without phage inspired AuNPs: no zone of growth inhibition. The phage inspired AuNPs shows zone of growth inhibition against b Salmonella paratyphi and c Pseudomonas aeruginosa (MTCC 728). Fluorescence microscopy images of biofilm formation by P. aeruginosa on the glass surface d without phage inspired AuNPs and with different concentrations of phage inspired AuNPs e 0.05 mM f 0.1 mM g 0.2 mM. Magnification at ×1000. Inset bar represents 10 μm
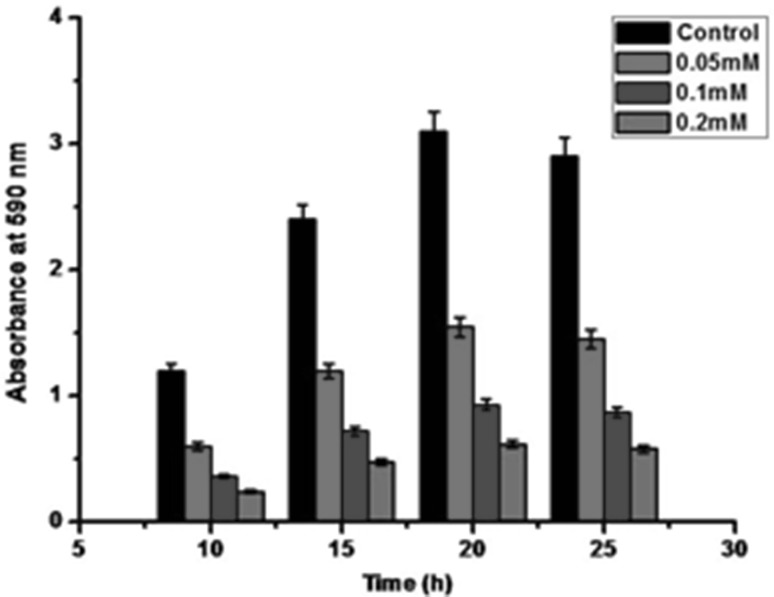
Biofilm inhibition by AuNPs was investigated by using 96 well plate assay performed in microtiter plates. AuNPs showed a remarkable reduction in the biofilm formation by P. aeruginosa with increasing AuNPs concentrations. This might be due to the toxicity of AuNPs towards the bacterial cells [19]. Thus, bacterial biofilm formation (%) was inhibited significantly (p < 0.05) as compared to biofilm formation (%) in control (without AuNPs). It was seen that about 70% biofilm formation was inhibited in presence of 0.1 mM AuNPs and 80% at 0.2 mM AuNPs as shown in Fig. 6. In previous study, it has been observed that bacterial mediated AuNPs have inhibited about 64.29, 71.43 and 76.79% of biofilm formation by P. aeruginosa at different concentrations of 0.5, 1.0 and 2.0 mM, respectively (17). Thus, phage inspired AuNPs at low concentrations (0.2 mM) showed higher anti-biofilm activity against P. aeruginosa as compared to bacterial mediated AuNPs. The anti-biofilm activity of AuNPs observed may be related due to the destabilisation of bacterial biofilms [24]. Thus, phage inspired AuNPs may serve as potential therapeutic agents against the biofilm forming bacterial pathogens.
Fig. 6.
Representative images of biofilm formation in wells of microtiter by P. aeruginosa with different concentrations of phage inspired AuNPs
Acknowledgements
All Authors are thankful to the UGC, New Delhi for financial support during the research work. Special thanks to Dr. H. W. Ackermann (Laval University, Canada) for assistance in electron microscopic studies of bacteriophages. AB is Thankful to BCUD, Savitribai Phule Pune University for financial support to Waghire College, Saswad, Pune, India.
Compliance with Ethical Standards
Conflict of interest
All authors declare that there is no conflict of interest.
References
- 1.Ackermann HW. Tailed bacteriophages: the order Caudovirales. Adv Virus Res. 1999;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duran AE, Muniesa M, Mendez X, Valero F, Lucena F, Jofre J. Removal and inactivation of indicator bacteriophages in fresh waters. J Appl Microbiol. 2002;92:338–347. doi: 10.1046/j.1365-2672.2002.01536.x. [DOI] [PubMed] [Google Scholar]
- 3.Leverentz B, Conway W, Ahvideze S, Janisiewier ZWJ. Examination of bacteriophage as a biocontrol agent method for Salmonella on fresh cut fruit: a model study. J Food Prot. 2001;64:1116–1121. doi: 10.4315/0362-028X-64.8.1116. [DOI] [PubMed] [Google Scholar]
- 4.Lee SK, Dong SY, Angela MB. Cobalt ion mediated self-assembly of genetically engineered bacteriophage for biomimetic Co–Pt hybrid material. Biomacromolecules. 2006;7:14–17. doi: 10.1021/bm050691x. [DOI] [PubMed] [Google Scholar]
- 5.Nam KT, Kim DW, Pil JY, Chiang CY, Meethong N, Hammond PT, Chiang YM, Angela MB. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312:885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Flynn CE, Hayhurst A, Sweeney R, Qi J, Georgiou G, Iverson B, Belcher AM. Viral assembly of oriented quantum dot nanowires. Proc Natl Acad Sci USA. 2003;100:6946–6951. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science. 2004;303:213–2017. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- 8.Ling T, Yu H, Shen Z, Wang H, Zhu J. Virus-mediated FCC iron nanoparticle induced synthesis of uranium dioxide nanocrystals. Nanotechnology. 2008;19:115608. doi: 10.1088/0957-4484/19/11/115608. [DOI] [PubMed] [Google Scholar]
- 9.Flynn CE, Lee SW, Peele BR, Belcher AM. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003;51:5867–5880. doi: 10.1016/j.actamat.2003.08.031. [DOI] [Google Scholar]
- 10.Lee SW, Mao C, Flynn CE, Belcher AM. Ordering of quantum dots using genetically engineered viruses. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 11.Lee SW, Lee SK, Belcher AM. Virus-based alignment of inorganic, organic, and biological nanosized materials. Adv Mater. 2003;15:689–692. doi: 10.1002/adma.200304818. [DOI] [Google Scholar]
- 12.Souza GR, Dawn RC, Staquicini FI, Michael GO, Evan YS, Richard LS, Miller JH, Wadih A, Renata P. Networks of gold nanoparticles and bacteriophage as biological sensors and cell-targeting agents. Proc Natl Acad Sci USA. 2006;103:1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar H, Yacoby I, Bencher I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 2008;8:37–50. doi: 10.1186/1472-6750-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everts M, Saini V, Leddon JL, Kok RJ, Stoff-Khalili M, Preuss MA, Millican CL, Perlins G, Brown JM, Bagaria H, Nikles DE, Johnson DT, Zharov VP, Curiel DT. Covalently linked Au nanoparticles to a viral vector: potential for combined photothermal and gene cancer therapy. Nano Lett. 2006;6:587–591. doi: 10.1021/nl0500555. [DOI] [PubMed] [Google Scholar]
- 15.Ahiwale SS, Bankar AV, Tagunde SN, Zinjarde S, Ackermann HW, Kapadnis BP. Isolation and characterization of a rare waterborne lytic phage of Salmonella enterica serovar Paratyphi B. Can J Microbiol. 2013;59:318–323. doi: 10.1139/cjm-2012-0589. [DOI] [PubMed] [Google Scholar]
- 16.Adams MH. Bacteriophages. New York: Interscience Publishers; 1959. pp. 1912–1956. [Google Scholar]
- 17.Bankar AV, Joshi B, Kumar AR, Zinjarde SS. Banana peel extract mediated synthesis of gold nanoparticles. Colloids Surf B Biointerfaces. 2010;80:45–50. doi: 10.1016/j.colsurfb.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Dusane DH, Rajput JK, Kumar AR, Nancharaiah YV, Venugopalan VP, Zinjarde SS. Disruption of fungal and bacterial biofilms by lauroyl glucose. Lett Appl Microbiol. 2008;47:374–379. doi: 10.1111/j.1472-765X.2008.02440.x. [DOI] [PubMed] [Google Scholar]
- 19.Rajput N, Bankar A. Bio-inspired gold nanoparticles synthesis and their anti-biofilm efficacy. J Pharm Investig. 2016 [Google Scholar]
- 20.Verma VC, Singh SK, Solanki R, Prakash S. Biofabrication of anisotropic gold nanotriangles using extract of endophytic Aspergillus clavatus as a dual functional reductant and stabilizer. Nanoscale Res Lett. 2011;6:16–22. doi: 10.1186/1556-276X-6-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauhan A, Zubair S, Tufail S, Sherwani A, Sajid M, Raman SC, Azam A, Owais M. Fungus-mediated biological synthesis of gold nanoparticles: potential in detection of liver cancer. Int J Nanomed. 2011;6:2305–2319. doi: 10.2147/IJN.S23195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sau TK, Rogach AL. Nonspherical noble metal nanoparticles: colloid-chemical synthesis and morphology control. Adv Mater. 2010;22:1781–1804. doi: 10.1002/adma.200901271. [DOI] [PubMed] [Google Scholar]
- 23.Nithya B, Jayachitra A. Improved antibacterial and antibiofilm activity of plant mediated gold nanoparticles using Garcinia cambogia. Int J Pure Appl Biosci A. 2016 [Google Scholar]
- 24.Chaw C, Manimaran M, Tay FEH. Role of silver ions in destabilization of intermolecular adhesion forces measured by atomic force microscopy in Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2005;49:4853–4859. doi: 10.1128/AAC.49.12.4853-4859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]