Abstract
In metazoan embryos, transcription is mostly silent for a few cell divisions, until release of a first major wave of embryonic transcripts by so-called zygotic genome activation (ZGA) 1. Maternally provided ZGA-triggering factors have been identified in Drosophila melanogaster and Danio rerio 2,3, but their mammalian homologues are still undefined. Here, we reveal that the DUX family of transcription factors 4,5 is essential to this process in human and mouse. First, human DUX4 and murine Dux are both expressed prior to ZGA in their respective species. Second, both orthologues bind the promoters and activate the transcription of ZGA genes. Third, Dux knockout in mouse embryonic stem cells (mESCs) prevents their cycling through a 2-cell-like state. Finally, zygotic depletion of Dux leads to impaired early embryonic development and defective ZGA. We conclude that DUX proteins are key inducers of zygotic genome activation in placental mammals.
Dux genes encode for double-homeodomain proteins and are conserved throughout placental mammals 4,5. Human DUX4, the intronless product of an ancestral DUXC, is nested within the D4Z4 macrosatellite repeat of chromosome 4 as an array of 10 to 100 units 6. DUX4, DUXC, and Dux genes from other placental mammals display the same repetitive structure, with DUX4 from primates and Afrotheria and DUXC from cow and other Laurasiatheria localizing at telomeric or pericentromeric regions, and murine Dux tandem repeats lying adjacent to a mouse-specific chromosomal fusion point that resembles a subtelomeric structure 4,5.
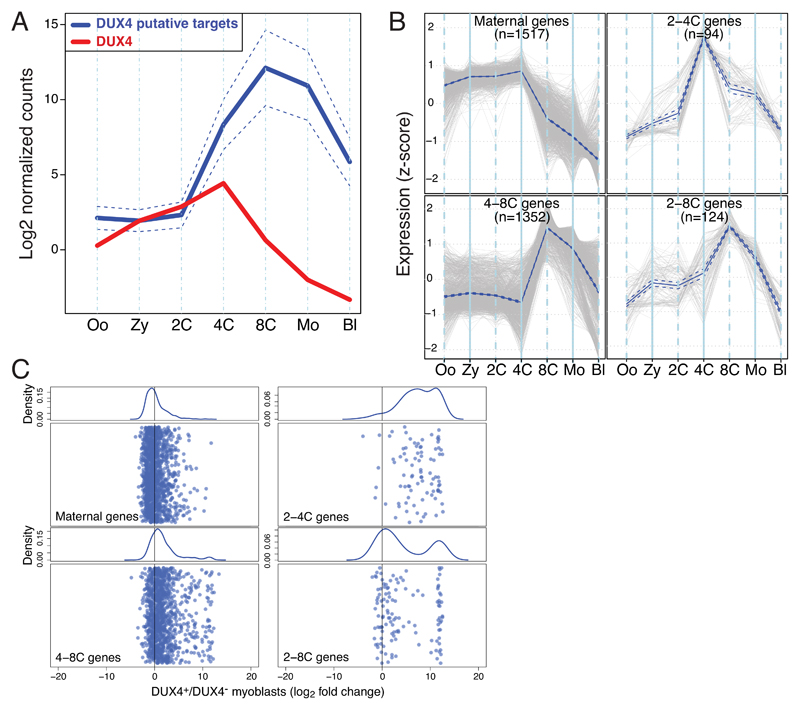
Overexpression-inducing mutations in DUX4 are associated with facio-scapulo-humeral dystrophy (FSHD), the third most common muscular dystrophy 7,8, and forced DUX4 production in human primary myoblasts leads to upregulation of genes active during early embryonic development 9. Based on this premise, we analyzed publicly available RNA-seq datasets corresponding to this period, focusing on DUX4 and the 100 genes most upregulated in DUX4-overexpressing muscle cells (Figure 1A, Table S1) 10,11. DUX4 RNA was detected from oocyte to 4-cell (4C) stage, while transcripts from its putative targets emerged on average at 2-cell (2C) and peaked at 8-cell (8C) stages, as previously defined for human ZGA 12. Transcripts upregulated in DUX4-overexpressing muscle cells 11 were also enriched at 8C stage (Supplementary Figure 1AB), and upon clustering genes according to their patterns of early embryonic expression (Figure 1B) we could delineate i) 1517 genes, the transcripts of which were already detected in oocytes, plateaued up to 4C and abruptly dropped afterwards (maternal gene cluster); ii) 94 genes and 124 genes, the expression of which started at 2C, and peaked at 4C and 8C, respectively, before decreasing briskly, consistent with early ZGA genes (2-4C and 2-8C gene clusters); and iii) 1352 genes expressed only from 4C, peaking at 8C, and then decreasing progressively, as expected for late ZGA genes (4-8C gene cluster). Only the two early ZGA clusters (2-4C and 2-8C) were highly enriched for genes upregulated in DUX4-overexpressing myoblasts (Figure 1C, Supplementary Figure 1C).
Figure 1. DUX4 promotes transcription of genes expressed during early ZGA.
(A) Comparative expression during early human embryonic development of DUX4 (red) and the top 100 genes upregulated upon DUX4 overexpression in human primary myoblasts (blue, full line average, dashed lines 95% confidence interval around the mean). Oo, oocyte; Zy, zygote; 2C, 4C, 8C, corresponding n-cell stage; Mo, morula; Bl, blastocyst. (B) Cluster of genes differentially expressed during early embryonic development were selected from the previously identified subsets of genes (Supplementary Figure 1A) based on high expression at 4C (upper panels) and 8C (lower panels). Blue and dotted line delineate mean and 95% confidence, respectively. (C) Expression of genes from each cluster illustrated in (B) when DUX4 is ectopically expressed in human primary myoblasts. Lower parts of the panels depict the fold change expression of genes within these clusters, all randomly distributed along the y-axes, with kernel density plotted in the upper part.
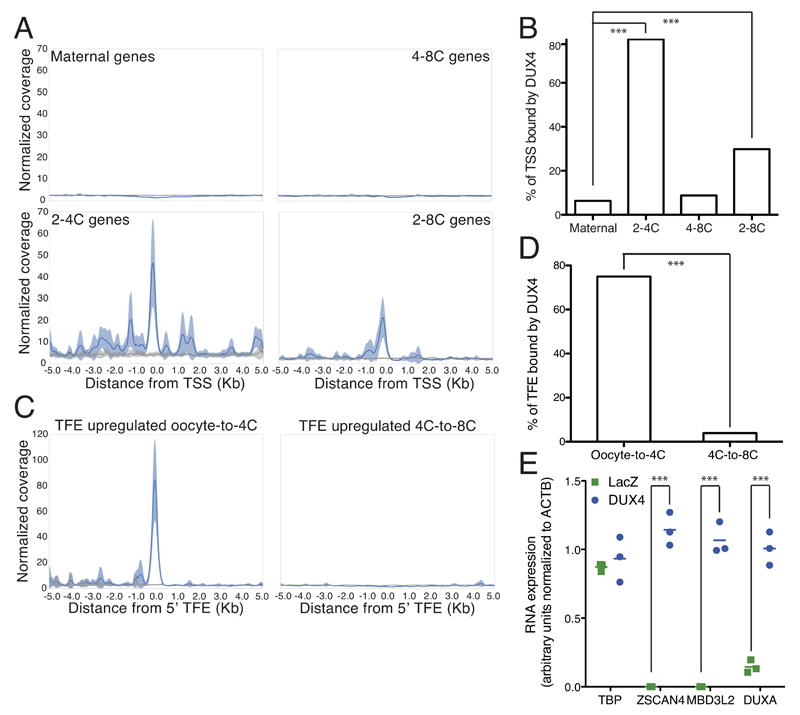
Chromatin cannot be reliably analyzed from the very low number of cells that make up an early embryo, but ChIP-seq data obtained in DUX4 overexpressing human embryonic stem cells (hESCs) (Figure 2AB, Supplementary Figure 2) and myoblasts 9 (Supplementary Figure 3) revealed a marked enrichment of the transcription factor around the annotated transcriptional start site (TSS) region of early ZGA genes (2-4C and 2-8C clusters), but not of zygotic (maternal) and late ZGA (4-8C) genes. Interestingly, several genes were not bound on their annotated TSS, but on neighboring sequences, and their transcription was found to start near this DUX4 binding site (Supplementary figure 4). It was previously demonstrated that DUX4 drives expression of many of its target genes from alternative promoters 11. Upon examining publicly available single-cell RNA sequencing data quantifying the far 5’-ends of transcripts (TFEs) in early human development 13, we correspondingly found that the TFE of 24 out of 31 early ZGA genes overlapped with DUX4 binding sites (Figure 2CD, Supplementary figure 3CD). DUX4 was also recruited to several groups of transposable elements (TEs), notably endogenous retroviruses such as HERVL, MER11B and C, the expression of which increased at ZGA (Supplementary figure 2BC). Furthermore, DUX4 overexpression in hESCs led to early ZGA genes induction, as previously observed in myoblasts (Figure 2E) 11.
Figure 2. DUX4 binds TSSs of genes expressed during early ZGA and activates their expression in hESCs.
(A) Average coverage normalized for sequencing depth of ChIP-seq signal of DUX4 (blue) when overexpressed in hESCs in a window of 5 kb from the annotated TSS of genes belonging to the 2-4C and 2-8C clusters from Figure 1B. Total input is represented in gray (line, average; shade, standard error of the mean). B) Fraction of genes belonging to each cluster from Figure 1B with a DUX4 peak within 5 kb of their annotated TSS. Fisher’s exact test was performed to compare maternal vs. 2-4C and 2-8C (p= 3.54e-61 and p= 2.23e-13 respectively) (C) Average coverage of ChIP-seq signal of DUX4 (blue) when overexpressed in hESCs within 5 kb of TFE of transcripts specifically upregulated at oocyte-to-4C and 4C-to-8C transitions. Total input is represented in gray (line, average; shade, standard error of the mean). (D) Fraction of TFE from oocyte-to-4C (n=32) and 4C-to-8C (n=128) transitions that have a DUX4 peak overlapping with their 5' end. Fisher’s exact test was performed to compare 4C-to-8C vs. oocyte-to-4C TFEs (p= 4.48e-17). (E) Comparative expression in hESCs of three genes activated at ZGA (ZSCAN4, MBD3L2 and DUXA) and two control housekeeping genes (ACTB and TBP) 24 hours after transfection with plasmids expressing LacZ (green squares) or DUX4 (blue circles). Expression was normalized to ACTB. Horizontal lines represent the mean. *** p ≤ 0.001, unpaired t-test.
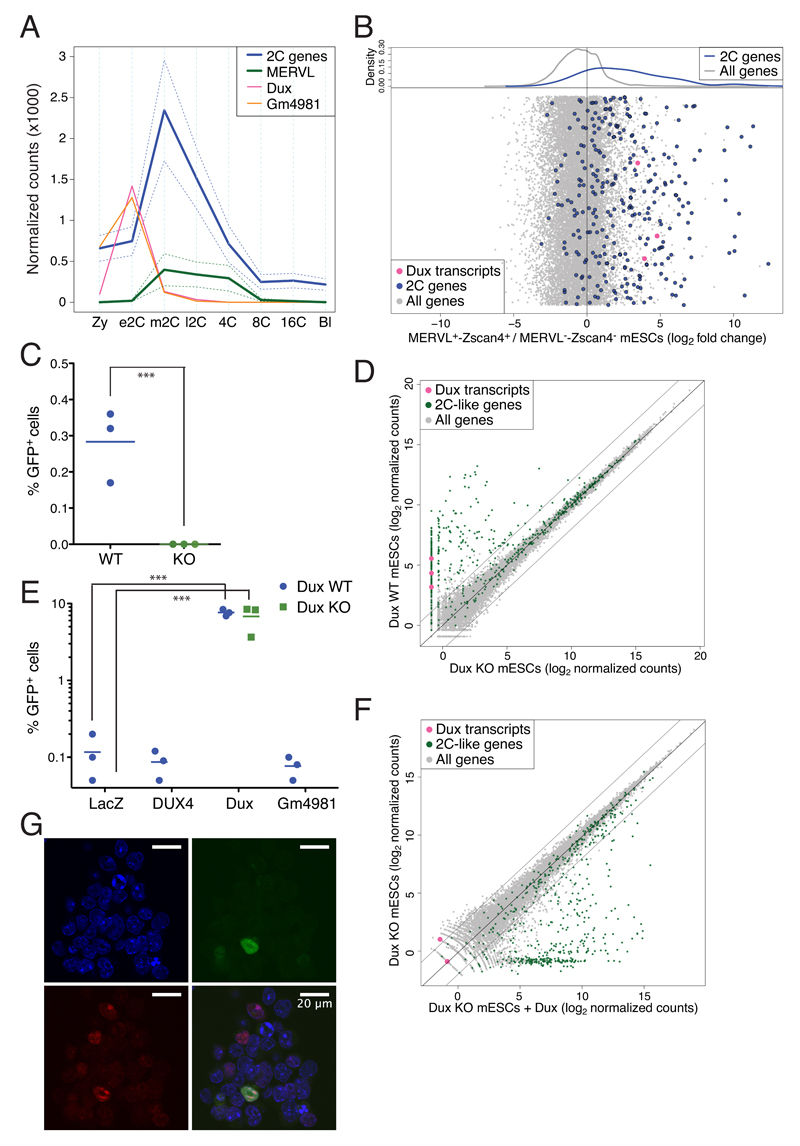
Dux and DUX4 have largely conserved amino acid sequences, in particular within the two DNA-binding homeodomains and the C-terminal region, previously described as responsible for recruiting p300/CBP (Supplementary Figure 5B) 14. The murine Dux tandem repeat encodes two main transcripts, full-length Dux (or Duxf3) and a variant named Gm4981 lacking the first homeodomain (Supplementary Fig. 5A). Both Dux and Gm4981 are expressed in mouse embryos prior to ZGA-defining genes and transposable elements (e.g. murine ERVL or MERVL) at the middle 2C stage, indicating that their products likely are functional homologues of DUX4 (Figure 3A) 15. To consolidate these results, we turned to mESCs, a small percentage of which displays at any given time a 2C-like transcriptome in culture, with expression of ZGA genes notably from the MERVL promoter 16,17. Upon analyzing single-cell RNA-seq data from 2C-like mESCs 18, we confirmed that Dux transcripts were markedly enriched, as were early ZGA RNAs such as Zscan4, Zfp352 and Cml2 (Figure 3B and Supplementary Figure 6). We used CRISPR/Cas9-mediated genome editing to delete the Dux-containing macrosatellite repeat in mESCs expressing a GFP reporter under control of a MERVL promoter. This resulted in a complete absence of GFP+ 2C-like cells, and in the loss of a large fraction of 2C-like cell-specific transcripts (Figure 3CD, Supplementary Figure 7). Overexpression of Dux but not DUX4 rescued the 2C-like state in the mESC KO clones (Figure 3EF, Supplementary Figure 8 and 9), albeit not in all cells where Dux was produced (Figure 3G). Interestingly, both murine Dux and human DUX4 were able to induce the transcription of ZGA genes in the human 293T cell line (Supplementary Figure 10).
Figure 3. Dux is necessary for formation of 2C-like mESCs.
(A) Comparative expression of the two alternative transcripts of Dux, Dux (pink) and Gm4981 (orange), with genes (blue) and transposable elements (MERVL; green) specifically expressed during murine ZGA. Full lines represent the average and dashed lines the 95% confidence interval around the mean (B) Single-cell RNA-sequencing comparison between mESCs sorted for expression of both tomato and GFP reporters driven by MERVL and Zscan4 promoters, respectively (revelators of 2C-like cells), and the double negative population. Average gene expression was quantified and fold change between positive and negative cells was plotted. Dots are randomly distributed along the y-axes. The upper plot represents the kernel density estimate of middle-2C stage (blue line) and the rest of the genes (gray line). The Dux macrosatellite repeat was deleted in mESCs carrying a MERVL-GFP reporter by CRISPR/Cas9-mediated excision. (C) Fraction of GFP+ cells in WT or Dux-deleted cells. (D) RNA sequencing analysis of WT and Dux KO mESC clones. The dot plot displays the average gene expression of three independent clones from each cell type. (E) GFP expression in Dux KO (blue circles) and WT (green squares) mESC clones carrying an integrated MERVL-GFP reporter, and transiently expressing LacZ, DUX4, Dux or Gm4981 transgenes. (F) RNA sequencing analysis of Dux KO mESC clones transiently expressing Dux or control. The dot plot displays the average gene expression of two independent clones from each cell type. (G) Dux KO mESCs carrying an integrated MERVL-GFP reporter and transiently expressing a HA-tagged form of Dux were stained for HA and immunofluorescence was detected by confocal microscopy. DAPI, blue; GFP, green; HA, red. Horizontal bars in (C) and (E) represent the mean. *** p ≤ 0.001, unpaired t-test.
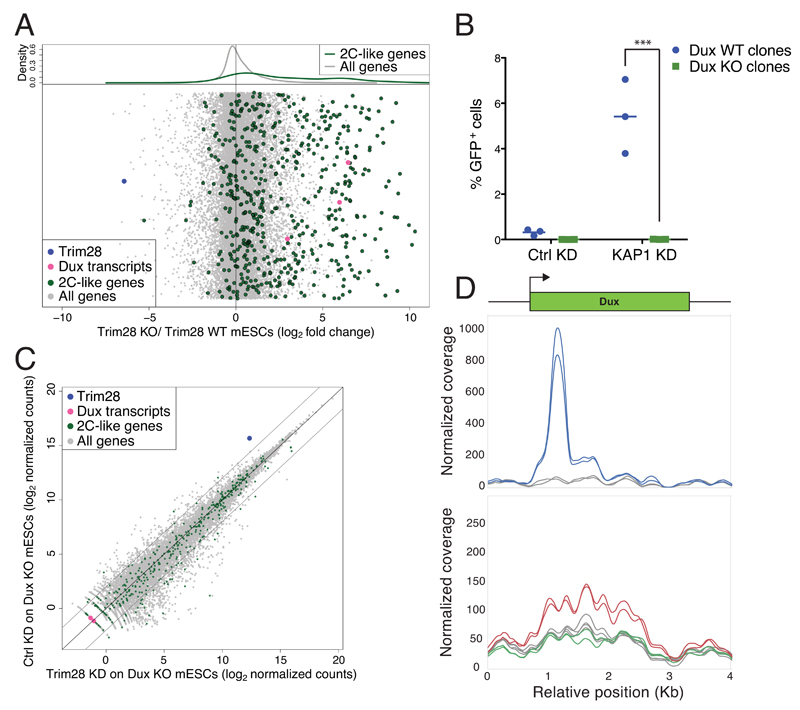
Upon depletion of the transcriptional repressor TRIM28 (tripartite motif-containing protein 28; KAP1) from mESCs, expression of 2C-specific genes increased as previously observed 17, as did levels of Dux transcripts (Figure 4A, Supplementary Figure 11BCD). Remarkably, this phenotype was completely abrogated in Dux-depleted mESCs (Figure 4BC, Supplementary Figure 9 and 11ABCD). Correspondingly, we found that TRIM28 associates with the 5’-end of the Dux gene and that tri-methylation of histone 3 lysine 9 (H3K9me3), a canonical marker of TRIM28-mediated repression, was enriched on the Dux locus and lost upon knockdown of the heterochromatin inducer (Figure 4D, Supplementary Figure 11EF).
Figure 4. TRIM28 regulates formation of 2C-like mESCs by repressing Dux expression.
(A) RNA sequencing analysis of WT and Trim28 KO mESCs. Average gene expression was quantified and fold change between KO and WT cells plotted. Dots are randomly distributed along the y-axes. The upper plot represents the kernel density estimate of genes specifically expressed in 2C-like mESCs (green line) and the rest of the genes (gray line). (B) WT (blue circles) and Dux KO (green squares) mESC clones carrying an integrated MERVL-GFP reporter were transduced with lentiviral vectors encoding for shRNAs targeting Trim28 or a control. 4 days later GFP expression was quantified. Horizontal lines represent the mean. *** p ≤ 0.001, unpaired t-test. (C) RNA sequencing of Trim28-depleted or control Dux KO mESC clones. The dot plot represents the average gene expression of three independent KO clones transduced with lentiviral vectors encoding for a control or a Trim28-specific shRNA. (D) Average coverage of ChIP-seq signal of Trim28 (top plot; blue lines; two replicates) and H3K9me3 (bottom plot; two replicates) in control (red lines) and Trim28 KD mESCs (green line) around the Dux gene. Total input is represented in gray. ChIP-seq reads were mapped on the genome, before focusing the analysis on a 500bp window around the main Dux gene. H3K9me3 peaks over the Dux macrosatellite repeat were only called in the control KD mESCs (Sicer; false discovery rate 0.05)
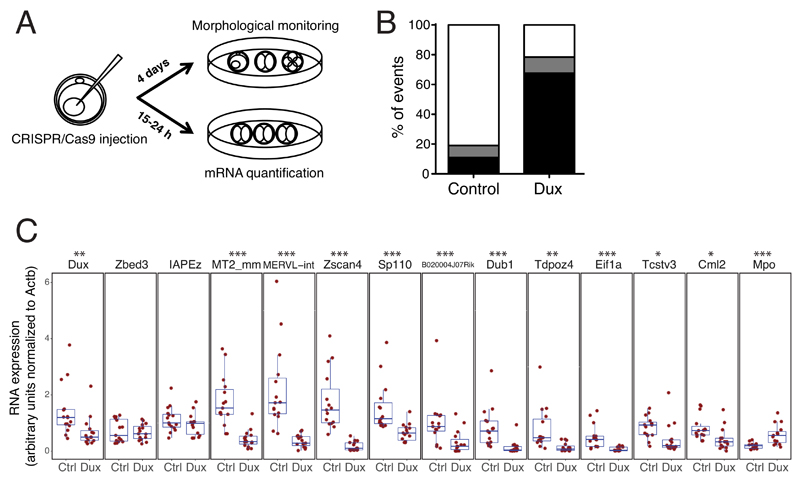
Finally, we addressed the role of Dux during murine early embryonic development. For this, we injected zygotes with plasmids encoding for the Cas9 nuclease and either the two guide RNAs (sgRNAs) used to generate Dux KO mESCs or a non-targeting sgRNA control. We then determined the RNA profile of 2C embryos around 7 hours after the first cell division or monitored their ex vivo development into blastocysts over 4 days (Figure 5A). We found that Dux–depleted embryos presented a major differentiation defect, most failing to reach the morula/blastocyst stage, and did not exhibit transcriptional changes typical of ZGA, such as induction of MERVL, Zscan4 and several other tested early ZGA genes, and drop in Mpo maternal transcript (Figure 5BC, Supplementary Figure 12).
Figure 5. Dux is necessary for mouse early embryonic development.
(A) Schematic of the Dux loss-of-function experiment in mouse pre-implantation embryos. Zygotes were first injected in the pronucleus with plasmids encoding for the Cas9 nuclease and sgRNAs targeting the flanking region of the Dux macrosatellite repeat or a non-targeting sgRNA, then were either (B) monitored for their ability to differentiate ex vivo or (C) collected at 2C-stage for mRNA quantification. (B) Average percent of embryos reaching the morula/blastocyst stages (white) or failing to differentiate (delayed/dead embryos, black; defective morula/blastocyst, grey) 4 days after pronuclear injection. The plot represents an average from 3 independent experiments with 16 to 23 embryos for each condition. Fisher’s exact test was performed to compare the embryonic stage of control against Dux KO (p= 1.54e-10) (C) Comparative expression of Dux, early ZGA genes (Zscan4, Sp110, B020004J07Rik, Dub1, Tdpoz4, Eif1a, Tcstv3, Cml2), 2C-restricted TE (MERVL, the LTR and int regions of which are detected with MT2_mm and MERVL-int primers, respectively), a gene (Mpo), the expression of which decreases at ZGA, 2 genes (Actb, Zbed3) stably expressed during pre-implantation embryonic development and a control TE (IAPEz) in 15 2C stage embryos (5 from each of 3 independent experiments) 15-24 hours after pronuclear injection with plasmids expressing Cas9 and control or Dux-specific sgRNAs. Boxes depict the 25 and 75 percentiles, line in the boxes represents the median. Expression was normalized to Actb. * p ≤ 0.05 ** p ≤ 0.01, *** p ≤ 0.001, Wilcoxon test.
In sum, our data reveal DUX genes as key regulators of early embryonic development. The demonstrated ability of DUX4 to recruit the p300/CBP complex and to induce local chromatin relaxation 14 as well as the mechanism of action of Zelda, a master inducer of ZGA in Drosophila 19,20, suggest that DUX proteins could act as pioneer factors for transcriptional activation, by opening chromatin around the TSS of early ZGA genes to facilitate access for other transcription factors. Still, the genomic recruitment of pioneer factors such as OCT4, NANOG and KLF4 can be hampered if heterochromatin marks are overly abundant at their target loci 21. Many murine ZGA genes are expressed from the LTR of endogenous retroviruses, which in mESC cells are typically enriched in repressive marks 17. It could be that, at any given time, these marks are relieved in only a small percentage of mESC in culture. What drives this fluctuation remains to be determined. As well, what controls expression of DUX genes themselves is yet to be defined, although the conserved genomic localization of all placental mammal DUX orthologs close to telomeric and subcentromeric regions suggests that this genomic context, characterized by high levels of repression, might be of primary relevance 4,5,22. DUX genes seem indeed to become expressed only during events associated with major chromatin relaxation, for instance in early embryos and upon loss of repression of the D4Z4 macrosatellite repeat in myoblasts of FSHD patients 23,24. Our data indicate that TRIM28 plays a major role in murine Dux repression, but the only mild increase in cells entering the 2C state when it is depleted (around 5% of mESCs) and the demonstrated ability of several other transcriptional modulators (e.g. SETDB1, EHMT2, HP1, CHAF1A/B, RYBP, KDM1A) to prevent cycling of mESCs through this state indicate that control of the Dux macrosatellite repeat is most likely multifactorial 16,25–29. Broad derepression of the human and murine DUX-containing repeats could similarly occur right after fertilization in either species. Future investigations of the chromatin state of these loci in early embryos will shed light on the epigenetic changes responsible for this process and on the nature of their molecular mediators.
Materials and methods
Cell lines and tissue culture
mESC WT and KO for Trim28 30, and E14 mESCs containing the MERVL regulatory sequence driving expression of a 3XturboGFP-PEST 16 were cultured in feeder-free conditions on 0.1% gelatin-coated tissue culture plates in 2i medium, a N2B27 base medium supplemented with the MEK inhibitor, PD0325901 (1 μM), the GSK3β inhibitor CHIR99021 (3 μM) and LIF. E14 mESCs express the main markers of pluripotency (RNA-seq). H1 ESCs (WA01, WiCell) were maintained in mTesRI (StemCell Technologies) on hES-qualified Matrigel (BD Biosciences). 293T cells were maintained in DMEM supplemented with 10% FCS. All cells were regularly checked for the absence of mycoplasma contamination.
Plasmids and lentiviral vectors
The MT2/gag sequence was amplified from the pGL3 plasmid 29, and the human PGK promoter from pRRLSIN.cPPT.R1R2.PGK-GFP.WPRE 30, to be cloned upstream of luciferase in pGL4.20. Table S2 shows the primers used to obtain truncations of the MT2/gag sequence. Single guide RNAs (sgRNAs) targeting sequences flanking the 5’ and 3’ of the Dux-containing macrosatellite repeat were cloned into px459 (version 2) using a standard protocol 31. Table S2 shows the primers used to clone the sgRNAs. The pLKO.1-puromycin shRNA vector was used for the Trim28 knock-down 30. The pLKO.1 vector was further modified to express blasticidin-S-deaminase drug resistance cassette in place of the puromycin N-acetyltransferase. The resulting pLKO.1-blasticidin backbone was used to clone shRNAs against the murine Dux transcript. The sequence of the primers used to clone the Dux shRNA is shown in Table S2. The Gm4981 cDNA was cloned from the genome of E13 mESCs while codon-optimized hDUX4 and mDux were synthesized (Invitrogen). Gm4981, DUX4, Dux and LacZ cDNAs were cloned in the pAIB HIV-1-based transfer vector encoding also for blasticidin resistance using the In-Fusion® HD Cloning Kit (Clontech) 32. pMD2-G encodes the vesicular stomatitis virus G protein (VSV-G). The minimal HIV-1 packaging plasmid 8.9NdSB carrying a double mutation in the capsid protein (P90A/A92E) was used to achieve higher transduction of the lowly permissive mESCs 33.
Production of lentiviral vectors, transduction and transfection of mammalian cells
Lentiviral vectors were produced by transfection of 293T cells using Polyethylenimine (PEI) (Sigma, Inc) 33. To generate stable KDs, mESCs were transduced with empty pLKO.1 vector or vectors containing the shRNA targeting Kap1 or Dux transcripts 30. Cells were selected with 1 μg⁄ml puromycin or 3 μg⁄ml blasticidin starting one day after transduction. hESCs expressing LacZ and DUX4 were generated by transfecting the corresponding AIB plasmids with TransIT®-LT1 Transfection Reagent (Mirus Bio LLC), while nucleofection (Amaxa™ P3 Primary Cell 4D-Nucleofector™ X Kit) was used to engineer mESC expressing LacZ, DUX4, Dux and Gm4981.
Creation of Dux KO mESC lines
E14 mESCs containing the MERVL regulatory sequence driving expression of a 3XturboGFP-PEST were co-transfected with px459 plasmids encoding for Cas9, the appropriate sgRNAs and puromycin resistance cassette by nucleofection (Amaxa™ P3 Primary Cell 4D-Nucleofector™ X Kit). 24 hours later, the cells were selected for 48 hours with 1 μg⁄ml puromycin, single-cell cloned by serial dilution, expanded and their DNA was extracted to detect the presence of WT and/or KO alleles. Three WT and three homozygous Dux KO clones were selected and used in this study.
Luciferase assay
293T or E14 mESCs were cotransfected with the various pGL4.20 derivatives, the renilla plasmid and the pAIB transfer vector encoding either for LacZ, Dux, Gm4981 or DUX4 using Lipofectamine 3000 (Invitrogen). Luciferase activity was quantified 24h after transfection. Firefly luciferase activity was normalized to the activity of Renilla luciferase. Light emission was measured on a luminescence plate reader.
Immunofluorescence assay
mESC clones expressing an HA-tagged Dux protein were fixed for 20 min with 4% paraformaldhyde, permeabilized for 5 min with 0.1% Triton-X 100, and blocked for 30 min with 1% BSA in PBS. Cells were then incubated for 1 hour with anti-HA.11 (Covance) or anti-NANOG (Active Motif) or anti-SOX2 (Active Motif) antibodies diluted in PBS with 1% BSA. After 3 washes, the cells were incubated with anti-mouse (HA) or anti-rabbit (NANOG, SOX2) Alexa Fluor 647-conjugated secondary antibodies for 1 hour and washed again three times. Every step until this point, was carried with cells in suspension. Pelleted cells were then resuspended in VECTASHIELD® Mounting Medium with DAPI (Vector Laboratories) and mounted on the coverslip. The slides were viewed with a Zeiss LSM700 confocal microscope.
Fluorescence-activated cell sorting (FACS)
FACS analysis was performed with a BD FACScan system. Trim28 knock-down mESCs containing the MT2/gag-GFP reporter were subjected to FACS sorting with AriaII (BD Biosciences).
Standard PCR, RT-PCR and RNA sequencing
For the genotyping of Dux WT and KO alleles, genomic DNA was extracted with DNeasy Blood & Tissue Kits (QIAGEN) and the specific PCR products were amplified using PCR Master Mix 2X (Thermo Scientific) combined with the appropriate primers (design in Supplementary Figure 6A; primer sequences in Table S2).
Total RNA from cell lines was isolated using the High Pure RNA Isolation Kit (Roche). cDNA was prepared with SuperScript II reverse transcriptase (Invitrogen). Ambion Single Cell-to-CT kit (Thermo Fisher) was used for RNA extraction, cDNA conversion and mRNA pre-amplification of 2C stage embryos. Primers listed in Supplementary Table S2 were used for SYBR green qPCR (Applied Biosystems). Library preparation and 150-base-pair paired-end RNA-seq were performed using standard Illumina procedures for the NextSeq 500 platform (GSE94325).
ChIP and ChIP sequencing
ChIP and library preparation were performed as described previously 30. DUX4-HA ChIP was done using the anti-HA.11 (Covance) antibody. Sequencing of Trim28 and H3K9me3 ChIP was performed with Illumina HiSeq 2500 in 100-bp reads run. Sequencing of DUX4 was performed with Illumina NextSeq 500 in 75-bp paired-end reads run.
RNA-seq datasets preprocessing
Single-cell RNA-Seq of human and mouse early embryo development (GSE36552 and GSE45719 respectively), single-cell RNA-Seq of 2C-like cells (E-MTAB-5058), DUX4 overexpression in human myoblasts (GSE45883), and KAP1 KO (GSE74278) datasets were downloaded from different repositories (GEO, and ArrayExpress) 34,35. Reads were mapped to the human genome (hg19) or mouse genome (mm9) using TopHat (v2.0.11) 36 in sensitive mode (the exact parameters are: tophat -g 1 --no-novel-juncs --no-novel-indels -G $gtf --transcriptome-index $ transcriptome --b2-sensitive -o $localdir $index $reads1 $reads2). Gene counts were generated using HTSeq-count. Normalization for sequencing depth and differential gene expression analysis was performed using Voom 37 as it has been implemented in the limma package of Bioconductor 38. TEs overlapping exons were removed from the analysis. Counts per TE integrant (genomic loci) were generated using the multiBamCov tool from the bedtools software 39. Normalisation for sequencing depth was performed using Voom, with total number of reads on genes as size factor. To compute total number of reads per TE family, counts on all integrants of each family were added up.
Analysis of single cell expression data from human and mouse embryonic stages
For every embryonic stage we performed a statistical test to find the genes that had a different expression level compared to the other stages 10, using a moderated F-test (comparing the interest group against every other) as implemented in the limma package of Bioconductor. Genes were selected as expressed in a specific stage if having a significant p-value (<0.05 after adjusting for multiple testing with the Benjamini and Hochberg method) and an average fold change respective to the other embryonic stages bigger than 10. We additionally removed all genes exhibiting a 1.1-fold higher expression in any of the embryonic stages compared to the stage analyzed (Suppl. Figure 1A). Note that with this approach a gene can be marked as expressed in more than one stage. Codes are available on demand.
Correspondence between DUX4 overexpression and single cell expression data from human embryonic stages
For every stage, we classified the genes in 4 patterns of expression by performing a hierarchical clustering (with Pearson correlation as distance and complete agglomeration method). Figure 1B shows the 2 most relevant patterns derived from the 4C and 8C stages.
Expression of the genes identified with this method was then compared between DUX4- and GFP-overexpressing human myoblast cells. For a gene to be considered differentially expressed, a p-value (after multiple testing correction with the Benjamini and Hochberg method) lower than 0.05 and a fold change bigger than 2 were imposed. A moderated t-test was used for the statistical test, as implemented in the limma package of Bioconductor.
ChIP-seq data processing
ChIP-seq dataset of DUX4 overexpressed in human myoblasts (GSE94325) was downloaded from GEO. Reads were mapped to the human genome assembly hg19 using Bowtie2 using the sensitive-local mode 40. SICER was used to call histone mark peaks 41. For the ones that are not histone marks, we used MACS (with default parameters) when the data was single-end and MACS2 (the exact parameters are: macs2 callpeak -t $chipbam -c $tibam -f BAM -g $org -n $name -B -q 0.01 --format BAMPE) when the data was paired-end 42. Both, SICER peaks with an FDR above 0.05 and MACS peaks with a score lower than 50 were discarded. RSAT was used for motif discovery and to compute motif abundance 43. To compute the percentage of bound TE integrants in each family, we used bedtools suite.
Coverage plots
ChIP-seq signals on features of interest were extracted from the bigWigs beforehand normalized for sequencing depth (reads per hundred millions). Each signal was then smoothed using a running average of window 75bp for DUX4, 250bp for Trim28, and 500bp for H3K9me3. Finally, the mean and standard error of the mean of the signals were computed and plotted for each set of features of interest. Scripts are available on demand.
Pronuclear injection of mouse embryos
Pronuclear injection was performed according to the standard protocol of the Transgenic Core Facility of EPFL. In summary, B6D2F1 mice were used as egg donors (5 weeks old). Mice were injected with PMSG (10 IU), and HCG (10 IU) 48 hours after. After mating females with B6D2F1 males, zygotes were collected and kept in KSOM medium pre-gassed in 5% CO2 at 37 °C. Embryos were then transferred to M2 medium and microinjected with 10 ng/μg of either a px459 plasmid containing a non-targeting sgRNA or a mix of the two plasmids used to obtain the KO in mESCs, in injection buffer (10mM Tris HCl pH7.5, 0.1mM EDTA pH8, 100mM NaCl). After microinjection, embryos were cultured in KSOM medium at 37 °C in 5% CO2 for 4 days. In each of three independent experiments, 5 embryos per condition were collected around 7 hours after first cell division (2C formation) for qPCR analysis, and differentiation of the remaining embryos was followed. At day 4, all the fertilized embryos (between 16 to 23 per condition) were classified for their developmental state. Randomization and blind outcome assessment were not applied. All animal experiments were approved by the local veterinary office and carried out in accordance with the EU Directive (2010/63/EU) for the care and use of laboratory animals.
Sample sizes and statistical tests
We used non-parametric statistical tests (2-sided Wilcoxon test), when we had enough sample size (low-cell number qPCR). Otherwise we used a 2-sided unpaired t-test (standard qPCR and FACS). Fisher’s exact test was used to test for differences in proportions in contingency tables.
Supplementary Material
Acknowledgments
We thank T. Macfarlan and M.E. Torres-Padilla for sharing reagents and for helpful discussions, the Transgenic and Gene Expression Core Facility (EPFL) and S. Offner for technical assistance. This work was financed through grants from the Swiss National Science Foundation, the Gebert-Rüf Foundation, the INGENIUM grant (FP7 MC-ITN INGENIUM 290123), and the European Research Council (ERC 268721 and ERC 694658) to D.T.
Footnotes
Data availability
RNA-seq and ChIP-seq data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE94325.
Author contributions
A.D.I and D.T. conceived the project, designed the experiments, analyzed the data and wrote the manuscript; A.D.I., A.C. and S.V. carried out the experiments; E.P. and J.D. performed the bioinformatics and statistical analyses.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annual review of cell and developmental biology. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidenroth A, et al. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma. 2012;121:489–497. doi: 10.1007/s00412-012-0380-y. [DOI] [PubMed] [Google Scholar]
- 5.Clapp J, et al. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. American journal of human genetics. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hewitt JE, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Human molecular genetics. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 7.Wijmenga C, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nature genetics. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 8.Gabriels J, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 9.Geng LN, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Developmental cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature structural & molecular biology. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 11.Young JM, et al. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS genetics. 2013;9:e1003947. doi: 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassena R, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138:3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tohonen V, et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nature communications. 2015;6:8207. doi: 10.1038/ncomms9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SH, et al. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic acids research. 2016 doi: 10.1093/nar/gkw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 16.Ishiuchi T, et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nature structural & molecular biology. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 17.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckersley-Maslin MA, et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell reports. 2016;17:179–192. doi: 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, et al. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome research. 2015;25:1703–1714. doi: 10.1101/gr.192542.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz KN, et al. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome research. 2015;25:1715–1726. doi: 10.1101/gr.192682.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrod S, Gasser SM. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cellular and molecular life sciences : CMLS. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends in molecular medicine. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 25.Maksakova IA, et al. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics & chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes & development. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D, Muniesa P. Regulation of Mouse Retroelement MuERV-L/MERVL Expression by REX1 and Epigenetic Control of Stem Cell Potency. Frontiers in oncology. 2014;4:14. doi: 10.3389/fonc.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter M, Teissandier A, Perez-Palacios R, Bourc'his D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife. 2016;5 doi: 10.7554/eLife.11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes & development. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ecco G, et al. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Developmental cell. 2016;36:611–623. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Iaco A, et al. TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology. 2013;10:20. doi: 10.1186/1742-4690-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Iaco A, Luban J. Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology. 2014;11:11. doi: 10.1186/1742-4690-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolesnikov N, et al. ArrayExpress update--simplifying data submissions. Nucleic acids research. 2015;43:D1113–1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas-Chollier M, et al. RSAT: regulatory sequence analysis tools. Nucleic acids research. 2008;36:W119–127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.