Abstract
Structural rearrangements have long been recognized as an important source of genetic variation with implications in phenotypic diversity and disease, yet their detailed evolutionary dynamics remain elusive. Here, we use long-read sequencing to generate end-to-end genome assemblies for 12 strains representing major subpopulations of the partially domesticated yeast Saccharomyces cerevisiae and its wild relative Saccharomyces paradoxus. These population-level high-quality genomes with comprehensive annotation allow for the first time a precise definition of chromosomal boundaries between cores and subtelomeres and a high-resolution view of evolutionary genome dynamics. In chromosomal cores, S. paradoxus exhibits faster accumulation of balanced rearrangements (inversions, reciprocal translocations and transpositions) whereas S. cerevisiae accumulates unbalanced rearrangements (novel insertions, deletions and duplications) more rapidly. In subtelomeres, both species show extensive interchromosomal reshuffling, with a higher tempo in S. cerevisiae. Such striking contrasts between wild and domesticated yeasts likely reflect the influence of human activities on structural genome evolution.
Introduction
Understanding how genetic variation translates into phenotypic diversity is a central theme in biology. With the rapid advancement of sequencing technology, genetic variation in large natural populations has been extensively explored for humans and several model organisms1–9. However, our current knowledge of natural genetic variation is heavily biased towards single nucleotide variants (SNVs). Large-scale structural variants (SVs) such as inversions, reciprocal translocations, transpositions, novel insertions, deletions, and duplications are much less well characterized due to technical difficulties in detecting them using short-read sequencing data. This is a critical problem to address given that SVs often account for a substantial fraction of genetic variation and can have significant implications in adaptation, speciation and disease susceptibility10–12.
The long-read sequencing technologies from Pacific Biosciences (PacBio) and Oxford Nanopore offer powerful tools for high-quality genome assembly13. Their recent applications provided highly continuous genome assemblies with many complex regions correctly resolved, even for large mammalian genomes14,15. This is especially important in characterizing SVs, which are frequently embedded in complex regions. For example, eukaryotic subtelomeres, which profoundly contribute to genetic and phenotypic diversity, are known hotspots of SVs due to rampant ectopic sequence reshuffling16–19.
The baker’s yeast Saccharomyces cerevisiae is a leading biological model system with great economic importance in agriculture and industry. Discoveries in S. cerevisiae have illuminated almost every aspect of molecular biology and genetics. It is the first eukaryote to have its genome sequence, population genomics and genotype-phenotype map extensively explored1,20,21. Here, we applied PacBio sequencing to 12 representative strains of S. cerevisiae and its wild relative Saccharomyces paradoxus and revealed striking interspecific contrasts in structural dynamics across their genomic landscapes. This is the first study in eukaryotes that brings long-read sequencing technologies to the field of population genomics and studies genome evolution using multiple reference-quality genome sequences.
Results
End-to-end population-level genome assemblies
We applied deep PacBio (100-300x) and Illumina (200-500x) sequencing to seven S. cerevisiae and five S. paradoxus strains representing evolutionarily distinct subpopulations of both species1,6 (Supplementary Tables 1-2). The raw PacBio de novo assemblies of both nuclear and mitochondrial genomes exhibited compelling completeness and accuracy, with most chromosomes assembled into single contigs and highly complex regions accurately assembled (Supplementary Fig. 1). After manual gap filling and Illumina-read-based error correction (See Methods), we obtained end-to-end assemblies for almost all the 192 chromosomes, with only the rDNA array on chromosome XII (chrXII) and 26 of 384 (6.8%) chromosome-ends remaining not fully assembled. We estimate that only 45-202 base-level sequencing errors remain across each 12 Mb nuclear genome (Supplementary Tables 3-4). For each assembly, we annotated centromeres, protein-coding genes, tRNAs, Ty retrotransposable elements, core X-elements, Y'-elements and mitochondrial RNA genes (Supplementary Tables 5-7). Chromosomes were named according to their encompassed centromeres.
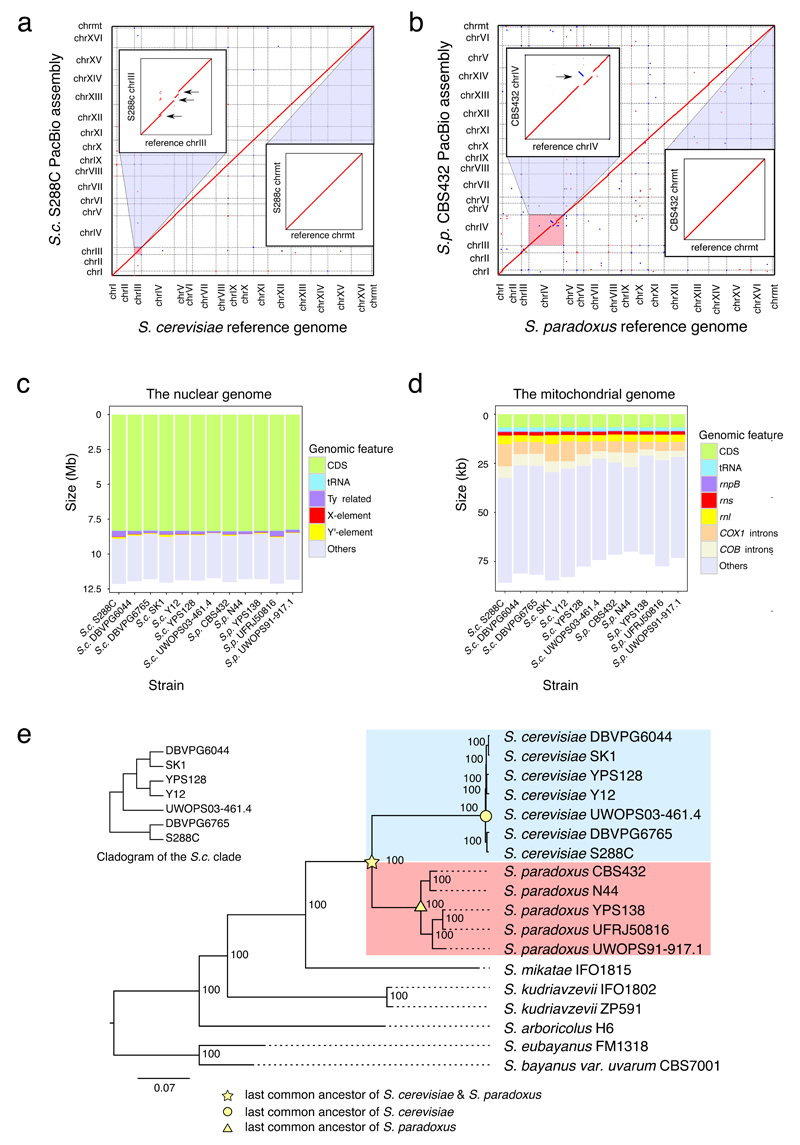
When evaluated against the current S. cerevisiae and S. paradoxus reference genomes, our PacBio assemblies of the same strains (S288C and CBS432 respectively) show clean collinearity for both nuclear and mitochondrial genomes (Figs. 1a-b), with only a few discrepancies at finer scales actually caused by assembly problems in the reference genomes. For example, we found five non-reference Ty1 insertions on chrIII in our S288c assembly (Fig. 1a, inset), which were corroborated by previous studies22–24 as well as our own long-range PCR amplifications. Likewise, we found a mis-assembly on chrIV (Fig. 1b, inset) in the S. paradoxus reference genome, which were confirmed by both Illumina and Sanger reads1. Moreover, we checked several known cases of copy number variants (CNVs) (e.g. Y’-elements25, the CUP1 locus6 and ARR6 gene clusters) and SVs (e.g. those in the Malaysian S. cerevisiae UWOPS03-461.426) and they were all correctly recaptured in our assemblies.
Fig. 1. End-to-end genome assemblies and phylogenetic framework.
(a) Dotplot for the comparison between the S. cerevisiae reference genome (strain S288C; X-axis) and our S288C PacBio assembly (Y-axis). Sequence homology signals were depicted in red (forward match) or blue (reverse match). The two insets show the zoomed-in comparison for chromosome III (chrIII) and the mitochondrial genome (chrmt) respectively. The black arrows indicate three Ty-containing regions (containing five full-length Ty1s) missing in the S. cerevisiae reference genome. (b) Dotplot for the comparison between the S. paradoxus reference genome (strain CBS432; X-axis) and our CBS432 PacBio assembly (Y-axis), color coded as in panel a. The two insets show the zoomed-in comparison for chromosome IV (chrIV) and the mitochondrial genome (chrmt) respectively. The black arrow indicates the mis-assembly on chrIV in the S. paradoxus reference genome. (c-d) The cumulative sequence lengths of different annotated genomic features relative to the overall assembly size of the nuclear (panel c) and mitochondrial genomes (panel d). (e) The phylogenetic relationship of the seven S. cerevisiae strains (highlighted in blue) and five S. paradoxus strains (highlighted in red) sequenced in this study. Six strains from other closely related Saccharomyces sensu stricto species were used as outgroups. All the internal nodes have 100% fast-bootstrap supports. The inset cladogram shows the detailed relationship of the seven S. cerevisiae strains.
The final assembly sizes of these 12 strains range from 11.73 to 12.14 Mb for the nuclear genome (excluding rDNA gaps) and from 69.95 to 85.79 kb for the mitochondrial genome (Fig. 1c-d and Supplementary Tables 8-9). The Ty and Y’-element abundance substantially contributed to the nuclear genome size differences (Fig. 1c and Supplementary Table 8). For example, we observed strain-specific enrichment of full-length Ty1 in S. cerevisiae S288C, Ty4 in S. paradoxus UFRJ50816 and Ty5 in S. paradoxus CBS432, whereas no full-length Ty was found in S. cerevisiae UWOPS03-461.4 (Supplementary Table 6). Similarly, >30 copies of the Y’-element were found in S. cerevisiae SK1 but none in S. paradoxus N44 (Supplementary Table 5). Mitochondrial genome size variation is heavily shaped by the presence/absence dynamics of group I and group II introns in COB1, COX1 and rnl (Fig. 1d and Supplementary Tables 9-10). Despite large-scale interchromosomal rearrangements in a few strains (S. cerevisiae UWOPS03-461.4, S. paradoxus UFRJ50816 and S. paradoxus UWOPS91-917.1), the 12 strains all maintained 16 nuclear chromosomes.
Molecular evolutionary rate and diversification timescale
To gauge structural dynamics in a well-defined evolutionary context, we performed phylogenetic analysis for the 12 strains and six Saccharomyces sensu stricto outgroups based on 4,717 one-to-one orthologs of nuclear protein-coding genes (Supplementary Data Set 1). The resulting phylogeny is consistent with our prior knowledge about these strains (Fig. 1e). Analyzing this phylogenetic tree, we found the entire S. cerevisiae lineage to have evolved faster than the S. paradoxus lineage as indicated by the overall longer branch from the common ancestor of the two species to each tip of the tree (Fig. 1e). We confirmed such rate differences by Tajima’s relative rate test27 for all S. cerevisiae versus S. paradoxus strain pairs, using S. mikatae as the outgroup (P < 1x10-5 for all pairwise comparisons). In contrast, molecular dating analysis reveals that the cumulative diversification time for the five S. paradoxus strains is 3.87-fold of that for the seven S. cerevisiae strains, suggesting a much longer time span for accumulating species-specific genetic changes in the former lineage (Supplementary Fig. 2a). This timescale difference is further supported by the synonymous substitution rate (dS) (Supplementary Fig. 2b).
Core-subtelomere chromosome partitioning
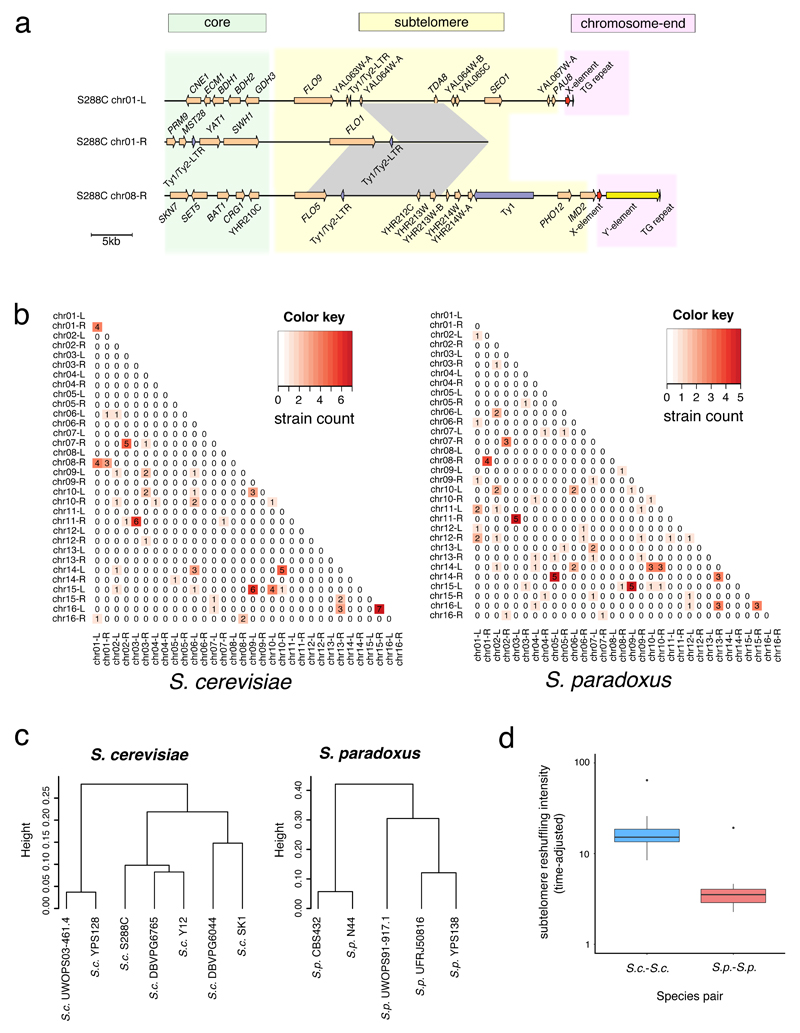
Conceptually, linear nuclear chromosomes can be partitioned into internal chromosomal cores, interstitial subtelomeres and terminal chromosome-ends. However, their precise boundaries are challenging to demarcate without a rigid subtelomere definition. Here, we propose an explicit way to pinpoint yeast subtelomeres based on multi-genome comparison, which can be further applied to other eukaryotic organisms. For each subtelomere, we located its proximal boundary based on the sudden loss of synteny conservation and demarcated its distal boundary by the telomere-associated core X- and Y'-elements (See Methods; Supplementary Fig. 3). The partitioning for the left arm of chrI is illustrated in Fig. 2a. The strict gene synteny conservation is lost after GDH3, thus marking the boundary between the core and the subtelomere for this chromosome arm (Fig. 2a). All chromosomal cores, subtelomeres, and 358 out of 384 chromosome-ends across the 12 strains could be defined in this way (Supplementary Tables 11-13 and Supplementary Data Sets 2-3). For the remaining 26 chromosome-ends, both X/Y'-elements and telomeric repeats (TG1-3) are missing. We assigned the orthology of subtelomeres from different strains based on the ancestral chromosomal identity of their flanking chromosomal cores (see Methods). Here, we used Arabic numbers to denote such ancestral chromosomal identities and the associated subtelomeres, which takes into account the large-scale interchromosomal rearrangements having occurred in some strains (Supplementary Fig. 4 and Supplementary Table 12). Such accurately assigned subtelomere orthology, together with explicit chromosome partitioning, allows for an in-depth examination of subtelomeric evolutionary dynamics.
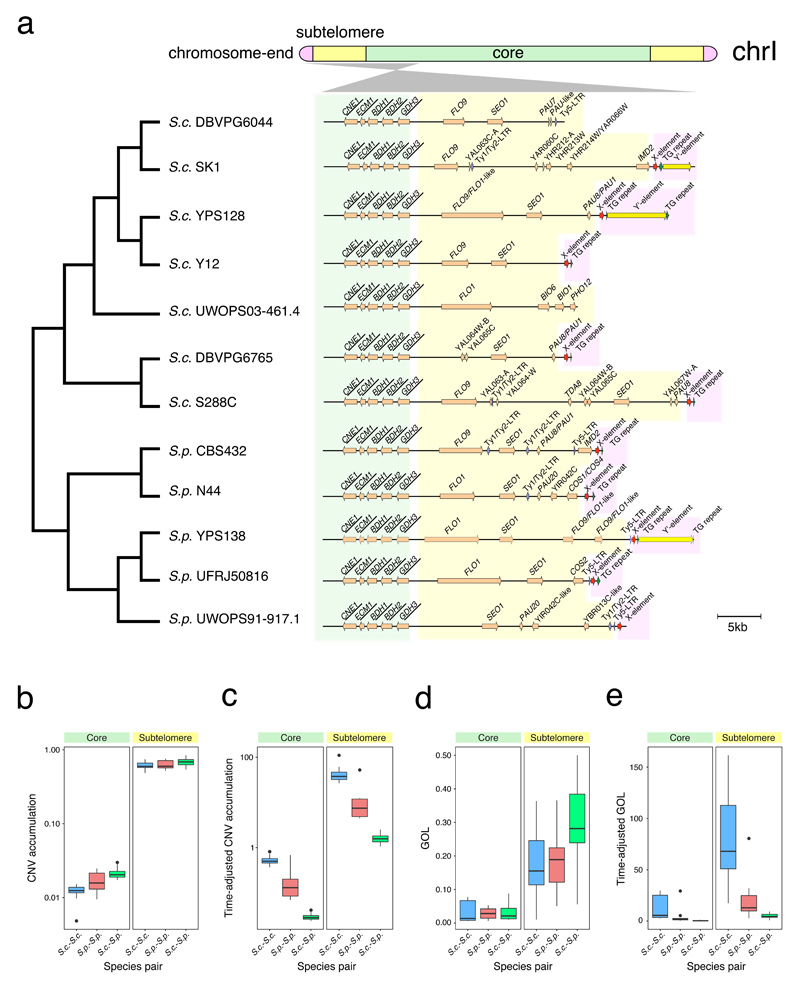
Fig. 2. Explicit nuclear chromosome partitioning.
(a) In this illustrated example, we partitioned the left arm of chromosome I into the core (green), subtelomere (yellow) and chromosome-end (pink) based on synteny conservation and the yeast telomere-associated core X- and Y’-elements. The cladogram (left side) depicts the phylogenetic relationship of the 12 strains, while the gene arrangement map (right side) illustrates the syntenic conservation profile in both the core and subtelomeric regions. The names of genes within the syntenic block were underlined. (b) Proportions of genes involved in copy number variants (CNVs). (c) Proportions of genes involved in CNVs adjusted by the diversification time of the compared strain pair. (d) The gene order loss index (GOL). (e) GOL adjusted by the diversification time of the compared strain pair. The Y-axes in panel b-c are in log-10 scales. In panel b-e, three comparison schemes were examined: within S. cerevisiae (S.c.-S.c.), within S. paradoxus (S.p.-S.p.) and between the two species (S.c.-S.p.). The middle line in the box shows the median value, while the bottom and top lines represent the first and third quartiles. The lengths of the whiskers extend to 1.5 times the interquartile range (IRQ). Data beyond the end of the whiskers are outliers represented by black dots.
Our analysis captures distinct properties of chromosomal cores and subtelomeres. All previously defined essential genes in S. cerevisiae S288C28 fell into the chromosomal cores, whereas all previously described subtelomeric duplication blocks in S288C (see URLs) were fully enclosed in our defined S288C subtelomeres. Furthermore, the genes from our defined subtelomeres show 36.6-fold higher level of CNV accumulation than those from the cores (one-sided Mann–Whitney U test, P < 2.2x10-16) (Figs. 2b-c). When only considering one-to-one orthologs, the subtelomeric genes show 8.4-fold higher level of gene order loss (GOL)29–31 than their core counterparts (one-sided Mann–Whitney U test, P < 2.2x10-16) (Figs. 2d-e). Additionally, subtelomeric one-to-one orthologs also show significantly higher nonsynonymous-to-synonymous substitution rate ratio (dN/dS) than those from the cores in the S. cerevisiae–S. cerevisiae and S. cerevisiae-S. paradoxus comparisons (one-sided Mann–Whitney U test, P < 2.2x10-16), although no clear trend was found in the S. paradoxus-S. paradoxus comparison (one-sided Mann–Whitney U test, P = 0.936). These observations fit well with known properties of cores and subtelomeres and provide the first quantitative assessment of the core-subtelomere contrasts in genome dynamics. Interestingly, aside from such core-subtelomere contrasts, we also observed clear interspecific differences in all three measurements. S. cerevisiae strains show faster CNV accumulation (one-sided Mann–Whitney U test; P = 6.7x10-5 for cores, P = 5.1x10-5 for subtelomeres) and more rapid GOL (one-sided Mann–Whitney U test, P = 5.5x10-5 for cores and P = 2.6x10-5 for subtelomeres) than S. paradoxus strains in both core and subtelomeres respectively (Fig. 2c and 2e). Similarly, S. cerevisiae subtelomeric genes also show higher dN/dS than their S. paradoxus counterparts (one-sided Mann–Whitney U test, P = 4.3x10-4), although their core genes appear to have similar dN/dS (one-sided Mann–Whitney U test, P = 1.000). These observations collectively suggest accelerated evolution in S. cerevisiae relative to S. paradoxus, especially in subtelomeres.
Structural rearrangements in chromosomal cores
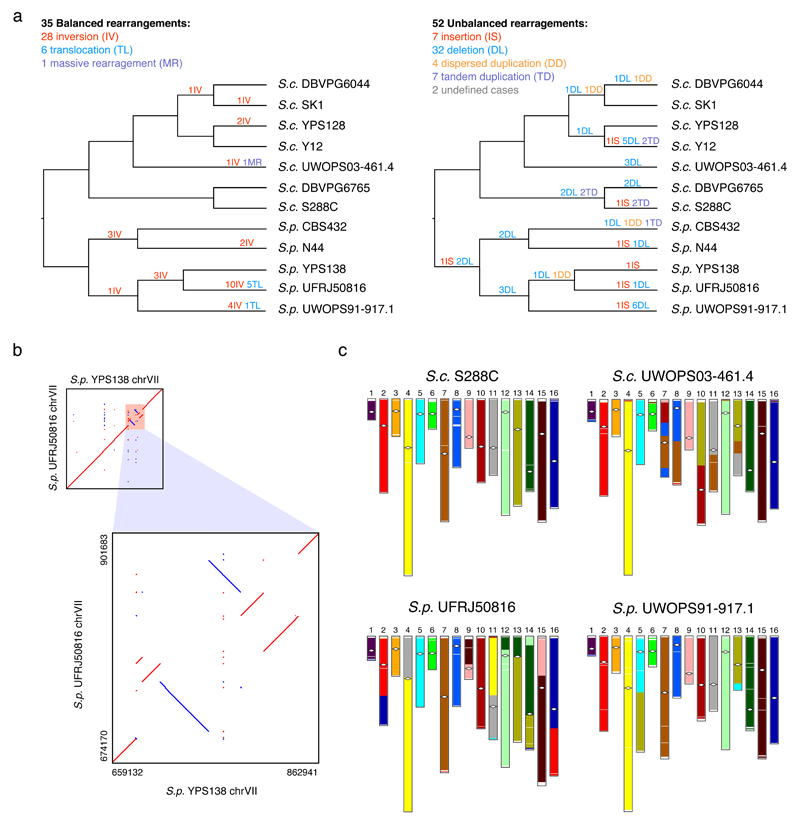
Structural rearrangements can be balanced (e.g. inversions, reciprocal translocations, and transpositions) or unbalanced (e.g. large-scale novel insertions, deletions, and duplications) depending on whether the copy number of genetic material is affected10. We identified 35 balanced rearrangements in total, including 28 inversions, six reciprocal translocations, and one massive rearrangement (Fig. 3a, Supplementary Figs. 5a-c, Supplementary Data Set 4). All events occurred during the species-specific diversification of the two species, with 29 events occurring in S. paradoxus and only six in S. cerevisiae. Factoring in the cumulative evolutionary time difference, S. paradoxus still shows 1.25-fold faster accumulation of balanced rearrangements than S. cerevisiae. Six inversions are tightly packed into a ~200 kb region on chrVII of the South American S. paradoxus UFRJ50816, indicating a strain-specific inversion hotspot (Fig. 3b). With regard to interchromosomal rearrangements, six of them are reciprocal translocations that occurred in two S. paradoxus strains (Fig. 3c and Supplementary Figs. 5a-b). The remaining one found in the Malaysian S. cerevisiae UWOPS03-461.4 is particularly striking: chrVII, chrVIII, chrX, chrXI, and chrXIII were heavily reshuffled, confirming recent chromosomal contact data26 (Fig. 3c and Supplementary Fig. 5c). We describe this as a “massive rearrangement” because it cannot be explained by typical independent reciprocal translocations. This is more likely to result from a single catastrophic event resembling the chromothripsis observed in tumor cells32,33. This massive rearrangement in the Malaysian S. cerevisiae and the rapid accumulation of inversions and translocations in the South American S. paradoxus resulted in extensively altered genome configurations, which explain the reproductive isolation of these two lineages34,35. As previously observed in yeasts on larger divergence scales36,37, the breakpoints of those balanced rearrangements are associated with tRNAs and Tys, highlighting the roles of these elements in triggering genome instability and suggesting non-allelic homologous recombination (NAHR) as the mutational mechanism.
Fig. 3. Structural rearrangements in the nuclear chromosomal cores.
(a) Balanced (left side) and unbalanced (right side) structural rearrangements occurred along the evolutionary history of the 12 strains. (b) The six clustered inversions on chrVII of the South American S. paradoxus UFRJ50816. (c) Genome organization of the strains UWOPS03-461.4, UFRJ50816 and UWOPS91-917.1 relative to that of S288C. The strain S288C is free from large-scale interchromosomal rearrangement, therefore could represent the ancestral genome organization. White diamonds indicate the position of centromeres.
Considering unbalanced structural rearrangements in chromosomal cores, we identified seven novel insertions, 32 deletions, four dispersed duplications and at least seven tandem duplications (Fig. 3a and Supplementary Data Set 5). There are two additional cases of which the evolutionary history cannot be confidently determined due to potentially multiple independent origins or secondary deletions (Supplementary Data Set 5). Although this is a conservative estimate, our identified unbalanced structural rearrangements clearly outnumbered the balanced ones, as recently reported in Lachancea yeasts38. We found that S. cerevisiae accumulated as many unbalanced rearrangements as S. paradoxus despite its much shorter cumulative diversification time. We noticed that the breakpoints of these unbalanced rearrangements (except for tandem duplications) were also frequently associated with Tys and tRNAs, mirroring our observation for balanced rearrangements. Finally, we found genes involved in unbalanced rearrangements to be significantly enriched for gene ontology (GO) terms related to the binding, transporting and detoxification of metal ions (e.g. Na+, K+, Cd2+ and Cu2+) (Supplementary Table 14), hinting that these events likely are adaptive.
Structural evolutionary dynamics of subtelomeres
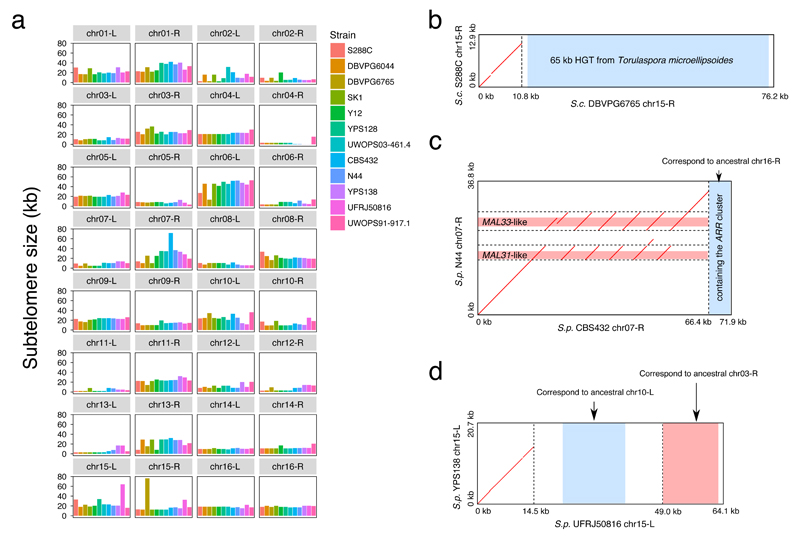
The complete assemblies and well-defined subtelomere boundaries enabled us to examine subtelomeric regions with unprecedented resolution. We found both the size and gene content of the subtelomere to be highly variable across different strains and chromosome arms (Fig. 4a and Supplementary Data Set 3). The subtelomere size ranges from 0.13 to 76 kb (median = 15.6 kb) while the number of genes enclosed in each subtelomere varies between 0 and 19 (median = 4) and the total number of subtelomeric genes varied between 134-169 (median = 146) per strain. While the very short subtelomeres (e.g. chr04-R and chr11-L) can be explained by an unexpected high degree of synteny conservation extending all the way to the end, some exceptionally long subtelomeres are instead the products of multiple mechanisms. For example, the chr15-R subtelomere of S. cerevisiae DBVPG6765 has been drastically elongated by a 65 kb horizontal gene transfer (HGT)39 (Fig. 4b and Supplementary Fig. 6a). The chr07-R subtelomere of S. paradoxus CBS432 was extended by a series of tandem duplications of MAL31-like and MAL33-like genes, as well as the addition of the ARR cluster (Fig. 4c and Supplementary Fig. 6b). The chr15-L subtelomere of S. paradoxus UFRJ50816 increased size by duplications of subtelomeric segments from two other chromosomes (Fig. 4d and Supplementary Fig. 6c). Inversions have also occurred in subtelomeres, including one affecting the HMRA1-HMRA2 locus in UFRJ50816 and another affecting a MAL11-like gene in CBS432 (Supplementary Fig. 7).
Fig. 4. Subtelomere size plasticity and structural rearrangements.
(a) Size variation of the 32 orthologous subtelomeres across the 12 strains. (b) Dotplot for the chr15-R subtelomere comparison between S. cerevisiae DBVPG6765 and S288C. The extended DBVPG6765 chr15-R subtelomere is explained by a previously reported eukaryote-to-eukaryote horizontal gene transfer (HGT) event. (c) Dotplot for the chr07-R subtelomere comparison between S. paradoxus CBS432 and N44. The chr07-R subtelomere expansion in CBS432 is explained by a series of tandem duplications of the MAL31-like and MAL33-like genes and an addition of the ARR-containing segment from the ancestral chr16-R subtelomere. (d) Dotplot for the chr15-L subtelomere comparison between S. paradoxus UFRJ50816 and YPS138. The expanded chr15-L subtelomere in UFRJ50816 is explained by the relocated subtelomeric segments from the ancestral chr10-L and chr03-R subtelomeres. Please note that the region coordinates in panel (b)-(d) are based on the defined subtelomeres rather than the full chromosomes.
The enrichment of segmental duplication blocks occurring via ectopic sequence reshuffling is a common feature of eukaryotic subtelomeres, however, incomplete genome assemblies have prevented population-level quantitative analysis of this phenomenon. Here, we identified subtelomeric duplication blocks based on pairwise comparisons of different subtelomeres within the same strain (Fig. 5a and Supplementary Data Set 6). In total, we identified 173 pairs of subtelomeric duplication blocks across the 12 strains, with 8-26 pairs for each strain (Supplementary Table 15). Among the 16 pairs of subtelomeric duplication blocks previously identified in S288C (mentioned above), all the 12 larger pairs passed our filtering criteria. Interestingly, the Hawaiian S. paradoxus UWOPS91-917.1 has the most subtelomeric duplication blocks and half of these are strain-specific, suggesting unique subtelomere evolution in this strain. The duplicated segments always maintained the same centromere-telomere orientation, supporting a mutational mechanism of double-strand break (DSB) repair as previously suggested in other species40,41. We further summarized those 173 pairs of duplication blocks based on the orthologous subtelomeres involved. This led to 75 unique duplicated subtelomere pairs, 59 (78.7%) of which are new compared to what was previously identified in S288C (Supplementary Data Set 7). We found 31 (41.3%) of these unique pairs to be shared between strains, or even between species with highly dynamic strain-sharing patterns (Fig. 5b and Supplementary Fig. 8a). Most (87.1%) of this sharing pattern could not be explained by the strain phylogeny (Supplementary Data Set 7). This suggests a constant gain and loss process of subtelomeric duplications throughout evolutionary history.
Fig. 5. Evolutionary dynamics of subtelomeric duplications.
(a) An example of subtelomeric duplication blocks shared among the chr01-L, chr01-R and chr08-R subtelomeres in S. cerevisiae S288C. The grey blocks denote their shared homologous regions with >= 90% sequence identity. (b) Subtelomeric duplication signals shared across the seven S. cerevisiae strains (left) and the five S. paradoxus strains (right). For each specific subtelomere pair, the number of strains showing strong sequence homology (BLAT score >= 5000 and identity >= 90%) was indicated in the heatmap. (c) Hierarchical clustering based on the proportion of conserved orthologous subtelomeres in cross-strain comparisons within S. cerevisiae and within S. paradoxus respectively. (d) Subtelomere reshuffling intensities within S. cerevisiae (S.c.-S.c.) and within S. paradoxus (S.p.-S.p.), which are adjusted by the diversification time of the compared strain pair. The Y-axis is in log-10 scale. The middle line in the box shows the median value, while the bottom and top lines represent the first and third quartiles. The lengths of the whiskers extend to 1.5 times the interquartile range (IRQ). Data beyond the end of the whiskers are outliers represented by black dots.
Given the rampant subtelomere reshuffling, we investigated to what extent the similarity in orthologous subtelomere composition reflects the intra-species phylogenies. We measured the proportion of conserved orthologous subtelomeres in all strain pairs within the same species and performed hierarchical clustering accordingly (Fig. 5c). While the clustering in S. paradoxus correctly recapitulated the true phylogeny, the clustering in S. cerevisiae revealed a quite different topology, with only the relationship of the most recently diversified strain pair (DBVPG6044 vs. SK1) being correctly recovered. Interestingly, the distantly related Wine/European (DBVPG6765) and Sake (Y12) S. cerevisiae strains were clustered together, suggesting possible convergent subtelomere evolution during their respective domestication for alcoholic beverage production. The proportion of conserved orthologous subtelomeres between S. cerevisiae strains (56.3%-81.3%) is comparable to that between S. paradoxus strains (50.0%-81.3%), despite the much smaller diversification timescales of S. cerevisiae. This translates into a 3.8-fold difference in subtelomeric reshuffling intensity between the two species during their respective diversifications (one-sided Mann–Whitney U test, P = 2.93x10-8) (Fig. 5d). The frequent reshuffling of subtelomeric sequences often has drastic impacts on gene content both qualitatively and quantitatively. For example, four genes (PAU3, ADH7, RDS1, and AAD3) were lost in S. cerevisiae Y12 due to a single chr08-L to chr03-R subtelomeric duplication event (Supplementary Fig. 8b). Therefore, the accelerated subtelomere reshuffling in S. cerevisiae is likely to have important functional implications.
Native non-canonical chromosome-end structures
S. cerevisiae chromosome-ends are characterized by two telomere associated sequences: the core X- and the Y'-element42. The core X-element is present in nearly all chromosome-ends, whereas the number of Y'-element varies across chromosome-ends and strains. The two previously described chromosome-end structures are (1) with a single core X-element and (2) with a single core X-element followed by 1-4 distal Y'-elements42. S. paradoxus chromosome-ends also contain core X- and Y’-elements43, but their detailed structures and genome-wide distributions have not been systematically characterized. Across our 12 strains, most (~85%) chromosome-ends have one of the two structures described above but we also discovered several novel chromosome-ends (Supplementary Table 13). We found several examples of tandem duplications of the core X-element in both species. In most cases, including the ones in the S. cerevisiae reference genome (chrVIII-L and chrXVI-R), the proximal duplicated core X-elements were degenerated. Nevertheless, we found two examples where intact duplicated copies were retained: the chrXII-R in S. cerevisiae Y12 and the chrIII-L in S. paradoxus CBS432. The latter case is especially striking, with six core X-elements (including three complete copies) tandemly arranged. Surprisingly, we discovered five chromosome-ends consisting of only Y'-elements (one or more copies) but no core X-elements. This is unexpected given the importance of core X-elements in maintaining genome stability44,45. The discovery of these non-canonical chromosome-end structures offers a new paradigm to investigate the functional role of core X-elements.
Mitochondrial genome evolution
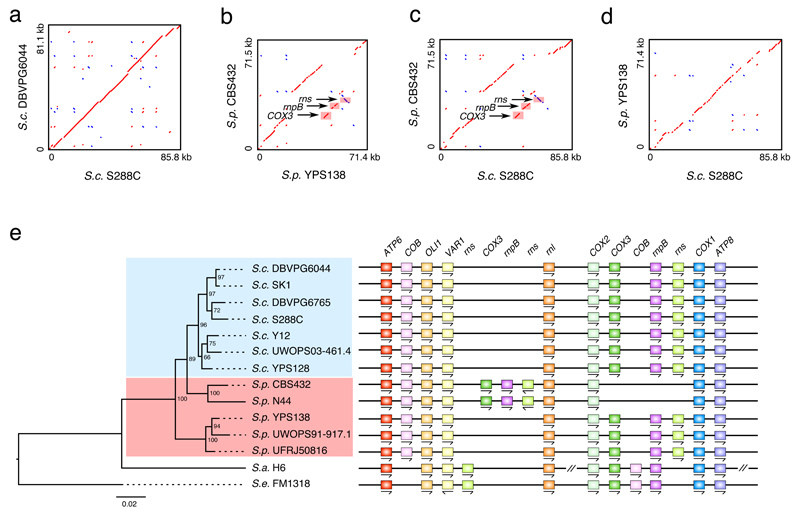
Despite being highly repetitive and AT-rich, we found the mitochondrial genomes of all S. cerevisiae strains show high degrees of collinearity (Fig. 6a). In contrast, S. paradoxus mitochondrial genomes show lineage-specific structural rearrangements. The two Eurasian strains (CBS432 and N44) share a transposition of the entire COX3-rnpB-rns segment, in which rns was further inverted (Fig. 6b-d). In addition, given the gene order in two outgroups, the COB gene was relocated in the S. cerevisiae-S. paradoxus common ancestor (Fig. 6e). The phylogenetic tree inferred from mitochondrial protein-coding genes show clear deviation from the nuclear tree (Fig. 6e). In particular, the Eurasian S. paradoxus lineage (CBS432 and N44) clustered together with the seven S. cerevisiae strains before joining with the other S. paradoxus strains, which supports the idea of mitochondrial introgression from S. cerevisiae46 (Fig. 6e). We found low topology consensus (normalized quartet score = 0.59 versus 0.92 for the nuclear gene tree) across different mitochondrial gene loci, suggesting heterogeneous phylogenetic histories. Together with the drastically dynamic presence/absence patterns of mitochondrial group I and group II introns (Supplementary Table 10), this reinforces the argument for extensive cross-strain recombination in yeast mitochondrial evolution47. In addition, we noticed that the COX3 gene in S. paradoxus UFRJ50816 and UWOPS91-917.1 started with GTG rather than the typical ATG start codon, which was further supported by Illumina reads. This suggests either an adoption of an alternative ATG start codon nearby (e.g. 45 bp downstream) or a rare case of near-cognate start codon48–50.
Fig. 6. Comparative mitochondrial genomics.
(a) Pairwise comparison for the mitochondrial genome of S288C and DBVPG6044 from S. cerevisiae. (b) Pairwise comparison for the mitochondrial genome of CBS432 and YPS138 from S. paradoxus. (c) Pairwise comparison for the mitochondrial genome of S. cerevisiae S288C and S. paradoxus CBS432. (d) Pairwise comparison for the mitochondrial genome of S. cerevisiae S288C and S. paradoxus YPS138. (e) Genomic arrangement of the mitochondrial protein-coding genes and RNA genes across the 12 sampled strains. The phylogenetic tree shown on the left is constructed based on mitochondrial protein-coding genes, with the number at each internal node showing rapid bootstrap support. The detailed gene arrangement map is shown on the right. Please note that there is a large inversion in S. arboricolus encompassing the entire COX2-ATP8 segment based on its original mitochondrial genome assembly, and here we inverted back this segment for better visualization.
Fully-resolved SVs illuminate complex phenotypic traits
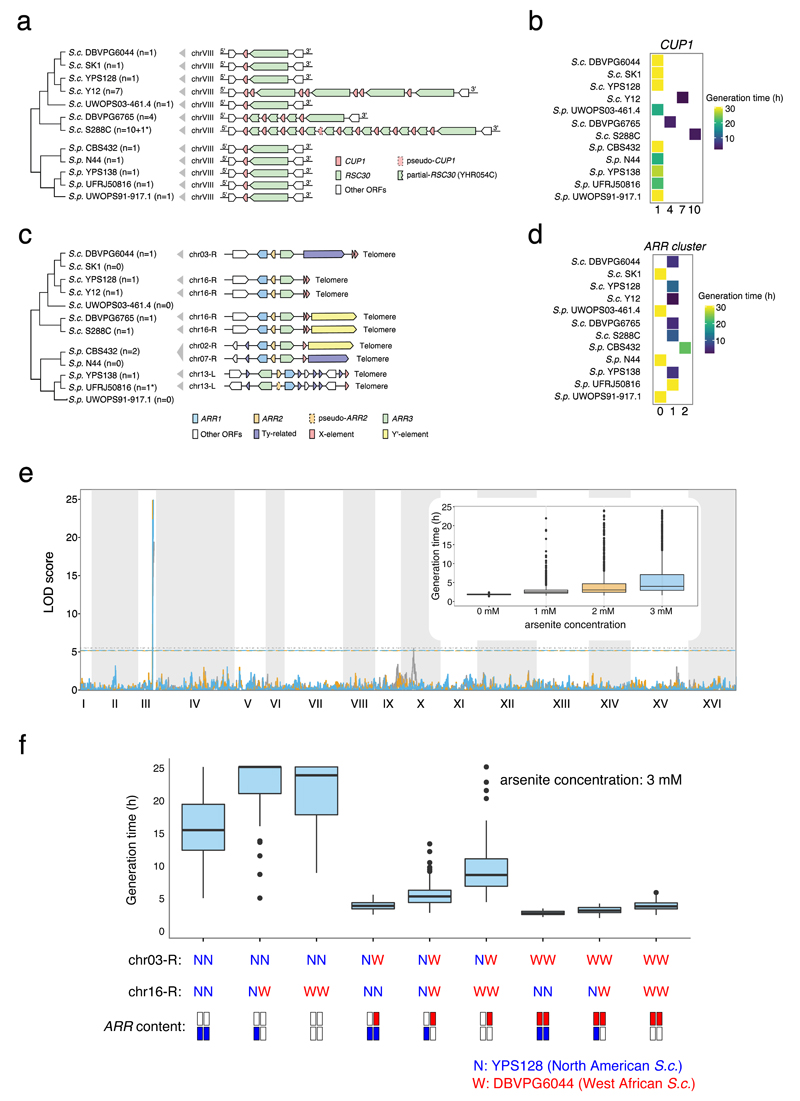
SVs are expected to account for a substantial fraction of phenotypic variation, therefore fully resolved SVs can be crucial in understanding complex phenotypic traits. Here, we used the copper-tolerance related CUP1 locus and the arsenic-tolerance related ARR cluster as two examples of associations between fully-characterized genomic compositions (i.e. copy numbers and genotypes) and conditional growth rates. The PacBio assemblies precisely resolve these complex loci and phenotype associations are consistent with previous findings based on copy number analysis6,21,51 (Fig. 7a-d and Supplementary Note). We further illustrated their phenotypic contributions via linkage mapping using 826 phased outbred lines (POLs) derived from crossing the North American (YPS128) and West African (DBVPG6044) S. cerevisiae52 (see Methods). The linkage analysis accurately mapped a large-effect quantitative trait locus (QTL) at the chr03-R subtelomere (the location of ARRs in DBVPG6044), but showed no arsenic resistance association with the YPS128 ARRs on the chr16-R subtelomere (Fig. 7e). This profile is consistent with the relocation of an active ARR cluster to the chr03-R subtelomere in DBVPG6044 and the presence of deleterious mutations predicted to inactivate the ARR cluster in YPS1286,35. Thus, a full understanding of the relationship between genome sequence and arsenic resistance phenotype is not provided by the knowledge of copy number alone, but rather requires the combined knowledge of genotype, genomic location, and copy number as provided by our end-to-end assemblies (Fig. 7f).
Fig. 7. Structural rearrangements illuminate complex phenotypic variation.
(a) Copy number and gene arrangement of the CUP1 locus across the 12 strains. The asterisk denotes the involvement of pseudogenes. (b) Generation time of the 12 strains in high copper condition (c) Copy number and gene arrangement of the ARR cluster. The asterisk denotes the involvement of pseudogenes. The subtelomere location of the ARR cluster is highly variable. (d) Generation time of the 12 strains in high arsenic condition. (e) The rearrangement that relocates the ARR cluster to the chr03-R subtelomere in the West African S. cerevisiae DBVPG6044 is consistent with the linkage mapping analysis using phased outbred lines (POLs) derived from the North American (YPS128) and West African (DBVPG6044) S. cerevisiae. (f) Phenotypic distribution of the 826 POLs for generation time in arsenic condition partitioned for genotype positions at the chr03-R and chr16-R subtelomeres and inferred copies of ARR clusters (underneath the plot). The middle line in the box shows the median value, while the bottom and top lines represent the first and third quartiles. The lengths of the whiskers extend to 1.5 times the interquartile range (IRQ). Data beyond the end of the whiskers are outliers represented by black dots.
Discussion
The landscape of genetic variation is shaped by multiple evolutionary processes, including mutation, drift, recombination, gene flow, natural selection and demographic history. The combined effect of these factors can vary considerably both across the genome and between species, resulting in different patterns of evolutionary dynamics. The complete genome assemblies that we generated for multiple strains from both domesticated and wild yeasts provide a unique dataset for exploring such patterns with unprecedented resolution.
Considering the evolutionary dynamics across the genome, eukaryotic subtelomeres are exceptionally variable compared to chromosomal cores40,53,54, with accelerated evolution manifested by extensive CNV accumulation, rampant ectopic reshuffling, and rapid functional divergence6,41,55–57. Our study provides the first quantitative comparison between subtelomeres and cores in structural genome evolution and a high-resolution view of the extreme evolutionary plasticity of subtelomeres. This rapid evolution of subtelomeres can substantially alter the gene repertoire and generate novel recombinants with adaptive potential57. Given that subtelomeric genes are highly enriched in functions mediating interactions with external environments (e.g. stress response, nutrient uptake, and ion transport)6,55,58, it is tempting to speculate that the accelerated subtelomeric evolution reflects selection for evolvability, i.e. the ability to respond and adapt to changing environments59.
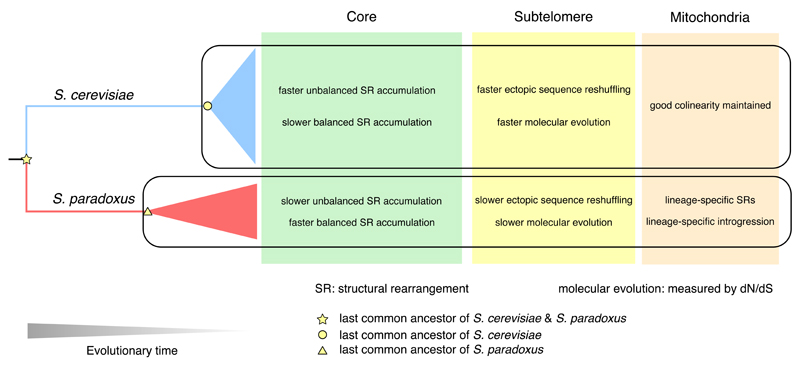
With regard to the genome dynamics between species, external factors such as selection and demographic history play important roles. The ecological niches and recent evolutionary history of S. cerevisiae have been intimately associated with human activities, with many strains isolated from human-associated environments, like breweries, bakeries and even clinical patients60. Consequently, this wide-spectrum of selection schemes could significantly shape the genome evolution of S. cerevisiae. In addition, human activities also promoted admixture and crossbreeding of S. cerevisiae strains from different geographical locations and ecological niches61, resulting in many mosaic strains with mixed genetic backgrounds1. In contrast, the wild-living S. paradoxus occupies very limited ecological niches, with most strains isolated from trees in the Quercus genus62. S. paradoxus strains from different geographical subpopulations are genetically well-differentiated with partial reproductive isolations34,63. Such interspecific differences in their life history could result in distinct evolutionary genome dynamics, which is captured in our study (Fig. 8). In chromosomal cores, S. cerevisiae strains show slower accumulation of balanced structural rearrangements compared with S. paradoxus strains. This pattern might be explained by the admixture between different S. cerevisiae subpopulations during their recent association with human activities, which would considerably impede the fixation of balanced structural rearrangements. In contrast, geographical isolation of different S. paradoxus subpopulations would favor relatively fast fixation of balanced structural rearrangements64. We observed an opposite pattern for unbalanced rearrangements in chromosomal cores. The S. cerevisiae strains accumulate such changes more rapidly than their S. paradoxus counterparts, which is likely shaped by selection considering the biological functions of those affected genes. Likewise, the more rapid subtelomeric reshuffling and higher dN/dS of subtelomeric genes in S. cerevisiae than in S. paradoxus are probably also driven by selection. As a consequence of such unbalanced rearrangements and subtelomeric reshuffling, we observed more rapid CNV accumulation and GOL in S. cerevisiae strains, which reinforce this argument. In addition, the mitochondrial genomes of S. cerevisiae strains maintained high degrees of collinearity, whereas those of S. paradoxus strains showed lineage-specific structural rearrangements and introgression, suggesting distinct modes of mitochondrial evolution. Taken together, many of these observed differences between S. cerevisiae and S. paradoxus likely reflect the influence of human activities on structural genome evolution, which shed new light on why S. cerevisiae, but not its wild relative, is one of our most biotechnologically important organisms.
Fig. 8. Contrasting evolutionary dynamics across the entire genomic landscape between S. cerevisiae and S. paradoxus.
The interspecific contrasts in nuclear chromosomal cores, subtelomeres and mitochondrial genomes were summarized respectively.
Online Methods
Strain sampling, preparation, and DNA extraction
Based on previous population genomics surveys1, we sampled seven S. cerevisiae and five S. paradoxus strains (all in the haploid or homozygous diploid forms) to represent major evolutionary lineages of the two species (Supplementary Table1). The reference strains for S. cerevisiae (S288C) and S. paradoxus (CBS432) were included for quality control. All strains were taken from our strain collection stored at -80°C and cultured on YPD plates. A single colony for each strain was picked and cultured in 5 mL YPD liquid at 30°C 220 rpm overnight. The DNA extraction was carried out using the MasterPure™ Yeast DNA Purification Kit (Epicentre, USA).
PacBio sequencing and raw assembly
The sequencing center at the Wellcome Trust Sanger Institute (Cambridge, UK) performed library preparation and sequencing using the PacBio Single Molecule, Real-Time (SMRT) DNA sequencing technology (platform: PacBio RS II; chemistry: P4-C2 for the pilot phase and P6-C4 for the main phase). The raw reads were processed using the standard SMRT analysis pipeline (v2.3.0). The de novo assembly was carried out following the hierarchical genome-assembly process (HGAP) assembly protocol with Quiver polishing65.
Assembly evaluation and manual refinement
We retrieved the reference genomes (Supplementary Note) for both species to assess the quality of our PacBio assemblies. For each polished PacBio assembly, we first used RepeatMasker (v4.0.5) (see URLs) to soft-mask repetitive regions (option: -species fungi -xsmall -gff). The soft-masked assemblies were subsequently aligned to the reference genomes using the nucmer program from MUMmer (v3.23)66 for chromosome assignment. For most chromosomes, we have single contigs covering the entire chromosomes. For the cases where internal assembly gaps occurred, we performed manual gap closing by consulting the assemblies generated in the pilot phase of this project. The only gap that we were unable to close is the highly repetitive rDNA array (usually consisting 100-200 copies of 9.1 kb unit) on chrXII. The S. cerevisiae reference genome used a 17,357 bp sequence of two tandemly arranged rDNA copies to represent this complex region. For our assemblies, we trimmed off the partially assembled rDNAs around this gap and re-linked the two contigs with 17,357 bp Ns to keep consistency. The mitochondrial genomes of the 12 strains were recovered by single contigs in the raw HGAP assemblies. We further circularized them and reset their starting position as the ATP6 gene using Circlator (v1.1.4)67. The circularized mitochondrial genome assemblies were further checked by consulting the raw PacBio reads and manual adjustment was applied when necessary.
Illumina sequencing, reads mapping, and error correction
In addition to the PacBio sequencing, we also performed Illumina 151 bp paired-end sequencing for each strain at Institut Curie (Paris, France). We examined the raw Illumina reads via FastQC (v0.11.3) (see URLs) and performed adapter-removing and quality-based trimming by trimmomatic (v0.33)68 (options: ILLUMINACLIP:adapters.fa:2:30:10 SLIDINGWINDOW:5:20 MINLEN:36). For each strain, the trimmed reads were mapped to the corresponding PacBio assemblies by BWA (v0.7.12)69. The resulting reads alignments were subsequently processed by SAMTools (v1.2)70, Picard tools (v1.131) (see URLs) and GATK (v3.5-0)71. Based on such Illumina reads alignments, we further performed error-correction with Pilon (v1.12)72 to generate final assemblies for our downstream analysis.
Base-level error rate estimation for the final PacBio assemblies
Eight of our 12 strains have previously been sequenced using Illumina technology with moderate-to-high depths6. We retrieved those raw reads and mapped them to our PacBio assemblies (both before and after the Pilon correction) following the same protocol described above. The SNPs and Indels were called by FreeBayes (v1.0.1-2)73 (option: -p 1) to assess the performance of the Pilon correction and estimate the remaining base-level error rate in our final assemblies. The raw SNP and Indel calls were filtered by the vcffilter tool from vcflib (see URLs) with the filter expression: “QUAL > 30 & QUAL / AO > 10 & SAF > 0 & SAR > 0 & RPR > 1 & RPL > 1”.
Assembly completeness evaluation
We compared our S288C PacBio assembly with three published S. cerevisiae assemblies generated by different sequencing technologies (i.e. PacBio, Oxford Nanopore and Illumina)74,75. We aligned these three assemblies as well as our S288C PacBio assembly to the S. cerevisiae reference genome using nucmer from MUMmer (v3.23)66. The nucmer alignments were filtered by delta-filter (from the same package) (option: -1). We converted the output file to the “BED” format and used bedtools (v2.15.0)76 to calculate the intersection between our genome alignment and various annotation features (e.g. chromosomes, genes, retrotransposable elements, telomeres, etc) of the S. cerevisiae nuclear reference genome. The percent coverages of these annotation features by different assemblies were summarized accordingly.
Annotation of the protein-coding genes, tRNA genes, and other genomic features
For nuclear genomes, we assembled an integrative pipeline that combines three existing annotation tools to form an evidence-leveraged protein-coding gene annotation. First, we used the RATT package77 for directly transferring the non-dubious S. cerevisiae reference gene annotations to our PacBio assemblies based on whole genome alignments. Furthermore, we used the Yeast Genome Annotation Pipeline (YGAP)78 to annotate our PacBio assemblies (default options without scaffolds reordering) based on gene sequence homology and synteny conservation. A custom Perl script was used to remove redundant, truncated, or frameshifted genes annotated by YGAP. Finally, we used the Maker pipeline (v2.31.8)79 to perform de novo gene discovery with EST/protein alignment support (Supplementary Note). As a by-product, tRNA genes were also annotated via the tRNAscan-SE (v1.3.1)80 module of the Maker pipeline. The gene annotations produced by RATT, YGAP, and Maker together with the EST/protein alignment evidences generated by Maker were further leveraged by EVidenceModeler (EVM)81 to form an integrative annotation. Manual curation was carried out for selected cases (e.g. the CUP1 and ARR clusters) and pseudogenes were manually labeled when verified. The same pipeline was used for upgrading the protein-coding gene annotation of S. arboricolus, for which the originally annotated coding sequences (CDSs) and protein sequences was used for initial EST/protein alignment. In addition, for the 12 strains, we systematically annotated other genomic features encoded in their nuclear genomes, such as centromeres, Ty retrotransposable elements, and telomere-associated core X- and Y’-elements (Supplementary Note). Protein-coding genes that overlap with truncated/full-length Tys, core X- or Y’-elements were removed from our final annotation.
As for mitochondrial genomes, the protein-coding genes, tRNA genes and other mitochondrial RNA genes such as RNase P RNA (rnpB), small (rns) and large (rnl) subunit rRNA were annotated by MFannot (see URLs). The exon-intron boundaries of annotated mitochondrial genes were manually curated based on BLAST and the 12-way mitochondrial genome alignment generated by mVISTA82.
Orthology group identification
For nuclear protein-coding genes, we used Proteinortho (v5.15)83 to identify gene orthology across the 12 strains and six other sensu stricto outgroups: S. mikatae (strain IFO1815), S. kudriavzevii (strain IFO1802), S. kudriavzevii (strain ZP591), S. arboricolus (strain H6), S. eubayanus (strain FM1318) and S. bayanus var. uvarum (strain CBS7001). The orthology identification took into account both sequence homology and synteny conservation (the PoFF feature84 of Proteinortho). For each annotated strain, the systematic names of non-dubious genes in the Saccharomyces Genome Database (SGD) (see URLs) were mapped to our annotated genes based on the orthology groups identified above.
Phylogenetic reconstruction
For nuclear genes, we performed the phylogenetic analysis based on those one-to-one orthologs that are shared across all 18 strains (seven S. cerevisiae + five S. paradoxus + six outgroups) using two complementary approaches: the concatenated tree approach and the consensus tree approach. For each one-to-one ortholog, we used MUSCLE (v3.8.1551)85 to align protein sequences and used PAL2NAL (v14)86 to align codons accordingly. For the concatenated tree approach, we generated a concatenated codon alignment across all orthology groups and fed it into RAxML (v8.2.6)87 for maximum likelihood (ML) tree building. Alignment partition was configured by the first, second, and third codon positions. The GTRGAMMA model was used for phylogenetic inference. The rapid bootstrapping method built in RAxML was used to assess the stability of internal nodes (option: -# 100). The final ML tree was visualized in FigTree (v1.4.2) (see URLs). For the consensus tree approach, we built individual gene trees with RAxML using the same method described above, which were further summarized into a coalescent-based consensus species tree by ASTRAL (v4.7.12)88. The normalized quartet score was calculated to assess the reliability of the final species tree given individual gene trees. For mitochondrial genes, we performed the same phylogenetic analysis based on the eight mitochondrial protein-coding genes.
Relative rate test
To test the rate heterogeneity between S. cerevisiae and S. paradoxus in molecular evolution, we constructed three-way sequence alignments by sampling one strain for each species together with S. mikatae as the outgroup. The sequences were drawn from the concatenated nuclear CDS alignment described above. The extracted sequences were fed into MEGA (v7.0.16)89 for Tajima’s relative rate test27. We conducted this test for all possible S. cerevisiae versus S. paradoxus strain pairs.
Molecular dating
Since no yeast fossil record can be used for reliable calibration, we performed the molecular dating analysis based on a relative time scale. We used the phylogenetic tree constructed from the nuclear one-to-one orthologs as the input and performed least-square based fast dating with LSD90 (options: -c -v -s). We specified S. bayanus var. uvarum CBS7001 and S. eubayanus FM1318 as outgroups for this analysis.
Conserved synteny block identification
We used SynChro from the CHROnicle package (version: January 2015)91,92 to identify conserved synteny blocks. We prepared the input files for SynChro with custom Perl scripts to provide the genomic coordinates of all annotated features together with the genome assembly and proteome sequences. SynChro subsequently performed exhaustive pairwise comparisons to identify synteny blocks shared in the given strain pair.
Subtelomere definition and chromosome partitioning
An often-used yeast subtelomere definition is 20-30 kb from the chromosome-ends. However, this definition is arbitrary in the sense that it treats all subtelomeres indiscriminately. In this study, we defined yeast subtelomeres based on gene synteny conservation profiles across the 12 strains. For each chromosome arm, we examined all syntenic blocks shared across the 12 strains and used the most distal one to define the distal boundary for the chromosomal core (Supplementary Table 11). Meanwhile, we defined the proximal boundary of the chromosome-end for this chromosome arm based on the first occurrence of core X- or Y’-elements. The region between these two boundaries was defined as the subtelomere for this chromosome arm with 400 bp interstitial transition zones on both sides (Supplementary Fig. 3).
Given that some strains (i.e. UWOPS03-461.4, UFRJ50816, and UWOPS91-917.1) are involved in large-scale interchromosomal rearrangements, the current chromosomal identities (determined by centromeres) might not necessarily agree with the ancestral chromosomal identities (determined by gene contents). Therefore, we used Roman and Arabic numbers to respectively denote these two identities for all 12 strains to avoid potential confusion when it comes to those interchromosomal rearrangements (Supplementary Fig. 4 and Supplementary Table 12). Each defined subtelomere was named according to the ancestral chromosomal identity of its flanking chromosomal core and denoted also using Arabic numbers (Supplementary Data Sets 2-3).
Identification of balanced and unbalanced structural rearrangements in chromosomal cores
To identify balanced rearrangements, we first used ReChro from CHROnicle (version: January 2015)91,92. We set the synteny block stringency parameter “delta=1” for the main analysis. A complementary run was performed with “delta=0” to identify single gene inversions. Alternatively, we started with the one-to-one ortholog gene pairs (identified by our orthology group identification) in chromosomal cores between any given strain pair and examined their relative orientation and chromosomal locations. If the two one-to-one orthologous genes locate on the same chromosome but with opposite orientations, an inversion should be involved. If they reside on different chromosomes, a translocation or transposition should be involved.
As for unbalanced rearrangements, we first generated whole genome alignment for every strain pair by nucmer66 (options: -maxmatch -c 500) and used Assemblytics93 to identify potential insertions, deletions and duplications/contractions. All candidates were further intersected with our gene annotations by bedtools intersect76 to only keep those encompassing at least one protein-coding gene. Alternatively, we started with all the genes enclosed in chromosomal cores of any given strain pair and filtered out those completely covered by unique genome alignment between this strain pair. All the remaining genes were classified as candidates potentially involved in unbalanced rearrangements.
All identified candidate cases were manually examined by dotplots using Gepard (v1.30)94. All verified rearrangements in chromosomal cores were further mapped to the phylogeny of the 12 strains to reconstruct their evolutionary histories based on the maximum parsimony principle. The corresponding genomic regions in those six outgroups were also checked by dotplots to provide further support for our evolutionary history inferences.
Gene ontology analysis
The CDSs of the S. cerevisiae non-dubious reference genes were BLAST against the NCBI non-redundant (nr) database using blastx (E-value = 1x10-3) and further annotated by BLAST2GO (v.3.2)95,96 to generate gene ontology (GO) mapping for each gene. We performed Fisher’s exact test97 to detect significantly enriched GO terms of our test gene set relative to the genome-wide background. False discovery rate (FDR) (cutoff: 0.05)98 was used for multiple correction. The significantly enriched GO terms were further processed by the “Reduce to most specific terms” function implemented in BLAST2GO to only keep child terms.
Molecular evolutionary rates, CNV accumulation, and GOL estimation
For the one-to-one orthologs in each strain pair, we calculate synonymous substitution rate (dS), nonsynonymous substitution rate (dN) and nonsynonymous-to-synonymous substitution rate ratio (dN/dS) using the yn00 program from PAML (v4.8a)99 based on Yang & Nielsen (2000) model100. We also measured the proportion of genes involved in CNVs (i.e. those are not one-to-one orthologs) in any strain pair. We denoted this measurement as PCNVs, a quantity analogous to the P-distance in sequence comparison. To correct for multiple changes at the same gene loci, the Poisson distance DCNVs can be given by −ln (1 − PCNVS). This value can be further adjusted with evolutionary time by dividing 2T, where T is the diversification time of the two compared strains obtained from our molecular dating analysis. To further capture evolutionary dynamics in terms of gene order changes, we further measured gene order loss (GOL) for those one-to-one orthologs using the method proposed by previously studies without allowing for intervening genes29–31. For GOL, we performed similar Poisson correction and evolutionary time adjustment as we did for CNV accumulation. The calculation values for dN/dS, CNV accumulation, and GOL were further summarized by “core genes” and “subtelomeric genes” based on our genome partitioning described above.
Subtelomeric homology search
For each defined subtelomeric region, we hard-masked all the enclosed Ty-related features (i.e. full-length Ty, truncated Ty and Ty solo-LTRs) and then searched against all the other subtelomeric regions for shared sequence homology. The search was performed by BLAT101 (options: -noHead -stepSize=5 -repMatch=2253 -minIdentity=80 -t=dna -q=dna -mask=lower -qMask=lower). We used pslCDnaFilter (options: -minId=0.9 -minAlnSize=1000 -bestOverlap -filterWeirdOverlapped) to filter out trivial signals and used pslScore to calculate sequence alignment scores for those filtered BLAT matches. Since the two reciprocal scores obtained from the same subtelomere pair are not symmetrical (depending on which sequence was used as the query), we took their arithmetic mean in our analysis. Such subtelomeric homology search was carried out for both within-strain and cross-strain comparisons and subtelomere pairs with strong sequence homology (BLAT alignment score >= 5000 and sequence identity >= 90%) were recorded.
Hierarchical clustering analysis and reshuffling rate calculation for orthologous subtelomeres
For all the strains within the same species, we performed pairwise comparisons of their subtelomeric regions to identify conserved orthologous subtelomeres in any given strain pairs based on homology search described above. For each strain pair, the proportion of conserved orthologous subtelomeres was calculated as a measurement of the overall subtelomere conservation between the two strains. Such measurements were converted into a distance matrix by the dist() function in R (v3.1)102, based on which the hclust() function was further used for hierarchical clustering. We gauged the reshuffling intensity of orthologous subtelomeres in a way similar to what we did for measuring CNV accumulation and GOL. For any given strain pair, we first calculated the proportion of the non-conserved orthologous subtelomeres in this strain pair as Preshuffling and then applied the Poisson correction and evolutionary time adjustment by −ln (1 − Preshuffling)/2T, in which T is the diversification time of the two compared strains.
Phenotyping the growth rates of yeast strains in copper- and arsenite-rich medium
The homozygous diploid versions of the 12 strains were pre-cultured in Synthetic Complete (SC) medium overnight to saturation. To examine their conditional growth rates in copper- and arsenite-rich environment, we mixed 350 µl conditional media (CuCl2 (0.38 mM) and arsenite (As[III], 3 mM) for the two environment respectively) with 10 µl saturated culture to the wells of Honeycomb plates. Oxygen permeable films were placed on top of the plates to enable a uniform oxygen distribution throughout the plate. The automatic screening was done with Bioscreen Analyser C (Thermic Labsystems Oy, Finland) at 30°C for 72 hours, measuring in 20 minute intervals using a wide-band filter at 420-580 nm103. Growth data pre-processing and phenotypic trait extraction were performed by PRECOG104.
Linkage analysis in diploid S. cerevisiae hybrids
A total of 826 phased outbred lines (POLs) were constructed and phenotyped in the same fashion as previously described52. Briefly, advanced intercrossed lines (AILs) were generated by successive rounds of mating and sporulation from the YPS128 and DBVPG6044 strains105. The resulting haploid AILs were sequenced106 and crossed in different combinations to yield the 826 POLs used for the analysis. The POL diploid genotypes can be accurately inferred from the haploid AILs. Effectively, these 826 POLs constitute a subset of the larger set of POLs in Hallin et al.52 but were constructed and phenotyped independently. Phenotyping of the POLs, each with four replicates, was performed using Scan-o-Matic107 on solid agar plates (0.14% Yeast Nitrogen Base, 0.5% ammonium sulphate, 2% (w/v) glucose and pH buffered to 5.8 with 1% (w/v) succinic acid, 0.077% Complete Supplement Mixture (CSM, Formedium™), 2% agar) supplemented with varying arsenite concentrations (0, 1, 2, and 3mM). Using the deviations between the POL phenotype and the estimated parental mean phenotype in the mapping to combat population structure issues52, quantitative trait loci (QTLs) were mapped using the scanone() function in R/qtl108 with the marker regression method.
Statistics
The Tajima’s relative rate test27 was performed in MEGA (v7.0.16)89. Fisher’s exact test97 with false discovery rate (FDR) correction98 was performed in BLAST2GO (v.3.2)95,96. The Mann–Whitney U test was performed in R (v3.1)102 using the wilcox.test() function, with one.sided alternative hypothesis. P < 0.05 was considered statistically significant in all statistical tests.
Supplementary Material
Acknowledgements
We thank G. Drillon for the help with using the program CHROnicle. We thank O. Croce and R. Marangoni for the help with maintaining the computing server and various bioinformatics tools. We thank Liti lab technician A. Llored technical help with yeast strains and DNA samples. This work was supported by ATIP-Avenir (CNRS/INSERM), Fondation ARC pour la Recherche sur le Cancer (grant PJA20151203273), Marie Curie Career Integration Grants (grant 322035), Agence Nationale de la Recherche (grants: ANR-16-CE12-0019, ANR-13-BSV6-0006-01 and ANR-11-LABX-0028-01), Cancéropôle PACA (AAP émergence 2015) and DuPont Young Professor Award to G.L., Wellcome Trust (grant WT098051) to R.D., and Vetenskapsrådet (The Swedish Research Council, grant 325-2014-4605) to J.W.. J.-X.Y. is supported by a postdoctoral fellowship from Fondation ARC pour la Recherche sur le Cancer (grant n°PDF20150602803). J.L. is supported by a postdoctoral fellowship from Fondation ARC pour la Recherche sur le Cancer (grant n°PDF20140601375). J.H. is supported by the Labex SIGNALIFE program from Agence Nationale de la Recherche (grant ANR-11-LABX-0028-01).
Footnotes
URLs
Previously identified subtelomeric duplication blocks in S. cerevisiae S288C: http://www2.le.ac.uk/colleges/medbiopsych/research/gact/images/clusters-fixed-large.jpg;
RepeatMasker: www.repeatmasker.org;
FastQC: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/;
Picard tools: http://broadinstitute.github.io/picard/;
vcflib: https://github.com/vcflib/vcflib;
MFannot: http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl;
The Saccharomyces Genome Database (SGD): http://www.yeastgenome.org;
Author Contributions
J.-X.Y. conceived, designed, and performed the bioinformatics analysis, wrote the manuscript; J.L. prepared DNA samples for sequencing, performed the experiment on verifying structural rearrangements, contributed to the manuscript; L.A. performed the PacBio sequencing and helped with diagnosing the assembly pipeline; J.H. performed experiments and data analysis for phenotyping, contributed to the manuscript; K.P. performed experiments and data analysis for phenotyping, contributed to the manuscript; K.O. performed the PacBio sequencing and ran the standard assembly pipeline; A.B. helped with discussion on data analysis and manuscript preparation; P.C. performed the PacBio sequencing for the pilot phase project; J.W. designed the phenotyping experiment and helped with data interpretation; M.C.L. helped with the analysis on measuring the sequence homology for subtelomeres; G.F. helped with study design, results discussion and manuscript writing; R.D. conceived and designed the study; G.L. conceived, designed, and guided the study, wrote the manuscript.
Competing Financial Interests
The authors declare that no competing interests exist.
Data Availability
All genome sequencing, assembly and annotation data that support the findings of this study have been deposited in public repositories. The PacBio sequencing reads for this project has been deposed in the European Nucleotide Archive (ENA) with the primary accession code “PRJEB7245” (http://www.ebi.ac.uk/ena/data/view/PRJEB7245). The Illumina sequencing reads for this project has been deposed in the Short Reads Archive (SRA) with the primary accession code “PRJNA340312” (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA340312). The genome assemblies and annotations generated by this study have been deposited at our dedicated website for this project:https://yjx1217.github.io/Yeast_PacBio_2016/data/. The genome assemblies have also been deposited in Genbank with the primary accession code “PRJEB7245” (https://www.ncbi.nlm.nih.gov/bioproject/PRJEB7245).
References
- 1.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 4.Mackay TFC, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W, et al. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergström A, et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol Biol Evol. 2014;31:872–888. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strope PK, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The 1001 Genomes Consortium. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallone B, et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell. 2016;166:1397–1410.e16. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 11.Rieseberg LH. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 12.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–38. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaisson MJP, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2014;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon D, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Current Opinion in Genetics and Development. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- 17.Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 18.Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 19.Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–24. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 20.Goffeau A, et al. Life with 6000 Genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 21.Warringer J, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheelan SJ, Scheifele LZ, Martínez-Murillo F, Irizarry Ra, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc Natl Acad Sci U S A. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata Y, Malhotra A, Bekiranov S, Dutta A. Yeast genome analysis identifies chromosomal translocation, gene conversion events and several sites of Ty element insertion. Nucleic Acids Res. 2009;37:6454–6465. doi: 10.1093/nar/gkp650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoang ML, et al. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 2010;6:1–18. doi: 10.1371/journal.pgen.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liti G, Peruffo A, James SA, Roberts IN, Louis EJ. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast. 2005;22:177–92. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- 26.Marie-Nelly H, et al. High-quality genome (re)assembly using chromosomal contact data. Nat Commun. 2014;5:5695. doi: 10.1038/ncomms6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 29.Rocha EPC. DNA repeats lead to the accelerated loss of gene order in bacteria. Trends in Genetics. 2003;19:600–603. doi: 10.1016/j.tig.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Rocha EPC. Inference and analysis of the relative stability of bacterial chromosomes. Mol Biol Evol. 2006;23:513–522. doi: 10.1093/molbev/msj052. [DOI] [PubMed] [Google Scholar]
- 31.Fischer G, Rocha EPC, Brunet F, Vergassola M, Dujon B. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2006;2:0253–0261. doi: 10.1371/journal.pgen.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C-Z, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–30. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liti G, Barton DBH, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cubillos FA, et al. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol. 2011;20:1401–1413. doi: 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- 36.Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–4. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 37.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–54. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 38.Vakirlis N, et al. Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Res. 2016;26:918–932. doi: 10.1101/gr.204420.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsit S, et al. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol Biol Evol. 2015;32:1695–1707. doi: 10.1093/molbev/msv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linardopoulou EV, et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairhead C, Dujon B. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 2006;6:428–41. doi: 10.1111/j.1567-1364.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 42.Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 43.Liti G, et al. Segregating YKU80 and TLC1 alleles underlying natural variation in telomere properties in wild yeast. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marvin ME, et al. The association of yKu with subtelomeric core X sequences prevents recombination involving telomeric sequences. Genetics. 2009;183:453–467. doi: 10.1534/genetics.109.106682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marvin ME, Griffin CD, Eyre DE, Barton DBH, Louis EJ. In Saccharomyces cerevisiae, yKu and subtelomeric core X sequences repress homologous recombination near telomeres as part of the same pathway. Genetics. 2009;183:441–451. doi: 10.1534/genetics.109.106674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu B, Hao W. A Dynamic Mobile DNA Family in the Yeast Mitochondrial Genome. G3 (Bethesda) 2015;5:1273–1282. doi: 10.1534/g3.115.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu B, Buljic A, Hao W. Extensive horizontal transfer and homologous recombination generate highly chimeric mitochondrial genomes in yeast. Mol Biol Evol. 2015;32:2559–2570. doi: 10.1093/molbev/msv127. [DOI] [PubMed] [Google Scholar]
- 48.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 49.Cole S, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 50.Abramczyk D, Tchórzewski M, Grankowski N. Non-AUG translation initiation of mRNA encoding acidic ribosomal P2A protein in Candida albicans. Yeast. 2003;20:1045–1052. doi: 10.1002/yea.1020. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Y, et al. Structures of naturally evolved CUP1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination. G3 (Bethesda) 2014;4:2259–69. doi: 10.1534/g3.114.012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hallin J, et al. Powerful decomposition of complex traits in a diploid model. Nat Commun. 2016;7:13311. doi: 10.1038/ncomms13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson JA, Song YS, Langley CH. Molecular population genetics of Drosophila subtelomeric DNA. Genetics. 2008;178:477–487. doi: 10.1534/genetics.107.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo H-F, Olsen KM, Richards EJ. Natural variation in a subtelomeric region of arabidopsis: Implications for the genomic dynamics of a chromosome end. Genetics. 2006;173:401–417. doi: 10.1534/genetics.105.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Louis EJ, Haber JE. Mitotic recombination among subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics. 1990;124:547–559. doi: 10.1093/genetics/124.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson MZ, Wigen LJ, Burrack LS, Berman J. Real-Time Evolution of a Subtelomeric Gene Family in Candida albicans. Genetics. 2015;200:907–919. doi: 10.1534/genetics.115.177451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ames RM, et al. Gene duplication and environmental adaptation within yeast populations. Genome Biol Evol. 2010;2:591–601. doi: 10.1093/gbe/evq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci. 1998;95:8420–7. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife. 2015;4:1–9. doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyma KE, Fay JC. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol Ecol. 2013;22:2917–2930. doi: 10.1111/mec.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borneman AR, Pretorius IS. Genomic insights into the Saccharomyces sensu stricto complex. Genetics. 2015;199:281–291. doi: 10.1534/genetics.114.173633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 64.Leducq J-B, et al. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat Microbiol. 2016;1:15003. doi: 10.1038/nmicrobiol.2015.3. [DOI] [PubMed] [Google Scholar]
- 65.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 66.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunt M, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walker BJ, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Prepr arXiv1207.3907. 2012;9 doi:arXiv:1207.3907 [q-bio.GN] [Google Scholar]
- 74.Kim KE, et al. Long-read, whole-genome shotgun sequence data for five model organisms. Sci data. 2014;1:140045. doi: 10.1038/sdata.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodwin S, et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25:1750–1756. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Proux-Wéra E, Armisén D, Byrne KP, Wolfe KH. A pipeline for automated annotation of yeast genome sequences by a conserved-synteny approach. BMC Bioinformatics. 2012;13:237. doi: 10.1186/1471-2105-13-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lechner M, et al. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lechner M, et al. Orthology detection combining clustering and synteny for very large datasets. PLoS One. 2014;9:e105015. doi: 10.1371/journal.pone.0105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirarab S, et al. ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–i548. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.To TH, Jung M, Lycett S, Gascuel O. Fast dating using least-squares criteria and algorithms. Syst Biol. 2016;65:82–97. doi: 10.1093/sysbio/syv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drillon G, Carbone A, Fischer G. Combinatorics of chromosomal rearrangements based on synteny blocks and synteny packs. Journal of Logic and Computation. 2013;23:815–838. [Google Scholar]
- 92.Drillon G, Carbone A, Fischer G. SynChro: A fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nattestad M, Schatz MC. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw369. btw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 95.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 96.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fisher R. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;85:87–94. [Google Scholar]
- 98.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 99.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 100.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 101.Kent WJ. BLAT - The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.R Developement Core Team. R: a language and environment for statistical computing. R Found Stat Comput. 2015;1:409. [Google Scholar]
- 103.Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez-Ricaud L, Kourtchenko O, Zackrisson M, Warringer J, Blomberg A. PRECOG: a tool for automated extraction and visualization of fitness components in microbial growth phenomics. BMC Bioinformatics. 2016;17:249. doi: 10.1186/s12859-016-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parts L, et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Illingworth CJR, Parts L, Bergström A, Liti G, Mustonen V. Inferring genome-wide recombination landscapes from advanced intercross lines: application to yeast crosses. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zackrisson M, et al. Scan-o-matic: high-resolution microbial phenomics at a massive scale. G3. 2016;6:3003–3014. doi: 10.1534/g3.116.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.