Abstract
Understanding how and why populations evolve is of fundamental importance to molecular ecology. Restriction site-associated DNA sequencing (RADseq), a popular reduced representation method, has ushered in a new era of genome-scale research for assessing population structure, hybridization, demographic history, phylogeography and migration. RADseq has also been widely used to conduct genome scans to detect loci involved in adaptive divergence among natural populations. Here, we examine the capacity of those RADseq-based genome scan studies to detect loci involved in local adaptation. To understand what proportion of the genome is missed by RADseq studies, we developed a simple model using different numbers of RAD-tags, genome sizes and extents of linkage disequilibrium (length of haplotype blocks). Under the best-case modelling scenario, we found that RADseq using six- or eight-base pair cutting restriction enzymes would fail to sample many regions of the genome, especially for species with short linkage disequilibrium. We then surveyed recent studies that have used RADseq for genome scans and found that the median density of markers across these studies was 4.08 RAD-tag markers per megabase (one marker per 245 kb). The length of linkage disequilibrium for many species is one to three orders of magnitude less than density of the typical recent RADseq study. Thus, we conclude that genome scans based on RADseq data alone, while useful for studies of neutral genetic variation and genetic population structure, will likely miss many loci under selection in studies of local adaptation.
Keywords: FST, genome scan, genome–environment association, genotyping by sequencing, local adaptation, outlier analysis
‘The moral of the story is: I chose a half measure, when I should have gone all the way. I’ll never make that mistake again. No more half measures, Walter’.
-Mike Ehrmantraut, Breaking Bad
Understanding how and why populations evolve is of fundamental importance to molecular ecology. Restriction site-associated DNA sequencing (RADseq) has dramatically decreased the cost of generating large numbers of polymorphic markers and has ushered in a new era of genome-scale research on the evolution of populations across a broad range of organisms. Several genotyping methods (RAD, ddRAD, GBS, MSG, etc.) developed in recent years, for convenience, can collectively be referred to as RADseq. The details of each of these methods have been reviewed recently by Andrews et al. (2016). Generally, RADseq methods produce DNA libraries for high-throughput sequencing using restriction enzymes that cut at specific motifs throughout the genome. RADseq markers come in the form of RAD-tags, which are short-read sequences adjacent to restriction enzyme cut sites. Because many polymorphic markers are produced by RADseq, it has frequently been used successfully for population genetic analyses, including assessment of population structure, hybridization, demographic history, phylogeography and migration (Catchen et al. 2013; Cavender-Bares et al. 2015; Combosch & Vollmer 2015; Qi et al. 2015). Markers generated by RADseq have also been quite useful for constructing linkage maps and identifying quantitative trait loci (QTL; Pfender et al. 2011; Houston et al. 2012; Weber et al. 2013; Laporte et al. 2015; Lowry et al. 2015).
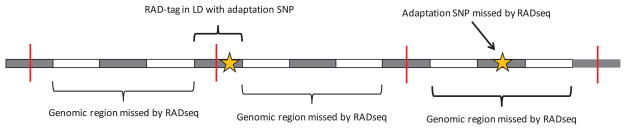
Many recent studies have used RADseq for genome scans to detect locally adapted loci (reviewed in Arnold et al. 2013; Tiffin & Ross-Ibarra 2014; Andrews et al. 2016). However, RADseq presents a major challenge for genome scan studies because the approach usually samples only a small proportion of the genome, despite generating hundreds to thousands of polymorphic markers (Fig. 1). This problem is particularly acute for organisms with large genome sizes and/or low levels of linkage disequilibrium (LD). While others have articulated the challenges that RADseq poses for genome scans (Arnold et al. 2013; Davey et al. 2013; Tiffin & Ross-Ibarra 2014), RADseq studies are still relatively common despite potentially major pitfalls.
Fig. 1.
Diagram depicting theoretical best-case scenario for RADseq, where all RAD-tags (red line) are polymorphic and evenly distributed across the genome among linkage disequilibrium (LD) blocks (grey and white). In the case illustrated, RAD-tags are not in LD with 75% of the genome, and thus, RADseq will miss many SNPs involved with local adaptation. [Colour figure can be viewed at wileyonlinelibrary.com]
Here, we analyse how much of the genome will be missed by genome scan studies that utilize RADseq as the source of genetic markers for analyses. We also survey the recent literature to establish how RADseq is being used for contemporary genome scan studies. Finally, we discuss potential alternative approaches for genome scans, including exome capture, RNA sequencing, pooled sequencing and whole-genome sequencing.
RAD approaches for identifying candidate loci underlying local adaptation
A major goal of molecular ecology is to identify the loci underlying local adaptation to important environmental factors in wild populations (Hereford 2009; Anderson et al. 2011; Barrett & Hoekstra 2011; Jones et al. 2012; Savolainen et al. 2013; Rausher & Delph 2015; Rellstab et al. 2015; Hoban et al. 2016). The two predominant genome scan approaches for identifying candidate loci underlying local adaptation are as follows: (i) differentiation outlier studies that scan the genome for differences in allele frequencies between locally adapted populations and (ii) genetic–environment association studies that evaluate associations between environmental variables and allele frequencies across a set of populations spread over geographic space (reviewed in Rellstab et al. 2015; Hoban et al. 2016). Both approaches require data for many loci across the genome. RADseq has recently been used in genome scans for adaptation loci because it is a cost-effective method for acquiring large numbers of polymorphic markers.
While RADseq data sets comprise several orders of magnitude more polymorphic markers than microsatellite data sets, RADseq will still not adequately sample haplotype blocks with enough markers to provide genomewide coverage (Fig. 1). It should be noted that even though multiple single-nucleotide polymorphisms (SNPs) within a single RAD-tag can be detected, those SNPs are usually redundant with other SNPs in the same haplotype block. Further, not all SNPs will be in complete linkage disequilibrium (R2 = 1) within haplotype blocks because new mutations occur at different times over the course of the evolution of haplotypes and because recombination does occur within blocks. Thus, studies that lack whole-genome sequencing coverage will miss potentially important adaptive SNPs because they are not in complete linkage disequilibrium with SNP markers detected by RADseq genotyping. In the next section, we quantify the portion of the genome that will be overlooked when surveyed by RADseq.
How bad is RAD for genomic scans of adaptation?
To understand potential limitations for RADseq, we developed a simple model using different numbers of RAD-tags, genome sizes and extents of LD (length of haplotype blocks) to calculate the percentage of the genome that could be studied using RADseq. The model depends on a number of assumptions: (i) recombination hot spots are equally distributed across the genome, and thus that the genome is divided into equal-length haplotype blocks; (ii) the optimal situation in which RAD-tag markers are distributed equally among these blocks; (iii) sequencing depth is uniform across RAD-tags; (iv) no more than one RAD-tag marker occurs on each linkage block; (v) RAD-tags are in perfect linkage disequilibrium with adaptive SNPs; (vi) all RAD-tags are polymorphic; (vii) if an adaptive locus is not linked to a polymorphic RAD-tag, genome scans would not be able to detect that locus as having a potential role in adaptation. Further, our analyses were conducted assuming the methodology of the original RAD protocol (Miller et al. 2007; Baird et al. 2008). Because this protocol digests DNA with a single enzyme, shears it and then attaches adaptors (Davey et al. 2013), it is the RAD method that will return the highest density of RAD-tags (Puritz et al. 2014). Finally, we assumed 50% GC content in the genome, and thus, for an enzyme cut site of length n, the frequency of cut sites could be estimated as 4n (higher or lower GC content will result in a difference in cut frequency; Schweyen et al. 2014). Note that all of the assumptions described above are for a best-case scenario because they are virtually never met in reality. Violations of any of these assumptions will make the problem of genome scans based on RADseq even worse.
It should be noted that for detecting signatures of selection, genomic regions affected by strong selection are expected to have higher LD, as neutral loci hitchhike along with a selected locus as it sweeps to fixation (i.e. hard sweep). Our assumption of equalsized haplotype blocks does not account for this scenario, which would increase the ability of RADseq-based genome scans to detect selective sweeps. However, hard sweeps on new mutations that lead to extended haplotypes are only one of several forms of adaptation. Adaptations can also occur through soft sweeps on standing genetic variation, can be highly polygenic and can occur without any adaptive alleles going to fixation (Pritchard et al. 2010). Soft sweeps have much weaker effects on linked sites than hard sweeps and are, thus, likely to be missed when markers are sparse (Ferrer-Admetlla et al. 2014). Additionally, the effect of selection on linked sites varies greatly among species (Cutter & Payseur 2013). Overall, genome scans conducted with RADseq can lead to a biased interpretation of how adaptation has occurred in a given system because often only recent hard sweeps from new mutations can realistically be detected.
Using our best-case scenario assumptions, we evaluated the relationship between different haplotype block sizes and the theoretical maximum proportion of the genome that would be sampled by RADseq (Fig. 2). We conducted the analysis for evenly spaced 6-base pair (bp) (every 46 = 4096 base pairs) and 8-bp (every 48 = 65 536 base pairs) restriction enzyme cutters, which are both commonly used in RADseq studies (Fig. 2). For a species with low LD (less than 1000 base pairs (bp), such as in insects, some marine species and many trees; Table 1), a 6-bp cutter would cover less than 25% of the genome, and an 8-bp cutter would cover less than 2% of the genome even in this best-case scenario (Fig. 2; R code provided in Supplemental Materials).
Fig. 2.
The theoretical maximum proportion of a genome that may be ascertained by RAD-tags from digestion with enzymes that recognize either a 6- or 8-bp recognition site, given the size of linkage disequilibrium blocks across the genome and assuming 50% GC content in the cut site and across the genome. Organisms with LD on the scale of: (A) a few hundred base pairs include honey bee Apis mellifera, mosquito Anopheles gambiae, purple sea urchin Strongylocentrotus purpuratus, Zebra finch Taeniopygia guttata, diverse maize Zea mays, wild tomato Solanum peruvianum, white spruce Picea glauca and European Aspen Populus tremula L., Salicaceae; (B) 1 kb include mosquito Anopheles arabiensis, fruit fly Drosphila melanogaster, zebra fish Danio rerio in the wild and coastal Douglas-fir Pseudotsuga menziesii var. menziesii, (C) a few kb include loblolly pine Pinus taeda, some populations of three-spined stickleback Gasterosteus aculeatus and round worm Caenorhabditis remanei; (D) greater than 10 kb include Collared Flycatcher Ficedula hypoleuca, Siberian jay Perisoreus infaustus, some populations of three-spined stickleback Gasterosteus aculeatus, European eel Anguilla anguilla, wild mouse Mus musculus domesticus, big horn sheep Ovis canadensis, Arabidopsis thaliana and wild yeast Saccharomyces paradoxus (See Table 1). [Colour figure can be viewed at wileyonlinelibrary.com]
Table 1.
Estimated length of average linkage disequilibrium for biological organisms
| Group | Species | Average LD | References |
|---|---|---|---|
| Invertebrates | |||
| Insect | Fruit fly, Drosophila melanogaster | <1 kb | Long et al. (1998) |
| Insect | Mosquito, Anopheles arabiensis | 1 kb | Marsden et al. (2014) |
| Insect | Mosquito, Anopheles gambiae | ≪1 kb | Harris et al. (2010) |
| Insect | Honey bee, Apis mellifera | 500 bp | Wallberg et al. (2014) |
| Insect | Bumble bee, Bombus impatiens | <10 kb | Sadd et al. (2015) |
| Marine invertebrate | Purple sea urchin, Strongylocentrotus purpuratus | Very low to none | Pespeni et al. (2010) |
| Worm | Round worm, Caenorhabditis elegans | Very large | Cutter (2006) |
| Worm | Round worm, Caenorhabditis remanei | 1–2kb | Cutter et al. (2006) |
| Vertebrates | |||
| Bird | Collared Flycatcher, Ficedula hypoleuca | 400 kb | Backström et al. (2006) |
| Bird | Siberian jays, Perisoreus infaustus | 6.28 Mb | Li & Merila (2010) |
| Bird | Zebra finch, Taeniopygia guttata | 300 bp | Balakrishnan & Edwards (2009) |
| Bird | Black-footed Albatrosses, Phoebastria nigripes | 75 bp | Dierickx et al. (2015) |
| Fish | Three-spined stickleback, Gasterosteus aculeatus | 1 kb | Roesti et al. (2015) |
| Fish | European eel, Anguilla anguilla | 10–20 kb | Pujolar et al. (2014) |
| Fish | Zebra fish (wild), Danio rerio | <1 kb | Whiteley et al. (2011) |
| Mammal | Humans, Homo sapiens | 50 kb | Pritchard & Przeworski (2001) |
| Mammal | Wild mice, Mus musculus domesticus | 50 kb | Laurie et al. (2007) |
| Mammal | Big horn sheep, Ovis canadensis | 4 Mb | Miller et al. (2011) |
| Plants | |||
| Flowering plant | Arabidopsis thaliana | 10 kb | Kim et al. (2007) |
| Flowering plant | Diverse maize, Zea mays | 400 bp | Tenaillon et al. (2001) |
| Flowering plant | Wild tomato, Solanum peruvianum | <150 bp | Arunyawat et al. (2007) |
| Flowering plant | Wild tomato, Solanum chilense | <750 bp | Arunyawat et al. (2007) |
| Flowering plant | Chickpea, Cicer arietinum | >1 Mb | Kujur et al. (2015) |
| Flowering plant | Barrelclover, Medicago truncatula | 5–10 kb | Branca et al. (2011) |
| Flowering plant | Wild soybean, Glycine soya | <500 kb | Wang et al. (2015) |
| Flowering plant | Wild rice, Oryza rufipogon | ≪40 kb | Mather et al. (2007) |
| Tree | Scots pine, Pinus sylvestris | 300 bp | Pyhäjärvi et al. (2007) |
| Tree | Coastal Douglas-fir, Pseudotsuga menziesii | 1.5 kb | Eckert et al. (2009) |
| Tree | European aspen, Populus tremula | <500 bp | Ingvarsson (2005) |
| Fungi | |||
| Fungus | Wild yeast, Saccharomyces paradoxus | 50–100 kb | Tsai et al. (2008) |
No RADseq study achieves the best-case assumptions. In reality, RAD-tags are distributed nonuniformly across the genome and LD actually varies throughout the genome because of recombination hot spots, proximity to centromeres and chromosomal rearrangements (Ortiz-Barrientos et al. 2016). Therefore, the ability to detect associations with linked markers with adaptive loci will vary depending on the genomic region analysed. Finally, any RADseq method that uses size selection on nonsheared DNA (MSG: Andolfatto et al. 2011; ddRAD: Peterson et al. 2012; RRLs: Greminger et al. 2014) or preferentially amplifies short fragments (GBS: Elshire et al. 2011; SBG: Truong et al. 2012) will further decrease the density of RAD-tags across the genome and therefore decrease the probability of detecting selection (Davey et al. 2013).
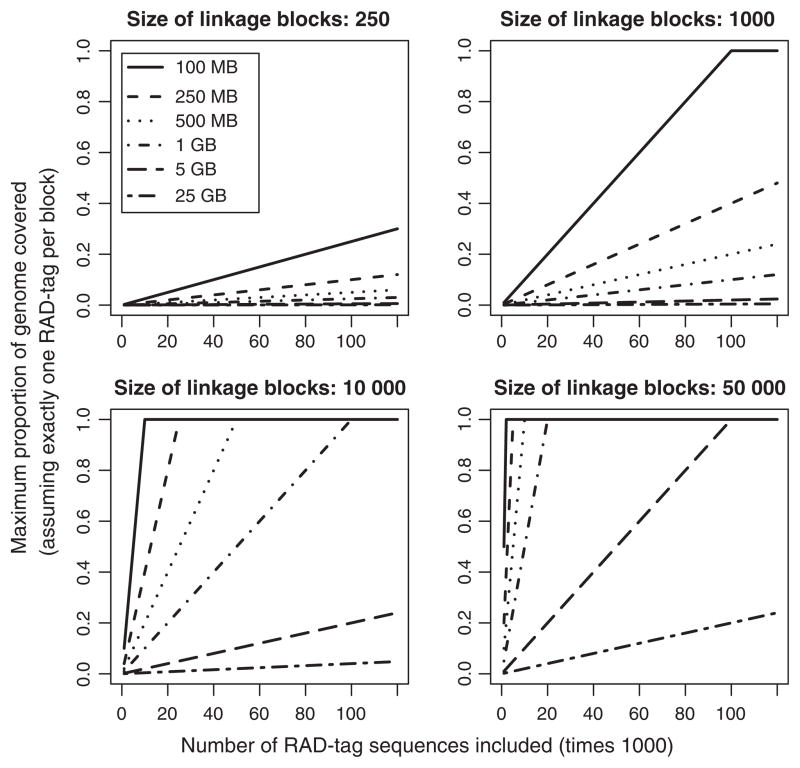
To more realistically assess the potential of RADseq, we quantified how different numbers of informative RAD-tags will influence genome scan studies. It is important to consider the number of informative RAD-tags generated in a study, as many tags will lack sufficient polymorphism for genome scans or be lost during bioinformatic filtering. Our analysis calculated how many haplotype blocks contained at least one informative RAD-tag for different genome sizes, lengths of LD and total numbers of informative RAD-tags (Fig. 3; R code provided in Supplemental Materials). The results of this analysis should have utility for researchers who wish to assess the potential of a proposed genome scan study. For example, a species with a 3-gigabase pair (Gbp) genome (approximately the size of the human genome) and haplotype block lengths of 10 kilobase (kb) (moderate linkage), 20 000 informative RAD-tags will only cover ~7% of the genome – even if they are evenly distributed. Thus, at best, ~93% of the genome will not contain an informative RAD-tag and effectively be ignored by a genome scan study with these parameters. Note the ‘fan’ pattern that we observe in Fig. 3 underscores the multiplicative effect of genome size and length of haplotype blocks. Overall, these results demonstrate that RADseq-based approaches will often only be able to evaluate a small portion of the genome for signals of local adaptation.
Fig. 3.
The proportion of the genome that will be sampled by RADseq (y-axes) for different numbers of RAD-tags (x-axes), genome sizes (line types) and lengths of linkage disequilibrium blocks.
Recent RAD genome scans
Based on the results of our theoretical analyses of the efficacy of RADseq for genome scans, we were interested in evaluating how much of the genome is sampled by RAD-seq for contemporary genome scan studies. To identify recent studies using RADseq for genome scans of local adaptation, we conducted a literature survey of articles from January 2015 to April 2016 (Table S1, Supporting information). We identified articles in two steps: (i) we examined the contents of the journals Molecular Ecology, Conservation Genetics, Genetics, Ecology and Evolution, Molecular Biology & Evolution and Tree Genetics and Genomics and (ii) we also examined all articles citing the major RADseq methods papers (Miller et al. 2007; Baird et al. 2008; Andolfatto et al. 2011; Elshire et al. 2011; Peterson et al. 2012) that were published since January 2015. In total, we identified 27 articles (with three studies using multiple species) that fit the following criteria: (i) the study used a genome scan to infer the genetic basis of adaptation in natural populations, (ii) the study used at least 1000 SNPs in the genome scan and (iii) the study was not based on an a priori list of candidate genes. Of those studies, 19 were conducted with single-digest RAD, and eight with double-digest RAD. For the following calculations, if the study examined multiple closely related species, the average values for the two species were used for the study. The distribution of the total number of individuals sampled was skewed, with most studies using fewer than 200 individuals, a few using from 200 to 700 and one study with nearly 2000 individuals (median = 144). There was a strong taxonomic bias among studies: 16 fish, two birds, five invertebrates, three mammals and one plant. The genome sizes of studied species were slightly skewed, with most between 0.5 and about 1 Gbp, and fewer species of 3–5 Gbp (median = 1.26 Gbp). Note that we used the species genome size, if available, or used the genome size of the closely related species to which reads were aligned. To gauge the breadth of possible values, we compiled the genome sizes and estimates of the length of linkage disequilibrium for a wide diversity of species (Table 1).
From our survey, we conclude that recent RADseq studies are typically too sparse to cover an appreciable amount of the genome and that they likely miss the vast majority of loci underlying adaptation. To estimate the number of markers per megabase pair (Mbp), we used the number of RAD-tags rather than number of SNPs, except in four cases, where the number of RAD-tags was not reported. The number of markers was skewed with most studies having fewer than 20 000, two studies with ~70 000 markers and one study with more than 160 000 makers (median = 9000). Excluding those species with no estimates of genome size, the number of markers per Mbp was also noticeably skewed (median of 4.08 RAD-tag markers per Mbp), with most studies having five or fewer RAD-tags per Mbp, a few having between six and 20 per Mbp, three having up to 110 per Mbp and only one study having more than a few hundred (362) RAD-tags per Mbp. The study (Roesti et al. 2015) with the highest density of RADtags (362 per Mbp) actually had a nearly threefold lower density of useful markers (136 SNPs per Mbp) because many RADtags were not polymorphic. Thus, the highest density of markers that is currently being achieved by RADseq is ~1 SNP per 7 kb. Given that RAD-tags and recombination rates are not evenly spaced throughout the genome, and not all RAD-tags are informative, the extent of linkage disequilibrium would have to be hundreds of kb in length for the average recent RADseq-based genome scan study (one marker per 245 kb) to be effective (Table S1, Supporting information). However, for most organisms, LD is far less than 100 kb (Table 1). For some plants and ocean animals having large population sizes and high levels of gene flow, where haplotype blocks are between 250 bp and 1 kb, the proportion of the genome sampled by the average RADseq density will be very small. Thus, it appears that many recent RADseq studies have only scanned a small fraction of the genome.
Beyond RAD
Although RADseq is an efficient and economical approach for generating genetic makers, our results suggest that other genotyping methods should be considered when conducting genome scan studies to search for putatively adaptive loci. We advise that researchers planning to use reduced representation approaches for genome scans to target genic regions, as genic regions are likely to be the location of much of the functional genetic changes involved in adaptation (Hoekstra & Coyne 2007; Stern & Orgogozo 2008). For example, transcriptome sequencing and exome capture can be used to target genic regions when no reference genome is available (Gugger et al. 2016). Genic regions captured by these methods are often in linkage with gene promoters, which also are likely locations of functional adaptive variation (Wray 2007; Stern & Orgogozo 2008). However, transcriptome sequencing will miss many genes because they have tissue- or condition-specific expression. Further, important genes underlying adaptation may be expressed at low levels, thus requiring more sequencing at deeper coverage. Allele-specific expression (Chen et al. 2016) can also lead to misinterpretation of allele frequency differences among populations. In contrast, exome capture sequencing of genomic DNA has a greater potential to genotype a larger set of genic regions, provided that the capture method is designed from a reference genome (Jones & Good 2016). Even so, both transcriptome sequencing and exon capture will likely fail to detect many polymorphisms in distant regulatory regions, such as in enhancers and insulators (Pennacchio et al. 2013).
Although still not feasible for large genomes, whole-genome sequencing is by far the best approach for genome scans because SNPs are quantified in most, if not all, haplotype blocks across the genome. An economical alternative to whole-genome sequencing of individuals is population pooled sequencing (pool-seq; Schlötterer et al. 2014). Pool-seq studies combine dozens to hundreds of individuals prior to sequencing to calculate allele frequencies of populations and conduct genome scans (Schlötterer et al. 2014; Wright et al. 2015; Kapun et al. 2016). It should be noted that genotype information generated from whole-genome sequencing and from pool-seq is no panacea and will contain significant levels of missing data and data artefacts. Some missing data and artefacts are caused by structural differences (insertions, deletions, inversions, etc.) between resequenced individuals and the reference genome to which they are aligned (Tiffin & Ross-Ibarra 2014; Hoban et al. 2016). Finally, it is important that researchers retain invariant sites in their data sets to accurately calculate population genetic summary statistics and demographic parameters. Invariant sites are often removed as part of bioinformatics processing of sequence data, but are necessary for coalescent analyses of demographic history.
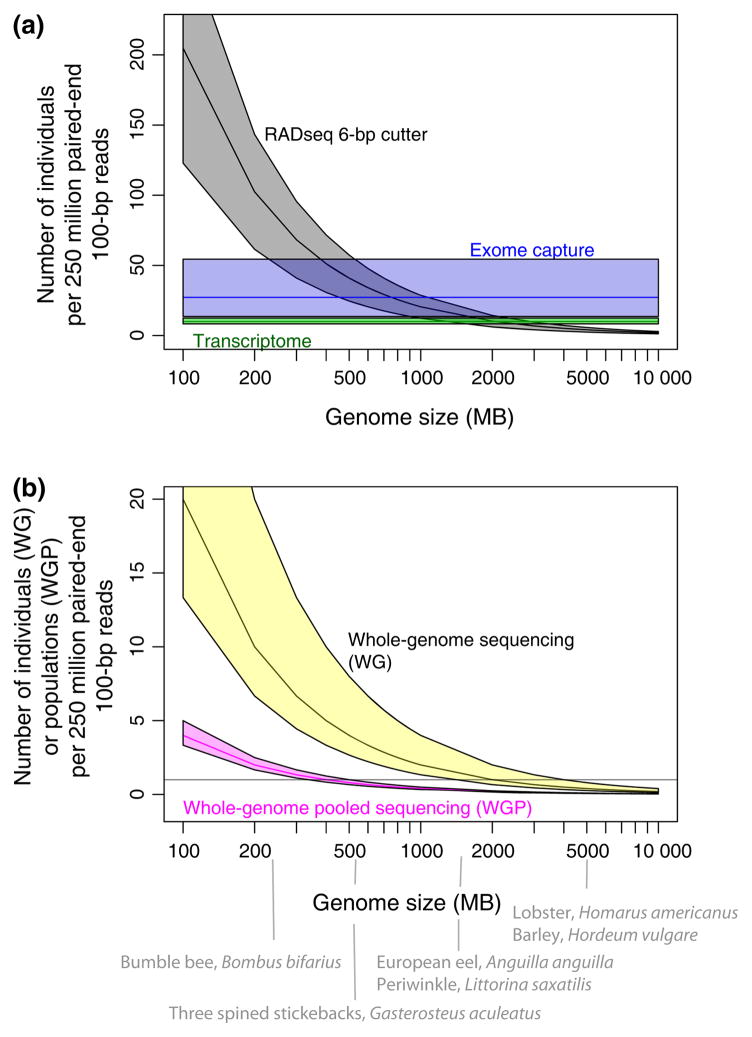
We evaluated the relative sequencing costs for these different approaches by calculating the number of individuals that could be multiplexed per 250 million Illumina paired-end reads (assuming a read length of 100 and with 20% of reads removed due to contamination, PCR duplicates and other quality control filtering). Currently, this corresponds to a single lane of Illumina HiSeq 2500, which yields ~250 million reads (04/2016 http://www.illumina.com). For RADseq, we assumed that a 6-base pair cutter (cut site 50% GC content) is used to digest the genome (50% GC content) and that a 20× depth of read coverage is needed per RAD-tag. Given these assumptions, an expected 40 individuals can be sequenced in a lane for genome sizes greater than 500 Mbp (e.g. stickleback fish), and an expected 10 individuals can be sequenced in a lane for genome sizes greater than 2 Gbp (e.g. bottle nose dolphin, Fig. 4 top). For transcriptome sequencing and exome capture, we assumed 30 000 genes with an average of seven exons per gene, one isoform and 150-bp per exon. For transcriptome sequencing, we also assumed that 20–30 million raw reads were needed per individual to ascertain genes expressed at low levels (The ENCODE Consortium). For sequence capture, we assumed that capture probes were designed near exon boundaries and would target the exon in addition to 100 bp of DNA on either side of the exon boundary, with a desired coverage of 20× per sequence. Based on these assumptions, the number of individuals multiplexed in a lane for RADseq is similar to exome capture for genome sizes in the range of 500 Mbp to 2 Gbp, and for transcriptome sequencing, in the range of 1.5–2.5 Gbp (Fig. 4 top). For genomes larger than 2.5 Gbp, more individuals can be multiplexed per lane for transcriptome sequencing and exome capture than for RADseq.
Fig. 4.
The number of individuals (or populations) that can be sequenced as a function of genome size. It is assumed that a 6-bp cutter will cut on average every 4096 base pairs (assuming 50% GC content in cut site and in genome). For RADseq, exome capture and whole-genome sequencing, we assume an average of 20× coverage. For transcriptome sequencing, we assume 20–30 million reads per individual, and for whole-genome pooled sequencing, we assume 100× coverage. See also supplementary R script. Shown on the figure are some species for which RAD-seq has been applied to identify genomic signatures of selection. For RADseq, CI were based on a more frequent cut site every 4096*0.6 = 2457.6 bp and less frequent cut site 4096*1.4 = 5734.4 bp. For exome capture, CI were based on half as many sequences capture and twice as many sequences captured. For transcriptome, we assume 25 million raw reads per individual, with confidence intervals from 20 to 30 million raw reads per individual. For whole-genome data, we assume 20× coverage per individual, with confidence intervals from 10× to 30× coverage. For whole-genome pool-seq, we assume 100× coverage per pool and CI based on 80–120× coverage per pool. [Colour figure can be viewed at wileyonlinelibrary.com]
We also conducted calculations for whole-genome sequencing of individuals (assuming a 20× depth of read coverage per individual) and whole-genome pool-seq of populations (assuming 100× coverage per population, Schlötterer et al. 2014). For genome sizes in the range of 500 Mbp to 2 Gbp, only a few individuals can be sequenced per lane at 20× coverage, which may be outside the budget for many laboratories (Fig. 4 bottom). Pool-seq appears to be a cost-effective alternative to sequencing individuals (Fig. 4 bottom). For investigators wishing to explore this parameter space further, we have included the R scripts used to generate the figures in the Supplementary Information online.
If you RAD
There are some situations where RADseq can have a high degree of utility for genome scans of local adaptation. For example, RADseq is very useful for studies of local adaptation where candidate genes have already been identified because RAD markers can be used to generate a null distribution of loci across the genome (Luikart et al. 2003). Natarajan et al. (2015) recently conducted an excellent study of this sort with candidate alpha- and beta-globin genes in ducks. They compared allele frequencies of these candidate genes to a null distribution generated by RADseq data for high- and low-altitude population pairs of three Andean duck species. For all three species, FST of the globin genes was greatly elevated above the null FST distribution generated by RADseq markers. We anticipate that many more researchers will use this approach in the near future.
RADseq might also be useful in some cases for genome scans that do not focus on candidate genes identified a priori, depending on the nature of the genome of the study organism. There are a few examples of RADseq genome scans recovering loci previously known to be involved in adaptive divergence (Hohenlohe et al. 2010; Nadeau et al. 2014). Further, some methylation-sensitive restriction enzymes do preferentially target genic regions of the genome (Pegadaraju et al. 2013). However, researchers thinking of using RADseq for naïve genome scans should first establish how much of the genome they can realistically expect to evaluate and make sure to report those estimates in publications. Pilot RADseq experiments with different enzymes can be used to estimate the number of informative RAD-tag markers that will be generated. Researchers should also try to establish the genome size of their focal organisms. Genome sizes for many organisms are listed in the ANIMAL GENOME SIZE Database (http://www.genomesize.com/), PLANT C VALUES Database (http://data.kew.org/cvalues/) or other resources (Garcia et al. 2014). If genome size is unknown, it can readily be established by flow cytometry. Unfortunately, it is more difficult to estimate the extent of LD for a given species. Rough estimates of LD can be made by comparisons to species with known LD estimates, although some closely related species may have very different genome sizes (e.g. Plethodontid salamanders; Sessions & Larson 1987). Additionally, LD is highly variable and even varies greatly among populations within species (e.g. haplotype blocksizes ranged from 8800 to 25 200 bp among human populations in one study; Hinds et al. 2005). Once the number of informative RAD-tags, genome size and LD has been estimated, researchers can conduct similar calculations to those that we described above to determine the maximum amount of the genome they can reasonably expect to evaluate in a genome scan study. Based on our survey of the literature, it appears that genome scans based upon RADseq will be informative for a minority of species.
Conclusions and closing remarks
The problem with the low density of markers produced by RADseq is just one of many challenges for studies aiming to identify loci involved in local adaptation. Other reviews have outlined these problems, which include the confounding effects of population structure, demographic history and issues with analytical methods (Lotterhos & Whitlock 2014; Tiffin & Ross-Ibarra 2014; Rellstab et al. 2015; Hoban et al. 2016). However, the problem with RADseq is with the marker data sets themselves, which are often far too sparse to have a reasonable chance of detecting the loci involved in adaptation. The issue of marker sparseness will be true for any other sort of genomic scan conducted without genome resequencing, including AFLPs and microsatellites (Nosil et al. 2009; Fischer et al. 2011). Marker sparseness is also an issue for genomewide association studies (GWAS) and caution must be taken for those studies as well when using RADseq (e.g. Stanton-Geddes et al. 2013).
Overall, RADseq should be viewed as one of many important tools to study local adaptation. Field experimentation, linkage and association mapping, forward and reverse genetics, candidate gene approaches, functional molecular genetics and modelling are also crucial approaches for understanding the genetic basis of local adaptation (Kawecki & Ebert 2004; Hereford 2009; Anderson et al. 2011; Savolainen et al. 2013; Pardo-Diaz et al. 2015). RADseq can be very useful for estimating important parameters that may affect interpretation of how adaptation occurs, including population structure, gene flow and demography. RADseq can also be useful for genomic scans of local adaptation when candidate genes have been identified a priori (e.g. Natarajan et al. 2015). However, genome scans based on RADseq data alone will often miss many loci involved in local adaptation, especially in species with short LD.
Supplementary Material
Acknowledgments
Comments from Jon Puritz, Kevin Wright, Joel Sharbrough, Dan Sloan, members of the Lowry Lab group and two anonymous reviewers helped improve this manuscript. Paul Hohenlohe, Julian Catchen and Eric Johnson identified errors in the original manuscript that are addressed in the correction note section. This work was conducted as a part of the Computational Landscape Genomic working group at the National Institute for Mathematical and Biological Synthesis, sponsored by the National Science Foundation through NSF Award #DBI-1300426, with additional support from The University of Tennessee, Knoxville. In addition, SH was supported during this working group as a postdoctoral fellow at NIMBioS. Partial support for individuals provided by NSF DEB-1316549 to AS; NIH R01GM098856 to LKR; Washington State University, Army Research Office W911NF-15-1-0175 and NSF IOS-1557795 to JLK; Michigan State University to DBL; and Northeastern University to KEL.
Footnotes
Correction note
The original advance online paper contained two errors associated with the calculation of the median density of RAD-seq tags in the survey of recent RAD-seq genome scan studies (Table S1). The first error was in the size of the assembled stickleback genome, which was reported as 0.53 Gbp, but should have been 0.46 Gbp. The second error was an accidental inversion of terms. These two mistakes contributed to an erroneous statement in the original abstract that the median density of RAD-tags across recent studies “was one marker per 3.96 megabases.” The statement has been revised to read: “was 4.08 RAD-tag markers per megabase.” Following these corrections, changes were made in the abstract and elsewhere in the paper to reflect a modified interpretation of the results, though we note the main arguments in the article are unaffected. Other minor modifications to the paper were made based upon suggestions by the editors of Molecular Ecology Resources. This version of the article, published as an “accepted article” on 12 November 2016 under DOI 10.1111/1755-0998.12635 replaces the original version of the article published on 12 September 2016 under DOI 10.1111/1755-0998.12596.
The idea for the manuscript was conceived collectively by all authors during an NSF National Institute for Mathematical and Biological Synthesis (NIMBioS) working group. All authors contributed to the writing of the manuscript.
Additional Supporting Information may be found in the online version of this article:
Appendix S1 Supplementary R scripts for Breaking RAD.
Table S1 Recent (January 2015 to April 2016) genome scan studies, which used RAD-seq for genotyping.
References
- Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends in Genetics. 2011;27:258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, Davison D, Erezyilmaz D, et al. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Research. 2011;21:610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nature Reviews Genetics. 2016;17:81–92. doi: 10.1038/nrg.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold B, Corbett-Detig RB, Hartl D, Bomblies K. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Molecular Ecology. 2013;22:3179–3190. doi: 10.1111/mec.12276. [DOI] [PubMed] [Google Scholar]
- Arunyawat U, Stephan W, Städler T. Using multilocus sequence data to assess population structure, natural selection, and linkage disequilibrium in wild tomatoes. Molecular Biology and Evolution. 2007;24:2310–2322. doi: 10.1093/molbev/msm162. [DOI] [PubMed] [Google Scholar]
- Backström N, Qvarnström A, Gustafsson L, Ellegren H. Levels of linkage disequilibrium in a wild bird population. Biology Letters. 2006;2:435–438. doi: 10.1098/rsbl.2006.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan C, Edwards S. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata) Genetics. 2009;181:645–660. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proceedings of the National Academy of Sciences USA. 2011;108:E864–E870. doi: 10.1073/pnas.1104032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Bassham S, Wilson T, et al. The population structure and recent colonization history of Oregon threespine stickleback determined using restriction-site associated DNA-sequencing. Molecular Ecology. 2013;22:2864–2883. doi: 10.1111/mec.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender-Bares J, González-Rodríguez A, Eaton DA, Hipp AA, Beulke A, Manos PS. Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Molecular Ecology. 2015;24:3668–3687. doi: 10.1111/mec.13269. [DOI] [PubMed] [Google Scholar]
- Chen J, Rozowsky J, Galeev TR, et al. A uniform survey of allele-specific binding and expression over 1000-Genomes-Project individuals. Nature Communications. 2016;7:11101. doi: 10.1038/ncomms11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combosch DJ, Vollmer SV. Trans-Pacific RAD-Seq population genomics confirms introgressive hybridization in Eastern Pacific Pocillopora corals. Molecular Phylogenetics and Evolution. 2015;88:154–162. doi: 10.1016/j.ympev.2015.03.022. [DOI] [PubMed] [Google Scholar]
- Cutter A. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Baird SE, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–913. doi: 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Cezard T, Fuentes-Utrilla P, Eland C, Gharbi K, Blaxter ML. Special features of RAD sequencing data: implications for genotyping. Molecular Ecology. 2013;22:3151–3164. doi: 10.1111/mec.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierickx EG, Shultz AJ, Sato F, Hiraoka T, Edwards SV. Morphological and genomic comparisons of Hawaiian and Japanese Black-footed Albatrosses (Phoebastria nigripes) using double digest RADseq: implications for conservation. Evolutionary Applications. 2015;8:662–678. doi: 10.1111/eva.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert AJ, Wegrzyn JL, Pande B, et al. Multilocus patterns of nucleotide diversity and divergence reveal positive selection at candidate genes related to cold hardiness in coastal Douglas Fir (Pseudotsuga menziesii var. menziesii) Genetics. 2009;183:289–298. doi: 10.1534/genetics.109.103895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Molecular Biology and Evolution. 2014;31:1275–1291. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MC, Foll M, Excoffier L, Heckel G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis) Molecular Ecology. 2011;20:1450–1462. doi: 10.1111/j.1365-294X.2011.05015.x. [DOI] [PubMed] [Google Scholar]
- Garcia S, Leitch IJ, Anadon-Rosell A, et al. Recent updates and developments to plant genome size databases. Nucleic Acids Research. 2014;42:D1159–D1166. doi: 10.1093/nar/gkt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greminger MP, Stölting KN, Nater A, et al. Generation of SNP datasets for orangutan population genomics using improved reduced-representation sequencing and direct comparisons of SNP calling algorithms. BMC Genomics. 2014;15:16. doi: 10.1186/1471-2164-15-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugger PF, Cokus SJ, Sork VL. Association of transcriptome-wide sequence variation with climate gradients in valley oak (Quercus lobata) Tree Genetics & Genomes. 2016;12:1–4. [Google Scholar]
- Harris C, Rousset F, Morlais I, Fontenille D, Cohuet A. Low linkage disequilibrium in wild Anopheles gambiae s.l. populations. BMC Genetics. 2010;11:81. doi: 10.1186/1471-2156-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. The American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, et al. Finding the genomic basis of local adaptation in non-model organisms: pitfalls, practical solutions and future directions. The American Naturalist. 2016;188:379–397. doi: 10.1086/688018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston RD, Davey JW, Bishop SC, et al. Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics. 2012;13:244. doi: 10.1186/1471-2164-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae) Genetics. 2005;169:945–953. doi: 10.1534/genetics.104.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Good JM. Targeted capture in evolutionary and ecological genomics. Molecular Ecology. 2016;25:185–202. doi: 10.1111/mec.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, Flatt T. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Molecular Biology and Evolution. 2016;33:1317–1336. doi: 10.1093/molbev/msw016. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Kim S, Plagnol V, Hu TT, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nature Genetics. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- Kujur A, Bajaj D, Upadhyaya HD, et al. Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Frontiers in Plant Science. 2015;6:162. doi: 10.3389/fpls.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte M, Rogers SM, Dion-Côté AM, et al. RAD-QTL Mapping reveals both genome-level parallelism and different genetic architecture underlying the evolution of body shape in Lake Whitefish (Coregonus clupeaformis) species pairs. G3: Genes, Genomes, Genetics. 2015;5:1481–1491. doi: 10.1534/g3.115.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Nickerson DA, Anderson AD, et al. Linkage disequilibrium in wild mice. PLoS Genetics. 2007;3:e144. doi: 10.1371/journal.pgen.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH, Merila J. Extensive linkage disequilibrium in a wild bird population. Heredity. 2010;104:600–610. doi: 10.1038/hdy.2009.150. [DOI] [PubMed] [Google Scholar]
- Long AD, Lyman RF, Langley CH, Mackay TF. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics. 1998;149:999–1017. doi: 10.1093/genetics/149.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos KE, Whitlock MC. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Molecular Ecology. 2014;23:2178–2192. doi: 10.1111/mec.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hernandez K, Taylor SH, et al. The genetics of divergence and reproductive isolation between ecotypes of Panicum hallii. New Phytologist. 2015;205:402–414. doi: 10.1111/nph.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Marsden CD, Lee Y, Kreppel K, et al. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3. 2014;4:121–131. doi: 10.1534/g3.113.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD. The extent of linkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177:2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JM, Poissant J, Kijas JW, Coltman DW International Sheep Genomics Consortium. A genome-wide set of SNPs detects population substructure and long range linkage disequilibrium in wild sheep. Molecular Ecology Resources. 2011;11:314–322. doi: 10.1111/j.1755-0998.2010.02918.x. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Ruiz M, Salazar P, et al. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Research. 2014;24:1316–1333. doi: 10.1101/gr.169292.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Projecto-Garcia J, Moriyama H, et al. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genetics. 2015;11:e1005681. doi: 10.1371/journal.pgen.1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Engelstädter J, Rieseberg LH. Recombination rate evolution and the origin of species. Trends in Ecology & Evolution. 2016;31:226–236. doi: 10.1016/j.tree.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Jiggins CD. Towards the identification of the loci of adaptive evolution. Methods in Ecology and Evolution. 2015;6:445–464. doi: 10.1111/2041-210X.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V, Nipper R, Hulke B, Qi L, Schultz Q. De novo sequencing of sunflower genome for SNP discovery using RAD (Restriction site Associated DNA) approach. BMC Genomics. 2013;14:556. doi: 10.1186/1471-2164-14-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nature Reviews Genetics. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pespeni MH, Oliver TA, Manier MK, Palumbi SR. Restriction site tiling analysis: accurate discovery and quantitative genotyping of genome-wide polymorphisms using nucleotide arrays. Genome Biology. 2010;11:R44. doi: 10.1186/gb-2010-11-4-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender WF, Saha MC, Johnson EA, Slabaugh MB. Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theoretical and Applied Genetics. 2011;122:1467–1480. doi: 10.1007/s00122-011-1546-3. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. American Journal of Human Genetics. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current Biology. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujolar JM, Jacobsen MW, Als TD, et al. Genome-wide single-generation signatures of local selection in the panmictic European eel. Molecular Ecology. 2014;23:2514–2528. doi: 10.1111/mec.12753. [DOI] [PubMed] [Google Scholar]
- Puritz JB, Matz MV, Toonen RJ, Weber JN, Bolnick DI, Bird CE. Demystifying the RAD fad. Molecular Ecology. 2014;23:5937–5342. doi: 10.1111/mec.12965. [DOI] [PubMed] [Google Scholar]
- Pyhäjärvi T, García-Gil MR, Knürr T, Mikkonen M, Wachowiak W, Savolainen O. Demographic history has influenced nucleotide diversity in European Pinus sylvestris populations. Genetics. 2007;177:1713–1724. doi: 10.1534/genetics.107.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi ZC, Yu Y, Liu X, et al. Phylogenomics of polyploid Fothergilla (Hamamelidaceae) by RAD-tag based GBS—insights into species origin and effects of software pipelines. Journal of Systematics and Evolution. 2015;53:432–447. [Google Scholar]
- Rausher MD, Delph LF. Commentary: when does understanding phenotypic evolution require identification of the underlying genes? Evolution. 2015;69:1655–1664. doi: 10.1111/evo.12687. [DOI] [PubMed] [Google Scholar]
- Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. A practical guide to environmental association analysis in landscape genomics. Molecular Ecology. 2015;24:4348–4370. doi: 10.1111/mec.13322. [DOI] [PubMed] [Google Scholar]
- Roesti M, Kueng B, Moser D, Berner D. The genomics of ecological vicariance in threespine stickleback fish. Nature Communications. 2015;6:8767. doi: 10.1038/ncomms9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biology. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nature Reviews Genetics. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals — mining genome-wide polymorphism data without big funding. Nature Reviews Genetics. 2014;15:749–763. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- Schweyen H, Rozenberg A, Leese F. Detection and removal of PCR duplicates in population genomic ddRAD studies by addition of a degenerate base region (DBR) in sequencing adapters. The Biological Bulletin. 2014;227:146–160. doi: 10.1086/BBLv227n2p146. [DOI] [PubMed] [Google Scholar]
- Sessions SK, Larson A. Developmental correlates of genome size in plethodontid salamanders and their implications for genome evolution. Evolution. 1987;41:1239–1251. doi: 10.1111/j.1558-5646.1987.tb02463.x. [DOI] [PubMed] [Google Scholar]
- Stanton-Geddes J, Paape T, Epstein B, et al. Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS ONE. 2013;8:e65688. doi: 10.1371/journal.pone.0065688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proceedings of the National Academy of Science USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Ross-Ibarra J. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution. 2014;29:673–680. doi: 10.1016/j.tree.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Truong HT, Ramos AM, Yalcin F, et al. Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PLoS ONE. 2012;7:e37565. doi: 10.1371/journal.pone.0037565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proceedings of the National Academy of Science USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Han F, Wellhagen G, et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nature Genetics. 2014;46:1081–1088. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shahid MQ, Huang H, Wang Y. Nucleotide diversity patterns of three divergent soybean populations: evidences for population-dependent linkage disequilibrium and taxonomic status of Glycine gracilis. Ecology and Evolution. 2015;5:3969–3978. doi: 10.1002/ece3.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JN, Peterson BK, Hoekstra HE. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature. 2013;493:402–405. doi: 10.1038/nature11816. [DOI] [PubMed] [Google Scholar]
- Whiteley AR, Bhat A, Martins EP, et al. Population genomics of wild and laboratory zebrafish (Danio rerio) Molecular Ecology. 2011;20:4259–4276. doi: 10.1111/j.1365-294X.2011.05272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wright KM, Arnold B, Xue K, Šurinová M, O’Connell J, Bomblies K. Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Molecular Biology and Evolution. 2015;32:944–955. doi: 10.1093/molbev/msu398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.