Abstract
It been shown that IL-6 modulates TGF-β1 expression in fibroblasts, however; what role IL-6 plays concerning TGF-βR expression and function in skin is unknown. Therefore, the aim of this study was to investigate the mechanism by which IL-6 might modulates TGF-β receptors in skin.
Skin from WT, IL-6 overexpressing mice, and IL-6 treated keratinocyte cultures were analyzed for TGF-βRI and TGF-βRII expression via histology, PCR, and flow cytometry. Receptor function was assessed by cell migration, bromodeoxyuridine (BrdU) proliferation assays, and Smad7 expression and Smad2/3 phosphorylation. Receptor localization within the membrane was determined by co-immunoprecipitation.
IL-6 overexpression and treatment increased TGF-βRII expression in the epidermis. IL-6 treatment of keratinocytes induced TGF-βRI and II expression, and augmented TGF-β1-induced function as demonstrated through increased migration and decreased proliferation. Additionally, IL-6 treatment of keratinocytes altered receptor activity as indicated by altered Smad2/3 phosphorylation and increased Smad7 and membrane localization.
These results suggest that IL-6 regulates keratinocyte function by modulating TGF-βRI and II expression and signal transduction via trafficking of the receptor to lipid raft pools.
Keywords: Skin, Receptor endocytosis, Smad, Migration, Caveolin
Introduction
Cytokines and growth factors are known to play critical roles in cellular responses and can ultimately control the function of the cell. These mediators can serve as molecular signals between cells to communicate changes in the environment or disruptions to homeostasis, which can occur following an injury. The wound environment is characterized by multiple cellular events within interdependent stages including inflammation, tissue formation, and tissue remodeling. Cutaneous homeostasis is maintained by permanent cross-talk between dermal fibroblasts, epidermal keratinocytes, and through the production of cytokines. A paradoxical partnership between cytokines such as interleukin (IL)-6 and transforming growth factor (TGF)-β, have long been suspected as a mediator of keratinocyte-fibroblast crosstalk in the wound (1). Yet, few studies characterizing the mechanism of this partnership exist.
IL-6 is a pleiotropic cytokine that plays a critical role in the inflammatory response as well as in growth and differentiation for numerous cell types (2). In the skin, IL-6 is produced by dermal and epidermal cells while other cells such as macrophages, B and T cells, and endothelial cells represent secondary sources (3). Dysregulation of IL-6 has been implicated in multiple disease states including rheumatoid arthritis, inflammatory bowel disease, and psoriasis (3). Wounds from IL-6 deficient mice demonstrate multiple healing defects including, delayed re-epithelialization, decreased granulation tissue, inhibited neovascularization and greatly increased time to closure, when compared to their wild type counterparts (4, 5). While the involvement of IL-6 in the inflammatory process has been well established, much is still unknown about its role in skin.
The TGF-β family consists of TGF-β1-3, bone morphogenic proteins (BMP), and activins. Of the 3 TGF-β isoforms, TGF-β1 predominates in cutaneous repair exerting pro- and anti-proliferative effects in numerous cell types, influencing migration, and stimulating the production of extracellular matrix proteins (6, 7). All isoforms are produced in wounds by macrophages, fibroblast, keratinocytes, and platelets. Mouse models deficient for TGF-β1 and its downstream signaling proteins, Smads, demonstrate multiple wound healing deficiencies (8). The consequences of TGF-β deficiency on re-epithelialization are of particular interest because of the molecule's ability to induce keratinocyte differentiation and subsequent migration, as these are essential processes in re-epithelialization (9). Indeed, late stage wounds from TGF-β1 knockout animals demonstrated severely impaired granulation tissue formation, re-epithelialization, and collagen deposition; however, these impairments are accompanied with an unchecked inflammatory response which leads to severe wasting (10). Thus, it is difficult to discern whether wound healing impairments are a direct consequence of TGF-β1 deficiency or overall physiological effects associated with this animal model.
TGF-β1 exerts is biological activity by binding to a heterotetrameric receptor complex composed of TGF-β type I receptor (TGF-βRI) and TGF-β type II receptor (TGF-βRII) dimers (7). Ligand binding leads to a formational change in which TGF-βRII phosphorylates and activates TGF-βRI. The activated TGF-βRI recruits R-Smads (Smad2/3 and Smad4) to the receptor where they are phosphorylated and then translocate to the nuclease to modulate transcription of target genes (11). In addition to inhibitory Smads, such as Smad7, TGF-β1 signaling can be governed by receptor localization within the membrane, specifically within the lipid raft domain vs. non-lipid raft. Di Guglielmo et al. previously reported that clathrin internalization into the early endosome antigen-1 (EEA-1) positive endosome promotes TGF-β signaling, while lipid raft-caveolin internalization pathway (with the help of Smad7) promotes receptor degradation (12).
Previous studies concerning the relationship between IL-6 and TGF-β have not provided a definitive mechanistic link between the mediators. Concerning its role in dermal-epidermal crosstalk, IL-6 has been shown to induce keratinocyte migration only when cultured in the presence of fibroblasts (13). This finding seems to indicate that IL-6 facilitates re-epithelialization through its partnership with a soluble effector produced by fibroblasts, such as TGF-β1. The present study aims to investigate the relationship of keratinocyte function induced by IL-6 and its association with TGF-β receptor expression and membrane localization. Herein it was found that IL-6 appears to indirectly modulate keratinocyte activity through functional augmentation of both TGF-β receptors.
Results
IL-6 induces TGF-βRI and TGF-βRII expression in keratinocytes
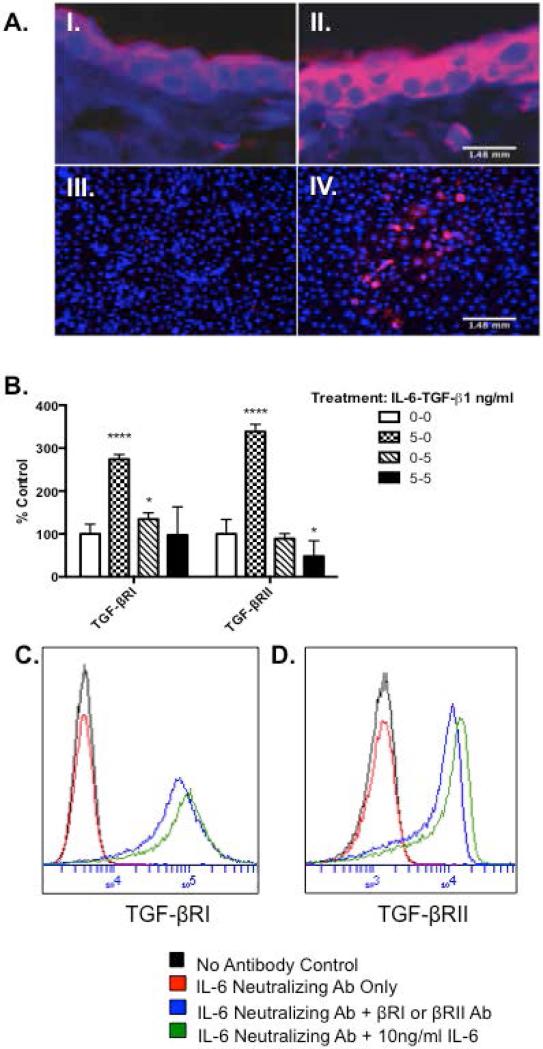
It was previously shown that intradermal injection of IL-6 induces TGF-βRII in IL-6 KO mice (16). To further confirm these findings, epidermal TGF-βRII expression was determined in mice that specifically over express IL-6 in skin (14). Immunohistochemistry revealed that epidermal TGF-βRII expression was increased as compared to control (Fig. 1A-I and II). To examine the effect of IL-6 treatment on isolated mouse epidermal keratinocytes, cells from IL-6KO mice (KO) were cultured as described in Methods and treated with IL-6. TGF-βRII was robustly increased in IL-6 treated KO keratinocytes when compared to control (Fig. 1A-III and IV). To determine whether IL-6 also modulates TGF-βRI expression, and see if this modulation translates in a human keratinocyte in vitro model, quantitative RT-PCR and flow cytometric analysis were utilized to examine mRNA and protein levels for both receptors in human epidermal keratinocytes (Ker-CT) after IL-6 +/− TGF-β1 treatment. PCR analysis revealed IL-6 significantly modulated both receptors, while TGF-β1 alone significantly increased TGF-βRI expression. Treatment with IL-6 and TGF-β1 significantly decreased TGF-βRII expression as compared to control (Fig. 1B). To minimize the effects of endogenously produced IL-6, cultures were incubated with IL-6 neutralizing antibody prior to treatment. Similarly, flow cytometry revealed IL-6 robustly increased TGF-βRI (Fig. 1C) and TGF-RII (Fig. 1D) when compared to control.
Figure 1. Interleukin (IL)-6 augments epidermal expression of transforming growth factor-β type I and II receptor (TGF-βRI and II).
Punch biopsies (4mm) obtained from wild type (A-I.) or transgenic IL-6 over-expressing mice (A-II.) were formalin-fixed, and paraffin-embedded for histology. IL-6 KO keratinocytes were treated with 0ng/ml (A-III) or 10ng/ml (A-IV) IL-6 for 16 hours, then fixed with paraformaldehyde. Slides were stained with monoclonal anti-TGF-βRII (red) followed by Alexafluor 546 conjugated secondary antibodies and DAPI (blue) counterstain. Slides were visualized at 20X objective. (B) Ker-CTs were cultured and treated as described. Treatments indicated as ng/ml of IL-6-TGF-β1. Expression of TGF-βRI and II mRNA were determined by SYBR Green real-time PCR. Data are expressed as percent negative control of average CT fold change ± percent SEM (*= p< 0.05, n=3 cultures per data point). (C-D) Ker-CTs were cultured as described, treated with IL-6 neutralizing antibody and +/− IL-6. Flow cytometry for TGF-βRI and II was performed and histograms for each are presented. (C) TGF-βRI (D) TGF-βRII.
IL-6 alters the function of TGF-β1 in IL-6 KO epidermal cells
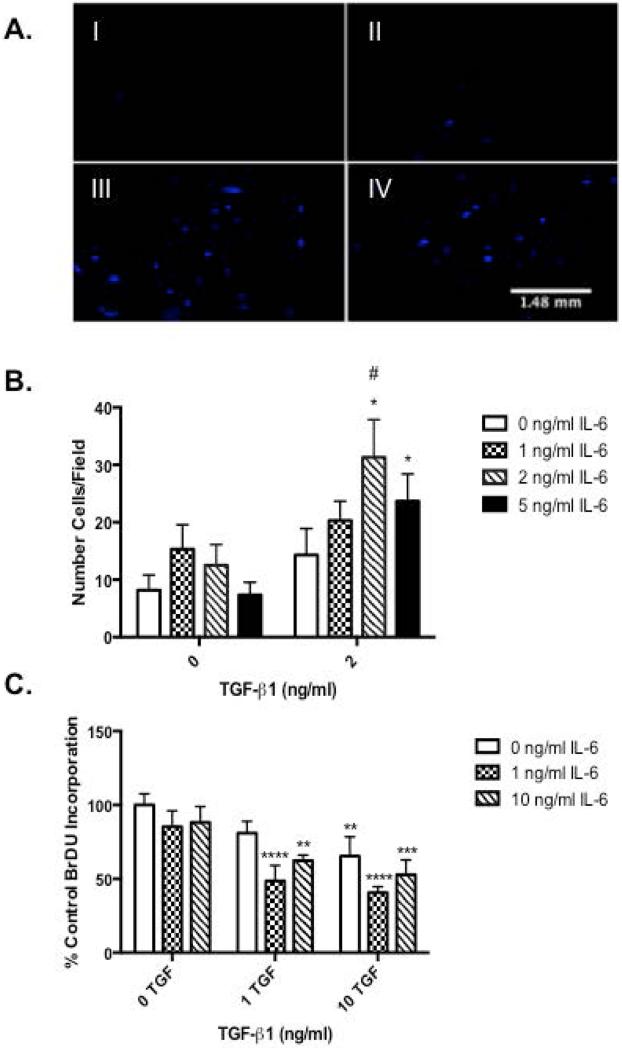
TGF-β1 is a known motogen of keratinocytes (6, 7, 9). To investigate the functional consequences of IL-6 modulation of TGF-βR expression, the migration and proliferation of IL-6 +/− TGF-β1-treated KO keratinocytes were assessed. Treatment with 2 ng/ml TGF-β1 alone showed a slight increase in migration, however this increase was not significant. Migration was significantly increased in a dose response manner with a maximal response following pre-treatment with IL-6 followed by TGF-β1 as determined by Transwell assay (Fig. 2A-I vs. III and B). Various studies have shown in respect to skin, TGF-β1 induces proliferation of dermal fibroblasts but suppresses growth of epidermal keratinocytes (17). Therefore, to assess whether IL-6 affects the anti-proliferative activity of TGF-β1, BrdU assays were performed on IL-6 and TGF-β1-treated KO keratinocytes. A significant reduction in BrdU incorporation was seen with 10ng/ml TGF-β1 alone as compared to control, while pre-treatment with IL-6 significantly augmented this response (Fig. 2C).
Figure 2. Interleukin (IL)-6 modulates transforming growth factor (TGF)-β1 induced migration and anti-proliferative effects.
(A) IL-6 knockout (KO) keratinocytes were seeded on a porous (8μm) membrane Transwell culture insert and incubated for 2 hours before 16-hour TGF-β1 and IL-6 treatment. Non-migratory cells were removed and remaining cells stained with DAPI. Migration was visualized using representative photographs under a fluorescent microscope. Treatments indicated as ng/ml of IL-6-TGF-β1: (I) 0-0ng/ml, (II) 0-2ng/ml, (III) 2-2ng/ml, (IV) 5-2ng/ml. Migration was quantified using Image J particle analysis (B) Data expressed as mean number of migratory cells ± SEM/field (*=p<0.05, n=6). (C) Isolated IL-6 knockout (KO) keratinocytes were cultured and treated with various concentrations of IL-6 and TGF-β1. Bromodeoxyuridine (BrdU) incorporation was quantified via commercial ELISA. Data expressed as percent of the negative control optical density (OD) value ± SEM (* = p< 0.05 different from 0ng/ml IL-6, # = p< 0.001 different from 0ng/ml TGF-β1, n=6).
IL-6 modulates TGF-β1 induced signal transduction through Smad2/3 phosphorylation and increases Smad7 expression
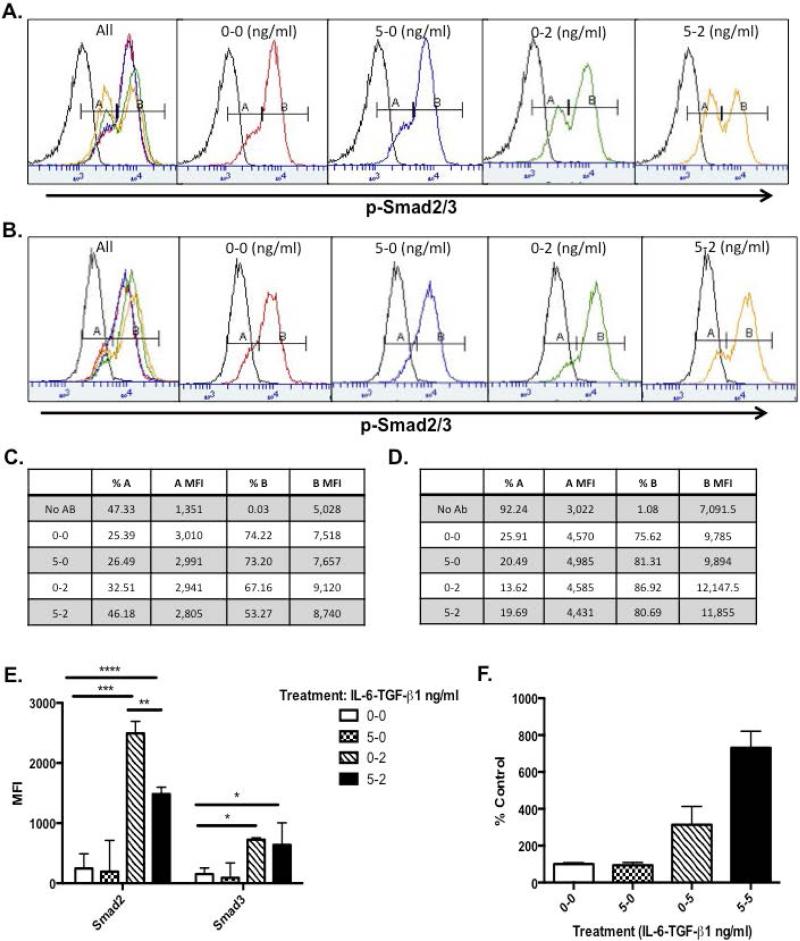
TGF-β1 mediated activation of the TGF-β receptor complex initiates signal transduction through the Smad2/3 and 4 pathway (7). To determine if IL-6 modulation of TGF-βRI and II expression alters signal transduction through the receptor, Ker-CT cells were pre-incubated with IL-6 and/or followed by treatment with TGF-β1. Flow cytometric analysis revealed two cell populations, p-Smadlow (peak A) and p-Smadhigh (peak B). After IL-6 treatment, overall p-Smad fluorescence intensity slightly decreased, while the percentage of p-Smadhigh population decreased and the p-Smadlow cells increased (Fig. 3A and 3C, Column 3 and 6). Following TGF-β1 treatment a similar change was seen in both peaks, albeit more pronounced (Fig. 3A and 3C, Column 4). When the treatments were applied in combination, there was a further enhancement of the shift from the high to low population (Fig. 3A and C, Column 5) indicating an additive effect. To further confirm this effect was modulated by IL-6, the Stat3 inhibitor, LLL-12, (18) was used. Upon inhibition of Stat3, a small increase in p-Smadlow was apparent after IL-6 treatment (Fig. 3B and D, Column 3), but decreased upon TGF-β1 only and IL-6 pre-treatment (Fig. 3B and D, Column 3). However, following TGF-β1 treatment, a profound increase in the p-Smadhigh cell population (peak B) was observed accompanied by increased intensity of staining of this population (Fig. 3B and D). Interestingly, the decrease in peak B cells that was seen in Fig. 3A with the combination treatment of IL-6 and TGF-β1 was no longer present when treated with LLL-12, indicating Stat3 involvement (Fig. 3 C vs. D, Row 6). Multiplex analysis also confirmed that upon IL-6 treatment alone, Smad2/3 phosphorylation was minimal, while TGF-β1 alone showed a marked increase in Smad2/3 phosphorylation (Fig. 3E). Similar to Fig. 3A, pre-treatment with IL-6 followed by TGF-β1 treatment, exhibited a significant decrease in Smad2/3 phosphorylation as compared to TGF-β1 treatment alone. IL-6 KO keratinocytes were also treated in the same manner and multiplex data showed very similar phosphorylation trends (data not shown). Furthermore, research has indicated that expression of the inhibitory Smad7 can also be directly correlated with the transcription from TGF-βR target genes in human keratinocytes and is associated with inhibition of Smad2/3 signaling (19). Ker-CT cultures were treated with IL-6 and/or TGF-β1 and Smad7 expression was measured via quantitative PCR. Pre-treatment with IL-6 and TGF-β1 treatment showed a much more pronounced increase in Smad7 mRNA expression as compared to IL-6 or TGF-β1 treatment alone further corroborating the link between IL-6 and TGF-β1 signaling (Fig. 3F).
Figure 3. Interleukin (IL)-6 modulates Smad2/3 phosphorylation and enhances transcription of Smad7 in epidermal keratinocytes.
(A) Ker-CTs were cultured and treated with IL-6 for 1 hour followed by TGF-β1 for 30 minutes. Treatments indicated as ng/ml of IL-6-TGF-β1 and p-Smad and p-Stat3 histograms are presented. (B) Ker-CT cells treated as in (A) with addition of LLL-12. (C-D) Mean fluorescent data of histograms in A and B. (E) nHEK protein lysates were isolated and multiplex analysis was performed using the Millipore TGF- β Signaling 6-plex kit. (F) Ker-CTs were cultured and treated and total RNA was isolated. Expression of Smad7 mRNA was determined by real-time PCR. Data are expressed as percent negative control average CT fold change ± percent SEM (*= p< 0.05, n=3).
IL-6 alters localization of TGF-βRI and II within the cell membrane following IL-6 treatment in human epidermal cells
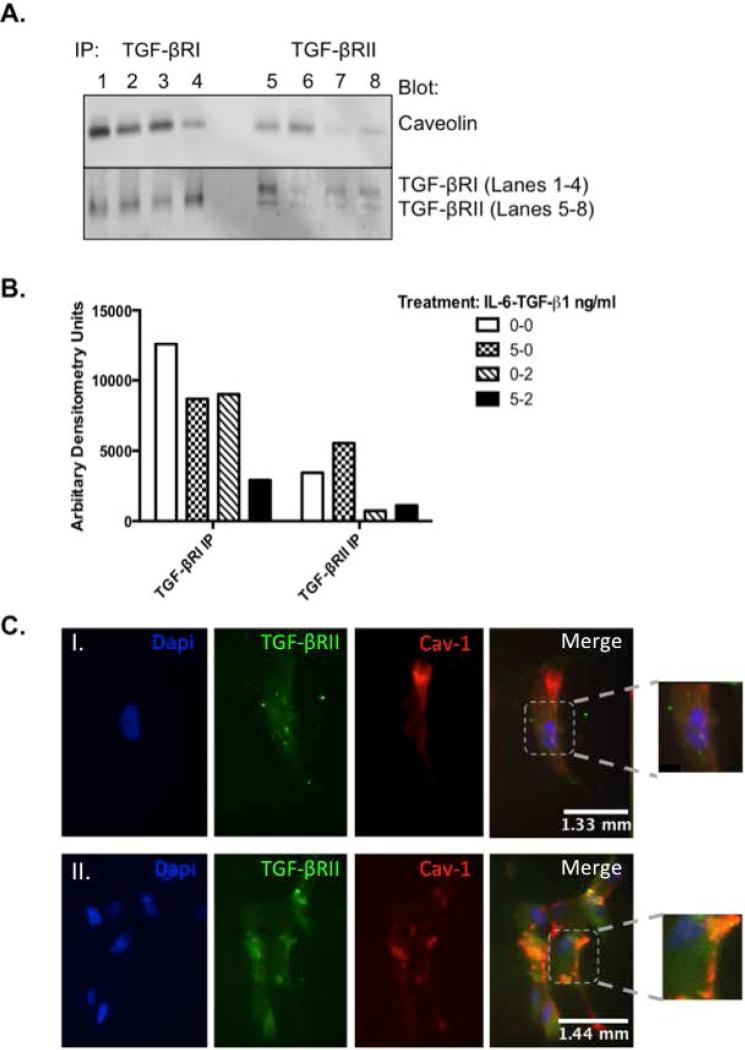
As the multi-functional role of TGF-β1 has been well documented, the spatial organization of the receptor and its signaling components is less clear. Receptor activation not only initiates signal transduction, but also triggers clathrin or non-clathrin mediated receptor endocytosis, which serves to further regulate TGF-β1 activity. Therefore, to determine if IL-6 plays a role in TGF-βR mediated endocytosis, Ker-CT cells were treated with IL-6 and/or TGF-β1 and immunoprecipitation and immunohistochemistry assays were performed. As shown in Figure 4A and B, treatment with 5ng/ml IL-6 induced TGF-βRII immunoprecipitation with caveolin as compared to no IL-6 treatment (Lane 5 vs. 6) but decreased the association with TGF-βRI (Lane 1 vs. 2). Activation of the receptor with 2ng/ml TGF-β1 significantly decreased this association of TGF-βRII with caveolin but had no effect on the TGF-βRI interaction. Pre-treatment with IL-6 followed by TGF-β1 significantly decreased both receptors interaction with caveolin compared to control (Lanes 1 vs. 4 and 5 vs. 8). Similarly, immunohistochemistry revealed diffuse, punctate staining of TGF-βRII (green) and little to no co-localization with caveolin (red) in the absence of IL-6 treatment (Fig. 4C-I). However, upon IL-6 treatment, TGF-βRII and caveolin expression increased and co-localized within the same area (Fig. 4C-II, orange). Co-localization with clathrin heavy chain and the early endosomal antigen (EEA-1) were also assessed and found that IL-6 treatment did not increase co-localization (Results not shown).
Figure 4. Interleukin (IL-6) alters localization of transforming growth factor (TGF)-βR within the cell membrane following IL-6 treatment in human epidermal cells.
(A) Ker-CTs were cultured as described and treated with IL-6 for 16 hours followed by TGF-β1. Cell lysates were immunoprecipitated with anti TGF-βRI or II (bottom panel) and blots were incubated with anti-caveolin (top panel). Treatments indicated as ng/ml of IL-6-TGF-β1: Lanes 1 and 5 = 0-0ng/ml, Lanes 2 and 6 = 5-0ng/ml, Lanes 3 and 7 = 0-2ng/ml, Lanes 4 and 8 = 5-2ng/ml. (B) nHEKs were cultured in 8-chamber slides and treated with: (I) 0ng/ml and (II) 10ng/ml IL-6. Slides were stained with anti-TGF-βRII (green) and anti-caveolin (red) and detected using Alexa-Fluor 488 or 546 conjugated secondary antibodies, respectively. Stained slides were visualized at 40x objective; photographs are representative of three replicate experiments.
Discussion
Regeneration of dermal and epidermal tissue following cutaneous wounding is a complex process involving the intricate coordination of many cell types in a sequential and predictable pattern. IL-6 and TGF-β are key factors in the normal procession through these events, and both knock out mice display multiple defects in repair (4, 5). Similar to IL-6, both TGF-βRI and II are minimally distributed in areas such as the epidermis, hair follicles, and sebaceous glands, all of which express TGF-β ligands (21) (22). Following injury, receptor expression increases in the epidermis adjacent to the wound, as does IL-6 expression and tends to persist in older wounds (6). Herein, it was demonstrated that IL-6 treatment of keratinocytes enhances expression of TGF-βRI and II as compared to controls (Fig. 1A-D). These results are similar to those found by Zhou, D. et al., which showed IL-6 increased the number of cells expressing TGF-β receptors in the spleen (23).
The low levels of TGF-β produced by monocultures of keratinocytes and fibroblasts can be increased substantially through co-culturing these two cell types (1). These findings and the motogenic properties of TGF-β are particularly interesting when considering that IL-6 has been shown to indirectly induce keratinocyte migration in conjunction with a soluble factor secreted by the dermal fibroblasts (13). Furthermore, IL-6 treatment has been shown to increase TGF-β1 mRNA expression in a dose-response manner in isolated fibroblasts and dermis (14). This evidence demonstrates a partnership between the epidermis and the dermis with regards to TGF-β expression and function seemingly driven by IL-6. While IL-6/TGF-β cooperation is well documented in the realm of collagen deposition and myofibroblast contraction (14, 24-26), few studies have characterized this partnership with respect to re-epithelialization. The present data demonstrates IL-6 pre-treatment elicits keratinocyte migration in response to TGF-β1 to a greater extent than TGF-β1 alone (Fig. 2A-B). These results support the notion of IL-6 mediating the keratinocyte-fibroblast interaction exhibited during wound healing through its regulation of the TGF-β ligand and receptor. While often conflicting, TGF-β has been demonstrated to modulate cell proliferation (6). Several studies have demonstrated an inhibitory effect on keratinocyte proliferation (27) (16) (28) whereas other have shown that overexpression of TGF- β1 increases the proliferative phenotype of keratinocytes (29) (30). In our cell system, it was anticipated that following IL-6 treatment, the anti-proliferative activity of TGF-β would be augmented if in fact it facilitates TGF-β mediated migration of these cells and that was what was demonstrated (Fig. 2C). Although still significantly different, the decrease in proliferation in response to TGF- β1 may seem lower than expected. However, with the exact modulatory nature of TGF-β1 conflicting as mentioned above, as well as variations in cell types and culture environment, these results are not necessarily surprising. This further illustrates the complexity of TGF-β signaling in wound healing and emphasizes the importance in which small changes in growth factor expression and timing may play a role in the cellular activity or function of a cell.
It is well established that TGF-β signal transduction is carried out through the intricate and highly regulated Smad2/3 signaling pathway, which is involved in repair by regulating migration, proliferation, and differentiation (30-32). In Smad3 null mutant mice, inflammatory infiltration into the dermis as well as keratinocyte proliferation are altered during repair (20). Unfortunately, little is known regarding the role of Smad2 in repair as the null genotype is embryonic lethal (33). Herein it is shown that pre-treatment with IL-6 and addition of TGF-β1 resulted in a marked increase in function of TGF-βR as evidenced by increased migration (Fig. 2B). Further, it was found that following treatment, keratinocyte cultures form two distinct populations, an apparent p-Smadlow and p-Smadhigh (Fig. 3A and B, peaks A and B, respectively). While not previously reported, this may indicate cells at different stages during the signaling process, where peak A is perhaps recycling (or post-signaling) and peak B is actively signaling as indicated by increased fluorescence intensity (Fig. 3C and D, Column 3 vs. 5). Inhibition of Stat3 seems to alter this population of cells indicating a potential mechanism for the shift from Smadhigh to the Smadlow population. Thus, this may be indicative of very active TGF-βR signaling in these cells. Interestingly, increased Smad2/3 expression as well as increased Smad2/3 phosphorylation has been shown to exhibit an inhibitory effect on keratinocyte migration during re-epithelialization (34) while increased Stat3 can induce migration and inhibit differentiation (35) (36). This may indicate that peaks A and B could be separate populations of differentiated and undifferentiated cells. However, this was not assessed directly herein and further study will be needed to determine this definitively.
Inhibitory (I)-Smads can terminate TGF-β signal transduction through a variety of complex mechanisms. Most notably, I-Smads aid in the degradation of Smad2/3 and Smad4 and exert feedback inhibitory effects on transcription of TGF-β induced genes (37-39). Herein, it was demonstrated that Smad7 mRNA expression appears to be directly correlated with TGF-β1 treatment in keratinocytes as indicated by a significant increase of mRNA expression (Fig. 3F). Furthermore, pre-treatment with IL-6 enhances TGF-β1 induced Smad7 mRNA expression supporting the observed decrease in TGF-βR signaling (Fig. 3). These results were similar to studies that identified a molecular link by which Smad7 mediates crosstalk with IL-6 mediated Stat3 hyperactivation and attenuation of TGF-βR signaling (40). It was shown that increased Stat3 activation in mouse embryonic fibroblasts treated with an IL-6/sIL-6r fusion protein resulted in impaired phosphorylation and nuclear translocation of Smad2 (40). Additionally, Stat3 directly interacts with Smad3 and interferes with the Smad3-Smad4 complex (43) and activated Stat3 also induces Smad7 expression to desensitize TGF-βR signaling (40). Indeed, IL-6 induced Stat3 expression and activation is increased in injured epidermis as compared to uninjured epidermis (42); therefore, Stat3 activation may be a modulatory link between IL-6R and TGF-βRII function during repair. Inhibition of Stat3 signaling, through the use of the novel small molecule inhibitor LLL-12, further supports this association in Figure 3B, where TGF-β1-induced signaling significantly increased upon inhibition of Stat3.
TGF-β receptors are constitutively internalized via clathrin dependent or lipid-raft (caveolin rich) dependent endocytic pathways (39) (43) (44) upon ligand binding to regulate receptor activity. The balance between the two internalization pathways is critical in modulating receptor activity. Data presented herein show an increased association between caveolin and TGF-βRII following IL-6 treatment but decreased TGF-βRI association with caveolin, although the interaction is still considerably higher than TGF-βRII association. Interestingly, the reciprocal immunoprecipitation found that only TGF-βRI showed association with caveolin and IL-6 and TGF- β1 increased this association (data not shown). Irrelevant IgG antibody was also used as a control and no bands were indicated confirming the specificity of our pull down (Data not shown). This suggests IL-6 may modulate TGF-β activity by trafficking the receptor to the lipid raft portion of the membrane (Fig. 4). These data support previous findings that a direct interaction occurs between both TGF-βRI and II, and caveolin (45) (46). These interactions have functional consequences as evidenced by a significant decrease in Smad2 phosphorylation upon receptor activation (Fig 3-4). Interestingly, a previous study in human renal cells, found that IL-6 increased compartmentalization of TGF-βRI and II into the non-lipid raft portion of the cell membrane, which further enhances TGF-β signaling (47). Thus, it might be concluded in the current model, IL-6 induced compartmentalization of TGF-βR into lipid rafts would decrease signaling and indeed, this is the case (Fig. 3). This would seem paradoxical when considering the apparent increase in TGF-βRI and II expression and function in keratinocytes when pre-treated with IL-6 (Figs. 1-2). However, it may be that this apparent IL-6 mediated shift into the lipid raft is a regulatory mechanism initiated to keep the resulting increase of TGF-βR expression and further or persistent signaling in check through increased receptor turnover. It was recently shown that TGF- βRII was detected in EEA-1 positive compartments and as inflammation progressed (by day 5), the receptor appeared in caveolin-1 positive intracellular structures as well, indicating a potential mechanism to control TGF- βR activity in inflammatory conditions (46). Perhaps once receptor expression attains a certain threshold or a particular Smad-regulated function is required (such as migration), TGF-βRII is directed into the lipid raft domain ultimately resulting in degradation of the receptor in an attempt to terminate any potential (over) signaling, and this may be represented by peak A in figure 3. Interestingly, kinase activation by bFGF and PDGF signaling was suppressed by manipulation of caveolin-1 expression itself, which is likely to influence trafficking of the receptor (48). Similarly, our studies found that in nHEKs treated with IL-6, total caveolin protein expression increased in a dose response manner (data not shown). It's also shown that the ratio of TGF-β binding to either TGF-βR can also determine the internalization pathway (11) and IL-6 may modulate the receptor expression and ultimately alter this ratio. Its also been shown in that 10% of IL-6R subunits associate with lipid rafts (49) (50) and treatment with a lipid raft inhibitor disrupted IL-6 signaling further supporting this link.
Herein, it was demonstrated that IL-6 plays a role in keratinocyte migration and proliferation through modulation of TGF-βR expression and function. The current findings, when viewed with previous results, illustrate a model of IL-6 involvement in skin TGF-β function where enhanced TGF-β1 secretion in the dermis (16) and upregulation of TGF-βR in the epidermis may facilitate the differentiation of epidermal keratinocytes to the pro-migratory phenotype observed during reepithelialization. However, further research into the mechanisms by which IL-6 mediates receptor upregulation and membrane localization is necessary for a more complete understanding of the IL-6/TGF-β interaction in skin and in the dynamic process of wound healing.
Materials and Methods
Mice
Experimental animals were treated in accordance with the criteria outlined in the PHS Policy on Humane Care and Use of Laboratory Animals and The Guide for the Care and Use of Laboratory Animals (NIH publication 86-23), in facilities accredited by AAALAC. IL-6 deficient mice were acquired from Jackson Laboratories (Bar Harbor, ME). Frozen embryos of mice that overexpressed IL-6 in epidermis via a K14-IL-6 promoter construct (14) were graciously provided by Dr. Elaine Fuchs (Rockefeller Institute). Embryos were revived by Jackson Laboratories, and maintained at the OUHSC rodent barrier facility.
Epidermal keratinocyte isolation and culture
IL-6 knockout (KO) epidermis was collected from neonatal mice and isolated as previously described (13). Neonatal human epidermal keratinocytes (Ker-CT, ATCC CRL-4048 and nHEKs, ThermoFisher, Waltham, MA) were cultured according to manufactures protocol. Cells were treated with IL-6 (overnight for protein expression studies or 1 hour for signaling assays) or in combination with TGF-β1 (30 minutes) or specified otherwise. IL-6 neutralizing antibody (Biolegend, San Diego, CA) was used at 500ng/ml and incubated overnight followed by treatment. LLL-12 was used at .5μM for 1 hour.
Transwell migration assay
Migration of IL-6 KO keratinocytes was assessed via Transwell assay as described (52) and quantified using Image J (NIH).
Proliferation assays
IL-6 KO keratinocytes were pre-treated with IL-6 for 6 hours followed by +/− TGF- β1. Bromodeoxyuridine (BrdU) (EMD, Gibbstown, NJ) was applied immediately following TGF-β1 treatment according to manufacturer's instructions and incubated overnight. Results were obtained via a 96-well plate reader (Molecular Devices, Sunnyvale, CA, U.S.A).
Semi-quantitative RT-PCR reaction
Total RNA from Ker-CT cultures was prepared and cDNA was synthesized as described previously (13). Primers for TGF-BRI and II, Smad7, and 28s rRNA were constructed from Genbank sequences utilizing Amplify for Mac 3.1.4 (Bill Engles, University of Wisconsin), and synthesized by Life Technologies (Carlsbad, CA). Quantitative real time (RT)-PCR utilizing SYBR green was performed on an ABI PRISM 7000 SDS. Expression differences were determined relative to 28S rRNA utilizing the ΔΔCt method.
Multiplex Assay
nHEKs were cultured as indicated and treated with IL-6 for 16-18hr followed by TGF-β1 for 30 minutes. TGF- β1, p-Smad2, and p-Smad3 expression was measured using Millipore Milliplex Map Kit (Cat# TGFBMAG-64K-03 and 48-614MAG) according to manufactures protocol and analyzed on a Bioplex 200 (Biorad, Hercules, CA)
Immunoprecipitation Assay
Ker-CT cultures were cultured and treated as described. Protein G Dynabeads Immunoprecipitation Kit was used according to manufactures protocol (ThermoFisher). Western blot was performed as described (25). Anti-caveolin (Cell Signaling, Danvers, MA), anti- TGF-βRII (Cell Signaling), anti-TGF-βRI (Abcam, Cambridge, MA), were used at the recommended dilution and IR labeled secondary antibody(s) (Licor, Lincoln, NE) were used at 1:10000 – 1:20,000. Blots were imaged via the Licor Oddyssey CLx imaging system (Licor).
Flow Cytometric Analysis
Ker-CT cells were cultured and treated with IL-6 and/or TGF-β1. Single cell suspensions were obtained and TGF-βRI (Abcam), p-Smad-PE (Cell Signaling) and p-Stat3-488 (Cell Signaling) antibodies were used according to recommended dilutions and analyzed via the Accuri C6 system.
Statistical Analysis
All experiments were replicated, and representative findings are shown. Statistical significance was determined by two-way analysis of the variance. In all statistical comparisons, a P-value of < 0.05 was used to indicate a significant difference.
Acknowledgements
Funding for the following research was graciously provided by NIH/NIGMS R01 GM67745-01A1 and CDC/NIOSH R01 OH010241-01.
Footnotes
Lerin R. Luckett-Chastain and Mackenzie L. Cottrell designed and performed the research, analyzed the data, and wrote the paper.
Bethany Kawar assisted with performing the research.
Michael A. Ihnat contributed essential reagents and tools.
Randle M Gallucci contributed to critically revising and providing final approval of the manuscript.
Conflict of Interest
The authors state no conflict of interest.
References
- 1.Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal PB. Interleukin-6: molecular pathophysiology. The Journal of investigative dermatology. 1990;94:2S–6S. doi: 10.1111/1523-1747.ep12874963. [DOI] [PubMed] [Google Scholar]
- 3.Paquet P, Pierard GE. Interleukin-6 and the skin. International archives of allergy and immunology. 1996;109:308–317. doi: 10.1159/000237257. [DOI] [PubMed] [Google Scholar]
- 4.Lin ZQ, Kondo T, Ishida Y, et al. Essential involvement of IL-6 in the skin wound-healing process as evidenced by delayed wound healing in IL-6-deficient mice. J Leukoc Biol. 2003;73:713–721. doi: 10.1189/jlb.0802397. [DOI] [PubMed] [Google Scholar]
- 5.Gallucci RM, Simeonova PP, Matheson JM, et al. Impaired cutaneous wound healing in interleukin-6-deficient and immunosuppressed mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:2525–2531. doi: 10.1096/fj.00-0073com. [DOI] [PubMed] [Google Scholar]
- 6.Barrientos S, Stojadinovic O, Golinko M, et al. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 7.Miyazawa K, Shinozaki M, Hara T, et al. Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 8.Grose R, Werner S. Wound-healing studies in transgenic and knockout mice. Molecular biotechnology. 2004;28:147–166. doi: 10.1385/MB:28:2:147. [DOI] [PubMed] [Google Scholar]
- 9.Seomun Y, Kim J, Joo C. MMP-14 mediated MMP-9 expression is involved in TGF-beta1-induced keratinocyte migration. J Cell Biochem. 2008;104:934–941. doi: 10.1002/jcb.21675. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu H, Miyajima H, Kondo M, et al. [A case report of variegate porphyria]. Rinsho shinkeigaku = Clinical neurology. 1995;35:1221–1224. [PubMed] [Google Scholar]
- 11.Huang F, Chen YG. Regulation of TGF-beta receptor activity. Cell & bioscience. 2012;2:9. doi: 10.1186/2045-3701-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Guglielmo GM, Le Roy C, Goodfellow AF, et al. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nature cell biology. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 13.Gallucci RM, Sloan DK, Heck JM, et al. Interleukin 6 indirectly induces keratinocyte migration. The Journal of investigative dermatology. 2004;122:764–772. doi: 10.1111/j.0022-202X.2004.22323.x. [DOI] [PubMed] [Google Scholar]
- 14.Turksen K, Kupper T, Degenstein L, et al. Interleukin 6: insights to its function in skin by overexpression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5068–5072. doi: 10.1073/pnas.89.11.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallucci RM, Lee EG, Tomasek JJ. IL-6 Modulates Alpha-Smooth Muscle Actin Expression in Dermal Fibroblasts from IL-6-Deficient Mice. J Invest Dermatol. 2006 doi: 10.1038/sj.jid.5700109. [DOI] [PubMed] [Google Scholar]
- 16.Luckett-Chastain LR, Gallucci RM. Interleukin (IL)-6 modulates transforming growth factor-beta expression in skin and dermal fibroblasts from IL-6-deficient mice. The British journal of dermatology. 2009;161:237–248. doi: 10.1111/j.1365-2133.2009.09215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellheyer K, Bickenbach JR, Rothnagel JA, et al. Inhibition of skin development by overexpression of transforming growth factor beta 1 in the epidermis of transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5237–5241. doi: 10.1073/pnas.90.11.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onimoe GI, Liu A, Lin L, et al. Small molecules, LLL12 and FLLL32, inhibit STAT3 and exhibit potent growth suppressive activity in osteosarcoma cells and tumor growth in mice. Invest New Drugs. 2012;30:916–926. doi: 10.1007/s10637-011-9645-1. [DOI] [PubMed] [Google Scholar]
- 19.Afrakhte M, Moren A, Jossan S, et al. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-beta family members. Biochem Biophys Res Commun. 1998;249:505–511. doi: 10.1006/bbrc.1998.9170. [DOI] [PubMed] [Google Scholar]
- 20.Ashcroft GS, Yang X, Glick AB, et al. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nature cell biology. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- 21.Bandyopadhyay B, Fan J, Guan S, et al. A “traffic control” role for TGFbeta3: orchestrating dermal and epidermal cell motility during wound healing. The Journal of cell biology. 2006;172:1093–1105. doi: 10.1083/jcb.200507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurnberg W, Haas N, Schadendorf D, et al. Interleukin-6 expression in the skin of patients with lupus erythematosus. Experimental dermatology. 1995;4:52–57. doi: 10.1111/j.1600-0625.1995.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D, Chrest FJ, Adler W, et al. Increased production of TGF-beta and Il-6 by aged spleen cells. Immunology letters. 1993;36:7–11. doi: 10.1016/0165-2478(93)90061-6. [DOI] [PubMed] [Google Scholar]
- 24.Tomasek JJ, Gabbiani G, Hinz B, et al. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 25.Vaughan MB, Howard EW, Tomasek JJ. Transforming growth factor-beta1 promotes the morphological and functional differentiation of the myofibroblast. Experimental cell research. 2000;257:180–189. doi: 10.1006/excr.2000.4869. [DOI] [PubMed] [Google Scholar]
- 26.Amendt C, Schirmacher P, Weber H, et al. Expression of a dominant negative type II TGF-beta receptor in mouse skin results in an increase in carcinoma incidence and an acceleration of carcinoma development. Oncogene. 1998;17:25–34. doi: 10.1038/sj.onc.1202161. [DOI] [PubMed] [Google Scholar]
- 27.Mazzieri R, Jurukovski V, Obata H, et al. Expression of truncated latent TGF-beta-binding protein modulates TGF-beta signaling. J Cell Sci. 2005;118:2177–2187. doi: 10.1242/jcs.02352. [DOI] [PubMed] [Google Scholar]
- 28.Bottinger EP, Letterio JJ, Roberts AB. Biology of TGF-beta in knockout and transgenic mouse models. Kidney international. 1997;51:1355–1360. doi: 10.1038/ki.1997.185. [DOI] [PubMed] [Google Scholar]
- 29.Zambruno G, Marchisio PC, Marconi A, et al. Transforming growth factor-beta 1 modulates beta 1 and beta 5 integrin receptors and induces the de novo expression of the alpha v beta 6 heterodimer in normal human keratinocytes: implications for wound healing. The Journal of cell biology. 1995;129:853–865. doi: 10.1083/jcb.129.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank S, Madlener M, Werner S. Transforming growth factors beta1, beta2, and beta3 and their receptors are differentially regulated during normal and impaired wound healing. The Journal of biological chemistry. 1996;271:10188–10193. doi: 10.1074/jbc.271.17.10188. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Chan T, Demare J, et al. Healing of burn wounds in transgenic mice overexpressing transforming growth factor-beta 1 in the epidermis. The American journal of pathology. 2001;159:2147–2157. doi: 10.1016/s0002-9440(10)63066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annual review of cell and developmental biology. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 33.Yagi K, Goto D, Hamamoto T, et al. Alternatively spliced variant of Smad2 lacking exon 3 Comparison with wild-type Smad2 and Smad3. The Journal of biological chemistry. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- 34.Hosokawa R, Urata MM, Ito Y, et al. Functional significance of Smad2 in regulating basal keratinocyte migration during wound healing. The Journal of investigative dermatology. 2005;125:1302–1309. doi: 10.1111/j.0022-202X.2005.23963.x. [DOI] [PubMed] [Google Scholar]
- 35.Orecchia V, Regis G, Tassone B, et al. Constitutive STAT3 activation in epidermal keratinocytes enhances cell clonogenicity and favours spontaneous immortalization by opposing differentiation and senescence checkpoints. Experimental dermatology. 2015;24:29–34. doi: 10.1111/exd.12585. [DOI] [PubMed] [Google Scholar]
- 36.Tokumaru S, Sayama K, Yamasaki K, et al. SOCS3/CIS3 negative regulation of STAT3 in HGF-induced keratinocyte migration. Biochemical and biophysical research communications. 2005;327:100–105. doi: 10.1016/j.bbrc.2004.11.145. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Fei T, Zhang L, et al. Smad7 antagonizes transforming growth factor-beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Molecular and Cellular Biology. 2007;27:4488–4499. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh S, ten Dijke P. Negative regulation of TGB-beta receptor/Smad signal transduction. Current Opinion in Cell Biology. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Yan X, Liu Z, Chen Y. Regulation of TGF-beta signaling by Smad7. Acta biochim biophys sin. 2009;41:263–272. doi: 10.1093/abbs/gmp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins BJ, Grail D, Nheu T, et al. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nature medicine. 2005;11:845–852. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Yu Y, Sun C, et al. STAT3 selectively interacts with Smad3 to antagonize TGF-beta. Oncogene. 2015 doi: 10.1038/onc.2015.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uto-Konomi A, Miyauchi K, Ozaki N, et al. Dysregulation of suppressor of cytokine signaling 3 in keratinocytes causes skin inflammation mediated by interleukin-20 receptor-related cytokines. PloS one. 2012;7:e40343. doi: 10.1371/journal.pone.0040343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YG. Endocytic regulation of TGF-beta signaling. Cell research. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- 44.Kardassis D, Murphy C, Fotsis T, et al. Control of transforming growth factor beta signal transduction by small GTPases. The FEBS journal. 2009;276:2947–2965. doi: 10.1111/j.1742-4658.2009.07031.x. [DOI] [PubMed] [Google Scholar]
- 45.Razani B, Zhang XL, Bitzer M, et al. Caveolin-1 regulates transforming growth factor (TGF)-beta/SMAD signaling through an interaction with the TGF-beta type I receptor. The Journal of biological chemistry. 2001;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- 46.Balogh P, Magyar M, Szabo A, et al. The subcellular compartmentalization of TGFbeta-RII and the dynamics of endosomal formation during the signaling events: An in vivo study on rat mesothelial cells. European journal of cell biology. 2015;94:204–213. doi: 10.1016/j.ejcb.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Zhang XL, Topley N, Ito T, et al. Interleukin-6 regulation of transforming growth factor (TGF)-beta receptor compartmentalization and turnover enhances TGF-beta1 signaling. The Journal of biological chemistry. 2005;280:12239–12245. doi: 10.1074/jbc.M413284200. [DOI] [PubMed] [Google Scholar]
- 48.Fujita Y, Maruyama S, Kogo H, et al. Caveolin-1 in mesangial cells suppresses MAP kinase activation and cell proliferation induced by bFGF and PDGF. Kidney international. 2004;66:1794–1804. doi: 10.1111/j.1523-1755.2004.00954.x. [DOI] [PubMed] [Google Scholar]
- 49.Guo GG, Patel K, Kumar V, et al. Association of the chaperone glucose-regulated protein 58 (GRP58/ER-60/ERp57) with Stat3 in cytosol and plasma membrane complexes. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2002;22:555–563. doi: 10.1089/10799900252982034. [DOI] [PubMed] [Google Scholar]
- 50.Sehgal PB, Guo GG, Shah M, et al. Cytokine signaling: STATS in plasma membrane rafts. The Journal of biological chemistry. 2002;277:12067–12074. doi: 10.1074/jbc.M200018200. [DOI] [PubMed] [Google Scholar]