Abstract
Background
Altered expression of partition-defective 3 (PARD3), a polarity-related gene associated with oncogenesis, has been identified in some cancers, but the role of PARD3 in esophageal squamous cell carcinoma (ESCC) remains unclear.
Material/Methods
PARD3 expression in Eca109 cells was silenced using siRNA and overexpressed using an expression vector. We investigated the role of PARD3 in ESCC growth and motility to evaluate its potential role in ESCC. Transwell assay was used to evaluated cell migration and invasion. PARD3 protein expression was assessed by Western blot.
Results
PARD3 overexpression promoted apoptosis, impaired proliferation, and inhibited cell migration and invasion in Eca109 cells, while PARD3 silencing promoted proliferation and increased migration and invasion. Overexpression of PARD3 exerted its antitumor activity in vitro by impairing cell proliferation, inducing apoptosis, and inhibiting migration and invasion of Eca109 cells, suggesting that PARD3 might play a tumor suppressor role in ESCC.
Conclusions
Overexpression of PARD3 could be a promising new therapeutic intervention against ESCC.
MeSH Keywords: Apoptosis; Carcinoma, Squamous Cell; Cell Migration Assays; Cell Proliferation
Background
Esophageal cancer is the eighth most common cancer worldwide, with 456 000 new cases and 400 000 deaths in 2012 alone [1]. In China, from 2003 to 2007, esophageal cancer was ranked as the sixth cause of new cancer and the fourth primary cause of cancer-related deaths [2]. Despite recent advances in the treatment of esophageal squamous cell carcinoma (ESCC), prognosis remains poor [3,4].
The development and maintenance of cell polarity play an indispensable role in the regulation of cell morphology and epithelium formation [5]. Loss of cell polarity is a prerequisite related to malignancy [6]. Epithelial cells adhere to one another through adherents and tight junctions, and they have 2 distinct apical and basolateral membranes [7]. The formation and maintenance of cell polarity is controlled by 3 protein complexes: PAR, Crumbs, and Scrib. The PAR complex promotes the formation of the apical-lateral membrane border, and requires PAR-3, PAR-6, and aPKC in mammalian epithelial cells [6,8]. Cell polarity proteins can also regulate the malignant progression of tumor cells, including epithelial-mesenchymal transition (EMT), invasion, and metastasis.
The partition-defective 3 (PARD3) protein is involved in the development of tight junctions implicated in the epithelial cell-cell contacts [9]. Previous studies revealed that epithelium polarity functions as a tumor suppressor due to its involvement in the three-dimensional organization of epithelial tissue [5,9,10]. Few studies are available on the relationship between polarity proteins and human tumors, and most of them are related to breast cancer. PARD3 low expression is related to lymph node metastasis and poor ESCC differentiation [11]. There are few reports regarding the biological role of PARD3 in ESCC. Zen et al. [11] showed that reduced PARD3 expression could be involved in the progression of ESCC. PARD3 is also involved in the pathogenesis of lung squamous cell carcinoma [12–14].
To characterize the biological roles of PARD3 in ESCC occurrence and development, PARD3 was overexpressed and silenced in cultured Eca109 cells to explore the effect of PARD3 on their proliferation, apoptosis, cell cycle, migration, and invasion.
Material and Methods
Cell culture and transfection
Eca109 cell lines were purchased from the cell collection center of Wuhan University (China). Cells were cultured in RPMI 1640 supplemented with 1% penicillin/streptomycin and 10% calf serum at 37°C in a 5% CO2 atmosphere. The PARD3 small interference RNA (siRNA) (5′-UUC AAA GUC ACC UCG GCA ACU GCG G-3′/5′-CAA CAG CUG GCU UCC UCA AGC AGA A) and scramble siRNA were synthesized by Invitrogen Inc. (Carlsbad, CA, USA). Cells (5×105 cells/well) were seeded in 6-well plates and transfected using Lipofectamine RNAiMAX (Invitrogen Inc., Carlsbad, CA, USA) in Opti-MEM I (Invitrogen Inc., Carlsbad, CA, USA), according to the manufacturer’s instructions. A fluorescence microscope was used to detect the transfection efficacy. The negative control group was transfected with the scrambled sequence and cultured under normal conditions. RNA and protein were extracted at 48 or 72 h after transfection, as previously described [15].
PARD3 eukaryotic expression vector
The PARD3 eukaryotic expression vector pcDNA3.1 (sense: 5′-CCC AAG CTT ATG AAA GTG ACC GTG TGC TTC G-3′; and antisense 5′-GCT CTA GAT CAG GAA TAG AAG GGC CTC CCT T-3′) was synthesized and purchased from Invitrogen Inc. (Carlsbad, CA, USA). PCR was used to identify the recombinant clones (Supplementary Figure 2). Sequencing was used to confirm the presence of the insert as well as the sequence and orientation of the insert. Cells (5×105 cells/well) were seeded in 6-well plates and transfected with empty pcDNA3.1 vector under the same conditions as the negative control group.
RNA extraction and qRT-PCR
Total RNA was extracted from Eca109 cells in both silenced and overexpressing groups using Trizol reagent (Invitrogen, CA, USA). RNA was reverse-transcribed into cDNA using the Prime-Script one-step qRT-PCR kit (C28025-032, Invitrogen). qRT-PCR PARD3 primers were the following: forward 5′-GGAGCGCGAGAGCTCCCAGCA-3′, reverse 5′-TTATTAGGAAAGGACAGTGGG-3′. qRT-PCR was performed using SYBR Select Master Mix (Applied Biosystems, USA). β-actin (primers were: forward 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′, reverse 5′-TGGCACCCAGCACAATGAA-3′) was used as internal reference [20]. Relative gene expression was determined using delta-delta CT method (2−ΔΔCt).
Protein isolation and western blotting
Protease inhibitors (Boster, Wuhan, China) were added to cell lysates on ice for 20 min. Lysates were centrifuged at 14 000 rpm for 10 min at 4°C. Proteins (50 μg) were boiled for 5 min and separated by SDS-PAGE. Separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane and blocked for 1 h at room temperature. Blots were incubated with rabbit polyclonal antibody against PARD3 (Abcam, Cambridge, MA, USA) or β-actin (Sangon, Shanghai, China) at 4°C overnight, followed by incubation with the secondary antibody (ZSGB-BIO, Beijing, China) at room temperature for 1 h. A chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA, USA) was added. Quantity One software was used to quantify the intensity of each band. β-actin was used as control.
Cell cycle and apoptosis detection
Eca109 cells were seeded in 6-well culture plates at 5×104 cells/well and transfected with siRNA and expression vector. After 48 h of trypsin digestion, cells were collected and washed with cold PBS twice and fixed with ethanol for 2 h at −20°C, followed by staining with PI/RNaseA/PBS (50 μg/mL PI and 50 μg/mL RNase A) and incubation in the dark for 20 min. To detect the distribution of cell in G0/G1, S, and G2/M phases, flow cytometry (3550UV; Bio-Rad, Hercules, CA, USA) was used. To evaluate apoptosis, cells were diluted in 1×binding buffer at 5×105 cells/ml. Annexin V-FITC (10 μL) and 10 μL of PI (1 mg/mL) were added in 190 μl of cell suspension and incubated at room temperature for 5 min. Then, a single-cell suspension was obtained and the apoptosis rate was detected by flow cytometry after 30 min.
MTT assay
Eca109 cells were seeded in 96-well plates at 4×103 cells/well in a volume of 0.2 mL/well and transfected with PARD3-siRNA, control-siRNA, and pcDNA3.1 plasmid. MTT (Sigma-Aldrich, St Louis, MO, USA) was used to measure cell proliferation [16]. Cells were incubated in 20 μL of MTT at 37°C for 4 h. The color was developed by incubating the cells in 150 μL of dimethyl sulfoxide (DMSO), and the absorbance was measured at 490 nm. Data were obtained from experiments performed in triplicate.
In vitro transwell migration and invasion assays
Migration was evaluated by Transwell assay. Cells transfected with PARD3-siRNA, control-siRNA, and pcDNA3.1 plasmid were suspended in 100 ml of serum-free medium and seeded into the upper compartment (Corning, NY, USA). Medium containing 10% calf serum was added to the lower chamber. After 10 h of incubation at 37°C, non-migrated cells were removed using a cotton swap. The number of cells that had migrated through the membrane was manually counted. Five random fields (ECLIPSE TS100, Nikon, Tokyo, Japan) were counted on each membrane.
For cell invasion, the membrane of the upper chamber of the Transwell was coated with Matrigel (BD, USA), and the invasion assay was performed as explained above for the migration assay.
Statistical analysis
Data are shown as means ±SD from experiments performed in triplicates and analyzed with the Student’s t-test. Statistical analysis was performed using SPSS 17.0 (IBM, Armonk, NY, USA). Two-tailed P<0.05 was considered statistically significant.
Results
PARD3 level in Eca109 after silencing or overexpression
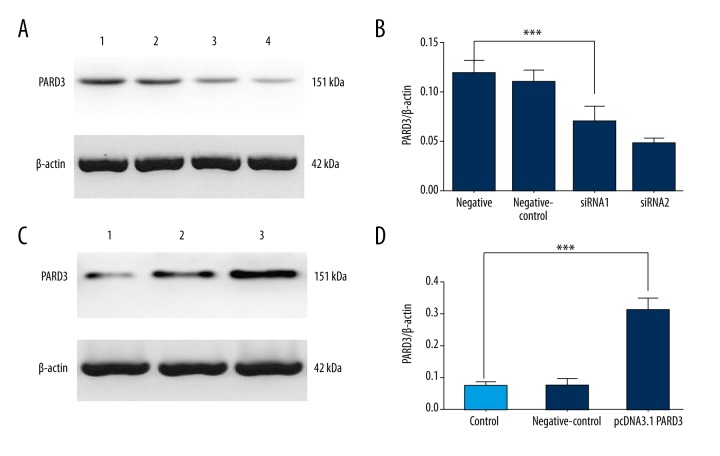
PARD3 mRNA and protein levels were significantly decreased after PARD3-siRNA transfection (Figure 1A, 1C and Supplementary Figure 1). Conversely, PARD3 expression was 4-fold higher after transfecting cells with pCDNA3.1-PARD3 (Figure 1B, 1D and Supplementary Figure 2).
Figure 1.
Expression of PARD3 protein. (A) The expression level of PARD3 protein was assessed by Western blotting after PARD3-siRNA transfection for 72 h. 1 was normal control, 2 was negative control, and 3 and 4 were the knockdown group. (B) The relative expression of PARD3 is shown, normalized to β-actin. The difference between the PARD3 siRNA and normal control groups was statistically significant. (C) The expression level of PARD3 protein assessed by Western blotting after transfecting cells with pCDNA3.1-PARD3. 1 was normal control, 2 was negative control, and 3 was the overexpression group. (D) PARD3 protein expression was significantly higher in the Eca109 cells compared with control cells. * P<0.05; ** P<0.01; *** P<0.001.
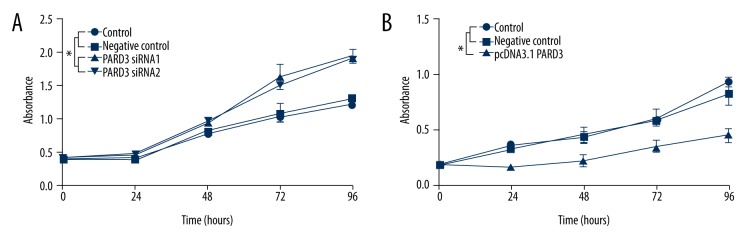
Effect of PARD3 on Eca109 cell proliferation in vitro
A significantly increased percentage of S phase cells (48.6±0.9%) was observed in the PARD3-siRNA group compared to controls (42.9±2.0%) (Table 1). PARD3 overexpression significantly increased the percentage of cells in G0/G1 at 48 h (63.0±2.8%) compared to the control group (52.1±4.9%). The percentage of PARD3-transfected cells in S phase was decreased (34.2±3.5%) compared to controls (47.3±3.3%) (Table 2). These results revealed that PARD3 overexpression decreased the basal proliferation rates and inhibited DNA replication of Eca109 cells. Cell proliferation was also measured by MTT. The results showed that PARD3 overexpression suppressed proliferation (Figure 2B), while PARD3 silencing promoted proliferation (Figure 2A).
Table 1.
Effect of PARD3 on cells cycle in Eca109 cell transfected with siRNA.
| G0/G1 (%) | S (%) | G2/M (%) | |
|---|---|---|---|
| Control | 54.5±3.0 | 41.6±1.6 | 3.9±3.2 |
| Negative control | 53.3±2.1 | 42.9±2.0 | 3.7±3.6 |
| PARD3 siRNA1 | 48.0±1.3 | 48.3±1.1* | 3.7±0.5 |
| PARD3 siRNA2 | 47.4±1.7 | 48.6±0.9* | 4.0±1.1 |
vs. the control group, P <0.01. Results are shown as mean ± standard deviation from 3 replicates.
Table 2.
Effect of PARD3 on cells cycle in Eca109 cell transfected with PcDNA3.1 PARD3 vector.
| G0/G1 (%) | S (%) | G2/M (%) | |
|---|---|---|---|
| Control | 51.7±2.1 | 46.9±1.5 | 1.3±2.0 |
| Negative control | 52.1±4.9 | 47.3±3.3 | 2.3±2.1 |
| pcDNA3.1 PARD3 | 63.0±2.8* | 34.2±3.5* | 2.8±0.9 |
vs. the control group, P<0.01. Results are shown as mean ± standard deviation from 3 replicates.
Figure 2.
Effects of silencing and overexpression of PARD3 on Eca109 cell proliferation. (A) MTT assay was used to measure proliferation every 24 h after transfection with siRNA-PARD3 and siRNA scramble into Eca109 cells. (B) MTT assay was used to measure proliferation every 24 h after transfection with pcDNA3.1-PARD3 and empty pcDNA3.1 vector into Eca109 cells. * P<0.05.
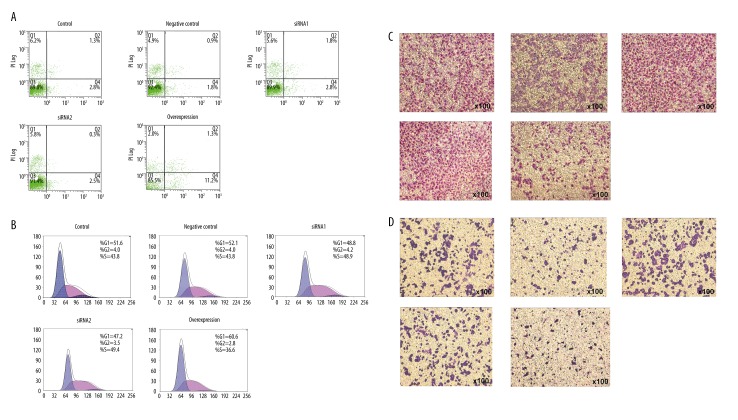
Effect of PARD3 on Eca109 cell apoptosis in vitro
PARD3 overexpression in Eca109 cells resulted in a remarkable and significant apoptosis increase (9.4±0.7%) compared to control cells (3.1±0.4%) 48 h after transfection (Table 3, Figure 3). On the other hand, PARD3 silencing did not significantly affect apoptosis (2.7±0.4% compared with 2.2±0.5% in controls; P>0.05, Table 4, Figure 3).
Table 3.
Effect of PARD3 on apoptosis in Eca109 cell transfected with PcDNA3.1 PARD3 vector.
| Apoptosis (%) | |
|---|---|
| Control | 2.9±0.4 |
| Negative control | 3.1±0.4 |
| pcDNA3.1 PARD3 | 9.4±0.7* |
vs. the control group, P<0.01. Results are shown as mean ± standard deviation from 3 replicates.
Figure 3.
PARD3 modulates ESCC cellular malignant phenotypes. Images of cell apoptosis, proliferation, migration, and invasion (A–D) in Eca109 cells (normal controls), PARD3 siRNA-transfected cells, and overexpression of PARD3, respectively. PARD3 siRNA-transfected cells showed significantly higher cell proliferation, invasion, and migration than controls, which significantly increased the malignant phenotype of Eca109 cells. Overexpression of PARD3 increased cell apoptosis and decreased cell proliferation, invasion, and migration in the Eca109 cell line.
Table 4.
Effect of PARD3 on apoptosis in Eca109 cell transfected with siRNA.
| Apoptosis (%) | |
|---|---|
| Control | 2.2±0.7 |
| Negative control | 2.2±0.5 |
| PARD3 siRNA1 | 2.4±0.4 |
| PARD3 siRNA2 | 2.7±0.4 |
Results are shown as mean ± standard deviation from 3 replicates.
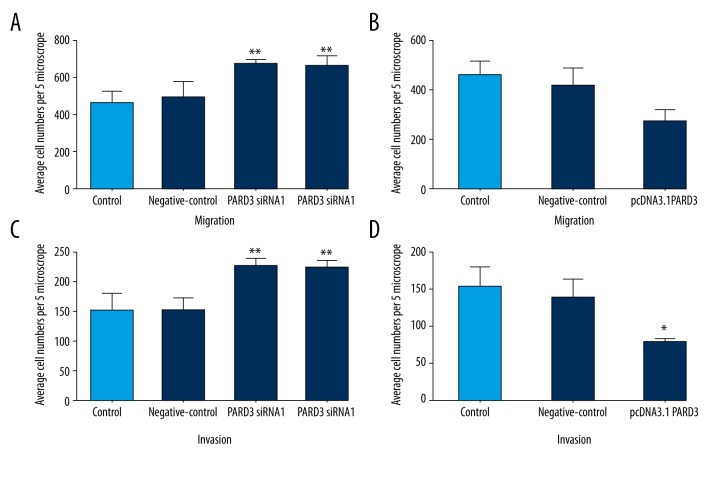
Effect of PARD3 on Eca109 cell migration and invasion
PARD3 overexpression resulted in a significant inhibition of the migratory and invasive abilities of Eca109 cells (Figures 3, 4). The number of migrated cells was significantly increased in siRNA-PARD3 Eca109 cells, compared to that of control cells (P<0.01, Figure 4A). On the other hand, PARD3 overexpression significantly reduced cell migration compared to control cells (P<0.05, Figure 4B). PARD3 silencing resulted in a significant promotion of Eca109 cell invasion (P<0.01, Figure 4C), while PARD3 overexpression significantly decreased cell invasion by approximately 50% compared to the control cells (P<0.05, Figure 4D).
Figure 4.
Effect of PARD3 on migration and invasion in Eca109 cells. PARD3 was overexpressed using a pcDNA3.1 vector and silenced using siRNA vectors. (A, B) PARD3 regulates migration of Eca109 cells. (C, D) PARD3 regulates invasion of Eca109 cells. * P<0.05; ** P<0.01.
Discussion
PARD3 is thought to be a polarity-related gene, and loss of epithelial cell polarity is a process that has been shown to be involved in oncogenesis [17]. This effect is considered a prerequisite for tumor formation and progression [18]. It is already known that cell polarity is mainly regulated by PAR proteins that regulate epithelial organization [19].
The role of PARD3 in ESCC initiation and progression is still unclear. Altered PARD3 expression has been identified in some cancer types, including lung squamous cell carcinoma [12–14,20] and ESCC [11]. PARD3 expression was reduced in ESCC compared to peri-cancerous tissue, and a strong negative correlation was found between PARD3 expression and aggressive cancer phenotypes, positive lymph node metastasis, and low differentiation [11]. Moreover, PARD3 expression was decreased in 90% of cell lines tested compared to normal esophageal cells [11]. Rothenberg et al. [21] observed that PARD3 deletions were limited to 2 distinct types of cancer: (1) squamous carcinomas including the esophagus, lung, and head and neck, and (2) glioblastoma, as supported by other studies [11–14]. Recurrent tumor-specific inactivating alterations of PARD3 were observed in 8% of lung squamous cell cancer (LSCC) [20]. However, it was surprising to find that PARD3 is highly expressed in hepatocellular carcinoma and is associated with extrahepatic metastasis and low survival [22]. PARD3 expression was reduced in human breast cancers [23] and in human keratoacanthomas [24], while PARD3 was amplified in radiation-transformed neoplastic retinal pigment epithelial cell lines [20]. Taken together, these results suggest that PARD3 is associated with carcinogenesis, but some discrepancies among primary cancer sites suggest that the role of PARD3 could be context-dependent and/or regulated by other factors. Additional studies are required to adequately determine these contexts and factors.
In the present study, PARD3 overexpression promoted Eca109 cell apoptosis and decreased their proliferation, while PARD3-siRNA enhanced proliferation, suggesting that PARD3 is a driver gene with significant cell growth effects in ESCC. These findings are in accordance with those of Rothenberg et al. [21], who reported that shRNA-mediated knockdown of PARD3 in some cancer cells with a disrupted wild type endogenous gene reduced the localization of ZO-1. Loss of PARD3 in mammary epithelial cells promotes proliferation [25]. Recent studies have shown that the PAR-3-PAR-6-aPKC complex is associated with the tumor suppressor von Hippel-Lindau (VHL) protein or with phosphatase and tensin homologue deleted on chromosome 10 (PTEN) [18], suggesting potential mechanisms used by PARD3 to modulate tumorigenicity. Loss of cell polarity can promote epithelial cell proliferation and apoptosis, but the mechanism described by Bilder et al. [26] is different from that described by McCaffrey et al. [27]. Additional studies are necessary to determine the mechanisms of PARD3 in tumorigenesis.
Loss of apical-basal polarity and cellular adhesions leads to EMT and induces metastasis [28]. Zen et al. [11] performed a monolayer wound-healing assay in knockdown T.T cells to investigate the role of PAR-3 in cell motility, but found that PARD3 suppression does not affect cell migration. In the present study, PARD3 inhibited Eca109 cells migration and invasion in vitro. These discrepancies could be due to the cell types. Bonastre et al. [20] defined PARD3 as a recurrently inactivated cell polarity manager in lung SCC and wtPAR3 restoration, preventing invasiveness and metastasis formation. A recent study reported that PARD3 suppression increases tumor invasion and metastasis [27]. The results of the present study suggest that PARD3 is involved in cell motility. Increased cell motility is important in tumor progression, particularly in the processes of invasion and metastasis [29].
The present study is mainly limited by the lack of a comprehensive analysis of multiple proteins that could be involved in the effects of PARD3 expression on carcinogenesis of ESCC, such as connective tissue growth factor [30], interacting protein kinase 1 [31], cofilin-1, and transgelin [32]. In addition, only the PARD3 protein was studied, and there could be some discrepancies between transcription and translation. Additional studies are needed to assess the exact role of PARD3 in ESCC.
Conclusions
The results of this study suggest that PARD3 plays a key role as a tumor suppressor due to its involvement in cell growth and motility, thus providing novel insights for understanding the potential mechanisms of ESCC. Although much more work is still required to determine the clinical relevance of its function, our work indicates that PARD3 could be a promising new target in ESCC therapy.
Supplementary Figures
The expression level of PARD3 mRNA was assessed by qRT-PCR after PARD3-siRNA transfection. # Compared with control group, P<0.05; * compared with negative control group, P<0.05.
The expression level of PARD3 mRNA was assessed by qRT-PCR after pCDNA3.1-PARD3 transfection. # Compared with control group, P<0.05; * compared with negative control group, P<0.05.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Source of support: This study is founded by Major Science and Technology Projects of the Xinjiang Uygur Autonomous Region (201430123-1)
References
- 1.Ferlay J, Soerjomataram I, Ervik M. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon: International Agency for Research on Cancer; 2013. [Google Scholar]
- 2.Zhang S. An analysis of incidence and mortality of esophageal cancer in China, 2003–2007. China Cancer. 2012;21:241–47. [Google Scholar]
- 3.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song H, Xu B, Yi J. Clinical significance of stanniocalcin-1 detected in peripheral blood and bone marrow of esophageal squamous cell carcinoma patients. J Exp Clin Cancer Res. 2012;31:35. doi: 10.1186/1756-9966-31-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer. 2011;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–80. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 7.Li XM, Wang H, Zhu LL, et al. Genes regulating epithelial polarity are critical suppressors of esophageal oncogenesis. J Cancer. 2015;6:694–700. doi: 10.7150/jca.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki A, Ohno S. The PAR-aPKC system: Lessons in polarity. J Cell Sci. 2006;119:979–87. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Lacoche S, Huang L, et al. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc Natl Acad Sci USA. 2013;110:163–68. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinck L, Nathke I. Changes in cell and tissue organization in cancer of the breast and colon. Curr Opin Cell Biol. 2014;26:87–95. doi: 10.1016/j.ceb.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zen K, Yasui K, Gen Y, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–18. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 12.Feigin ME, Muthuswamy SK. Polarity proteins regulate mammalian cell-cell junctions and cancer pathogenesis. Curr Opin Cell Biol. 2009;21:694–700. doi: 10.1016/j.ceb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene. 2015;34:939–50. doi: 10.1038/onc.2014.59. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Wang S, Wang S, et al. Dual function of partitioning-defective 3 in the regulation of YAP phosphorylation and activation. Cell Discov. 2016;2:16021. doi: 10.1038/celldisc.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma JQ, Tuersun H, Jiao SJ, et al. Functional role of NRF2 in cervical carcinogenesis. PLoS One. 2015;10:e0133876. doi: 10.1371/journal.pone.0133876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng S, Yang C, Liu T, et al. Clinicopathological significance of p38beta, p38gamma, and p38delta and its biological roles in esophageal squamous cell carcinoma. Tumour Biol. 2016;37:7255–66. doi: 10.1007/s13277-015-4610-9. [DOI] [PubMed] [Google Scholar]
- 17.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–31. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 18.Wodarz A, Nathke I. Cell polarity in development and cancer. Nat Cell Biol. 2007;9:1016–24. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 19.Li P, Mao X, Ren Y, Liu P. Epithelial cell polarity determinant CRB3 in cancer development. Int J Biol Sci. 2015;11:31–37. doi: 10.7150/ijbs.10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonastre E, Verdura S, Zondervan I, et al. PARD3 inactivation in lung squamous cell carcinomas impairs STAT3 and promotes malignant invasion. Cancer Res. 2015;75:1287–97. doi: 10.1158/0008-5472.CAN-14-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg SM, Mohapatra G, Rivera MN, et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 2010;70:2158–64. doi: 10.1158/0008-5472.CAN-09-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan YJ, Ko BS, Liu TA, et al. Expression of partitioning defective 3 (Par-3) for predicting extrahepatic metastasis and survival with hepatocellular carcinoma. Int J Mol Sci. 2013;14:1684–97. doi: 10.3390/ijms14011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22:601–14. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iden S, van Riel WE, Schafer R, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Archibald A, Mihai C, Macara IG, McCaffrey L. Oncogenic suppression of apoptosis uncovers a Rac1/JNK proliferation pathway activated by loss of Par3. Oncogene. 2015;34:3199–206. doi: 10.1038/onc.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilder D. Epithelial polarity and proliferation control: Links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–25. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 27.McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–35. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Ellenbroek SI, Iden S, Collard JG. Cell polarity proteins and cancer. Semin Cancer Biol. 2012;22:208–15. doi: 10.1016/j.semcancer.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Han Q, Zhang HY, Zhong BL, et al. MicroRNA-145 inhibits cell migration and invasion and regulates epithelial-mesenchymal transition (EMT) by targeting connective tissue growth factor (CTGF) in esophageal squamous cell carcinoma. Med Sci Monit. 2016;22:3925–34. doi: 10.12659/MSM.897663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan B, Ma F, Wang M, Xu X. Down-regulating receptor interacting protein kinase 1 (RIP1) promotes oxaliplatin-induced Tca8113 cell apoptosis. Med Sci Monit. 2015;21:3089–94. doi: 10.12659/MSM.894184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Liao R, Li H, et al. Expression of cofilin-1 and transgelin in esophageal squamous cell carcinoma. Med Sci Monit. 2015;21:2659–65. doi: 10.12659/MSM.895242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression level of PARD3 mRNA was assessed by qRT-PCR after PARD3-siRNA transfection. # Compared with control group, P<0.05; * compared with negative control group, P<0.05.
The expression level of PARD3 mRNA was assessed by qRT-PCR after pCDNA3.1-PARD3 transfection. # Compared with control group, P<0.05; * compared with negative control group, P<0.05.