Abstract
The CBL-CIPK pathway is a plant-specific Ca2+ sensor relaying pathway that has been shown to be involved in plant response to salt stress. Over-expression of CBL-CIPK network genes has been reported to increase salt tolerance in many studies. The studies on the overexpression of CBL-CIPK network genes, however, have used various indices to evaluate the effect of these genes on salt tolerance and have indicated a variety of roles for the major CBL-CIPK pathway genes. Therefore, it is of great interest to analyze the various effects resulting from the overexpression CBL-CIPK pathway genes and their relation to salt tolerance. The meta-analysis conducted in the present study investigated how over-expression of CBLs or CIPKs in transgenic plants affects the response to salt stress and identified the increase or decrease that occurs in these experimental variables when foreign CIPK or CBL genes are overexpressed in transgenic plants. The data from the collective studies on over-expression of CIPKs indicated that 6 of the 11 examined parameters (main effects) increased by 22% or more, while two of the six examined parameters increased by at least 78% in transgenic plants overexpressing CBL genes. In addition to analyzing the impact of overexpression on the main effects, eight different modifying parameters were also analyzed. Results indicated that several moderators impact the extent to which overexpression of CBLs and CIPKs affect the main parameters. The majority of CBLs have been obtained from dicotyledonous plants and most of the CBLs and CIPKs have been expressed in dicotyledonous plants. In comparison to homologous expression, the meta-analysis indicated that heterogeneous expression of CBLs resulted in greater increases in seed germination. The results of the meta-analysis provide information that could be useful in designing research to examine the mechanisms by which CBL-CIPK pathway genes increase salt tolerance in plants.
Keywords: CBL gene family, CIPK gene family, over-expression, salt tolerance, transgenic, meta-analysis
Introduction
Salinity is one of the major abiotic stresses that limits the production and yields of agricultural crops worldwide (Cramer et al., 2011). Salt stress impacts plants at the molecular, physiological, and cellular levels during their life cycle (Ruiz-Lozano et al., 2012). Elucidating the plant parameters that respond to salt stress, and understanding the mechanisms involved in salt tolerance, can improve efforts to genetically engineer crops with improved levels of salt tolerance.
Many studies have examined the functional role of the CBL-CIPK pathway in plants in relation to plant response to abiotic stress (Xiang et al., 2007; Chaves-Sanjuan et al., 2014; Zhang et al., 2014; Pandey et al., 2015; Wang et al., 2016). The CBL-CIPK pathway is a Ca2+ sensor relaying pathway that has been demonstrated, along with the SOS signaling pathway, to be involved in plant response to salt stress. External stresses induce an increase in the concentration of intracellular Ca2+, and a CBL, a plant-specific sensor relay (Liu and Zhu, 1998; Kudla et al., 1999), senses the change in the Ca2+ level. CBL, accompanied by a CIPK then activates SOS1 by phosphorylation. SOS1, which belongs to the CPA1 (cation/proton antiporter 1) family of genes acts as a Na+/H+ antiporter to compartmentalize or exclude Na+ and maintain the balance of K+/Na+ (Zhu, 2002).
In addition to the SOS pathway, which is the classical pathway in the CBL-CIPK signaling network, also interacts with other signaling pathways to help plants cope with salt stress. Various members of the CBL and CIPK gene families are involved in salt tolerance. In Arabidopsis, CBL1, CBL9, CBL4, and CIPK6 regulate AKT-type K channels such as AKT1 or AKT2 (Li et al., 2006; Lee et al., 2007; Held et al., 2011; Nieves-Cordones et al., 2012). Studies have also reported that, in response to salinity stress, the CBL-CIPK system interacts with the ABA signaling pathway via the ABI2 protein (Jia et al., 2002; Ohta et al., 2003).
In response to salinity stress, plants maintain the ionic balance by excluding Na+ from cells or sequestrating Na+ through compartmentalization. Many studies have reported that overexpression of CBL or CIPK genes can increase salt tolerance and numerous members of these gene families have been overexpressed in different plant species (39 papers listed in Appendix S1). It has been difficult to distinguish, however, the precise mechanisms and different physiological processes that have been activated or suppressed by the overexpression of CBLs or CIPKs in transgenic plants subjected to non-stress or salt stress conditions because so many parameters are affected.
Meta-analysis represents a statistical synthesis of the results obtained in a collection of studies that have been systematically collected and appraised. The conformance and magnitude of a treatment effect is estimated much more accurately in a meta-analysis than it is from any single study alone. A meta-analysis is also able to describe the range of the impact of an effect and test which factors affect the magnitude of the effect size if the treatment effect is inconsistent in different studies. In the present study, a meta-analysis was conducted on the over-expression of CIPK-CBL pathway genes in order to characterize the effect of overexpression on the response of transgenic plants subjected to salt stress. In addition, several moderator variables that may potentially affect the size of the foreign genes effect on plant salt tolerance were also examined. A meta-analysis of the effect of CPA1 overexpression has been previously published (Ma et al., 2017). Therefore, the main focus of the present study pertained to the overexpression of the CIPK and CBL gene family members. A meta-analysis was conducted on the impact of CIPKs and CBLs overexpression in transgenic plants subjected to salt stress across many studies. The effect of experimental variables on the impact of CBL or CIPK overexpression was also examined, and differences between CBL and CIPK effects on plants under salt-stress and non-stress conditions was also clarified. The results of the meta-analysis enabled us to identify areas of research requiring further investigation and increased our understanding of the function of CBLs or CIPKs in improving salt tolerance in plants.
Materials and Methods
Data Collection
Using the ISI Web of Science (Thompson Reuters; include 12 electronic databases), 633 unique articles on CBL and CIPK were located up through April, 2016. A total of 48 search terms, such as “CBL” “CIPK” AND (“overexpress” or “stress”), etc. were used to identify relevant studies (additional details provided in Appendix S1).
After closer examination, 591 of the articles were excluded because they did not conform to our inclusion standard. Reasons for exclusion included: data were unrelated to plants (421); data did not include the effect of CBLs or CIPKs overexpression on salt stress (108); they were review articles (12), they were patents (20) and figures or datasets (18); data were related but did not include the parameters on which we were focused (13), and; the same data had been reported in another article (1). This exclusion process resulted in retaining 40 articles for the meta-analysis. Of these, 15 articles pertained to the overexpression of CBL genes; 24 articles reported on the overexpression of CIPK genes, and only one article investigated the individual overexpression of both CBL and CIPK genes. The 39 articles were written in both English and Chinese (details provided in Appendix S1).
Treatment and control means, with sample sizes, were obtained for each study. If the sample size was given as a range, the smallest value in the range was used. If sample size was not reported, we defined it as n = 1 when no mean statistics were provided, or as n = 3 when standard errors or an LSD were provided. If the study included data from multiple time points, only the final time point was used in the meta-analysis. When the data were only presented in graphs, the numerical data was extracted using GetData Graph Digitizer1 software.
If multiple treatments were used in one article, each treatment was used as an independent study and represented as an individual unit in the meta-analysis. For example, He et al. (2013) examined both transformed and non-transformed Arabidopsis plants subjected to four different NaCl concentrations, which was represented as four individual studies in the meta-analysis. Hu et al. (2016) used two kinds of media in their study, which was then treated as two studies in the meta-analysis. Although the approach of breaking down a single study into multiple studies and designating them as independent studies may potentially result in an increase in the disadvantage of the statistical dependence among studies, which in the meta-analysis are assumed to be independent (Gurevitch and Hedges, 1999), the larger number of studies maximizes the statistical power of the meta-analysis (Lajeunesse and Forbes, 2003). Importantly, this approach has been commonly used in meta-analyses of plant biology (Holmgren et al., 2013; Klümper and Qaim, 2014), especially when the meta-analysis includes the evaluation of moderators (experimental variables) (Lehmann and Rillig, 2015).
Effect Size and Moderators
Each response variable was incorporated into the analysis as the ratio of transgenic to non-transgenic (control) plant means. This response ratio is referred to as the effect size, and its natural logarithm, ln R, is used in the meta-analysis (Borenstein et al., 2009), in the equation:
where YTC and YNC are the means of plants that were transformed (TC), or non-transformed (NC) (or transformed with an empty vector), with a CBL gene (Figure 1) or a CIPK gene (Figure 2), respectively. Response ratios refer to a standardized, unit-less expression of treatment-induced changes, that are commonly used in meta-analyses of plant response (Schaeffer et al., 2013; Worchel et al., 2013). The log transformation balances positive and negative treatment effects across response ratios and maintains symmetry in the analysis (Borenstein et al., 2009). In other words, summary effect sizes within a meta-analysis indicates the “response ratio” of transformed (TC)/non-transformed (NC) plants over all studies, and the natural logs of the response ratio reflects the relative magnitude of the treatment effect.
FIGURE 1.
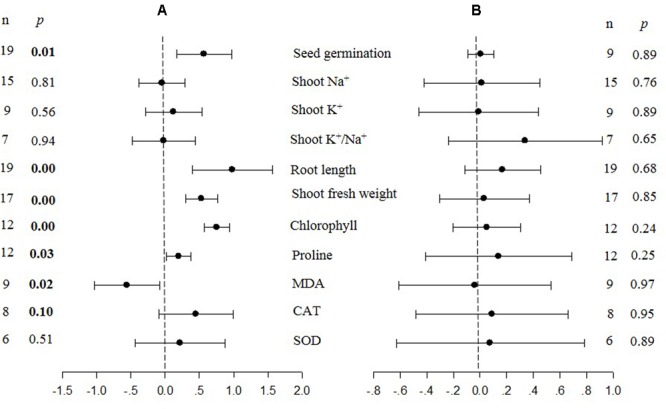
Weighted summary effect sizes (ln R) and 95% confidence intervals (CIs) for the effect of CIPK overexpression in transgenic plants subjected to NaCl stress conditions (A) and non-stress conditions (B). A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
FIGURE 2.
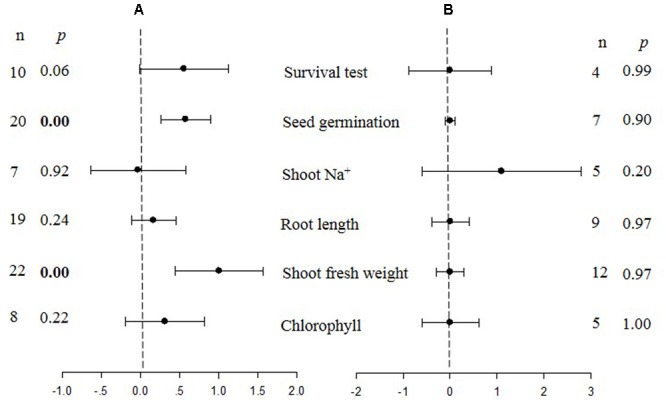
Weighted summary effect sizes (ln R) and 95% CIs for the effect of CBL overexpression in transgenic plants subjected to NaCl stress conditions (A) and non-stress conditions (B). A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
When Ln R-values are above 0, this indicates a TC-induced increase in the parameter, while values below 0 indicate a TC-induced decrease in the parameter; and a value of 0 signifies a lack of an effect by the overexpression of the transgene on that parameter.
Among the 39 articles, the only parameters that were selected for the determination of the summary effect size were those for which at least five studies from more than one article existed. On this basis, 11 summary effect sizes were computed for CIPK overexpression and 6 summary effect sizes were computed for CBL overexpression.
In addition to effect size, data for eight experimental attributes or “moderators” were collected from each study that may have impacted the response to salinity stress in the transgenic plants. Moderators were used to test for heterogeneity within effect sizes and were of two varieties: (1) experimental conditions: promoter, treatment medium, stress severity, and stress duration; (2) experimental materials: type (monocot vs. dicot), genus of gene donor and recipient, and whether the genera for the donor/recipient combination were the same or different. Each moderator included at least two levels (categories). The definition criteria for the moderators and categories were based on defined categories that were previously used in our laboratory (Ma et al., 2017).
In some cases, one experiment contained more than one level of salt stress severity. Therefore, we defined stress severity in three levels “low,” “medium,” and “high,” in order to evaluate whether or not the summary effects were impacted by different levels of stress severity. For example, when an experiment utilized 50, 100, 150, and 200 mM NaCl to expose plants to salt stress, we defined the 50 mM treatment as low, the 200 mM treatment as high, and 100 and 150 mM treatments as medium. Details on the defined and selected moderators are provided in Appendix S1.
Meta-Analysis
Meta-analyses were conducted using Comprehensive Meta-Analysis (CMA) v.3 (Biostat, Englewood, NJ, United States; 2014) software and the weight of individual studies was carried out using non-parameter variance:
where Vln R is the variance of the natural log of the response ratio R and nTC and nNC are the sample sizes of the TC and NC treatments (Borenstein et al., 2009). A detailed description of meta-analysis was previously described (Ma et al., 2017).
Heterogeneity
The Q statistic, which is based on weighted squared deviations, was used to analyze the heterogeneity and I2, which is a descriptive index that estimates the ratio of true heterogeneity to total heterogeneity across the observed effect sizes (Borenstein et al., 2009). In other words, the Q statistic was used to quantify heterogeneity.
Publication Bias
Studies that report relatively high effect sizes are more likely to be published than studies that report lower effect sizes. In the meta-analysis, any bias in the literature is likely to be reflected. This issue is known as publication bias (Borenstein et al., 2009). Potential publication bias was statistically examined using three kinds of general techniques designed to evaluate publication bias. These were the symmetry/asymmetry of a funnel plot (Viechtbauer, 2007), the Begg and Mazumdar rank (Kendall) correlation (Begg and Mazumdar, 1994; Borenstein et al., 2009), and the Egger’s linear regression method (Sterne and Egger, 2005).
Results
Summary Effects
Eleven different species (Appendix S1) served as gene donor plants among the 125 studies conducted on CIPK-overexpression. The most widely used CIPK genes were obtained from Populus (35 studies), and the majority (87 studies) of gene donor plants were dicotyledons. There were eight different genera of recipient plants, with the majority of recipient plants being represented by Arabidopsis (54 studies) and the overall majority of recipient plants were dicots (117 studies). The data set obtained from the 125 studies included the use of three different promoters, with the 35s promoter being the most utilized (115 studies), followed by Ubi (8 studies), and lastly rd29A (2 studies).
Seven plant species served as gene donor plants among the 97 studies conducted on CBL-overexpression in plants (Appendix S1). Similar to CIPK studies, Populus was the most highly represented donor of CBL genes (39 studies), and dicots were the most highly represented among all the CBL donor species (93 studies). Gene recipient plants were represented by three species, with Arabidopsis being the most highly represented (95 studies). The CBL data set included the use of two promoters, with 35s being used most often (91 studies), followed by rd29A (2 studies). Four of the studies did not include the type of promoter that was used to drive CBL expression.
Figures 1–7 and Supplementary Figures S1–S3 are forest plots that illustrate the summary (main) effects of CIPK and CBL overexpression, in transgenic plants, relative to non-transgenic plants, subjected to non-stress and NaCl stress conditions. The relative magnitude of each treatment effect and their CIs are presented in the forest plots, in which Ln R-values of the summary effect above 0 indicate that CIPK or CBL overexpression had a positive effect (increased) on that attribute. Values below 0 indicate that the overexpression had a negative (decreased) effect on that attribute. Summary effects are considered significant if their 95% CIs did not overlap zero. The raw percentage of the change resulting from either CIPK or CBL overexpression are listed in Tables 1, 2, respectively.
FIGURE 7.
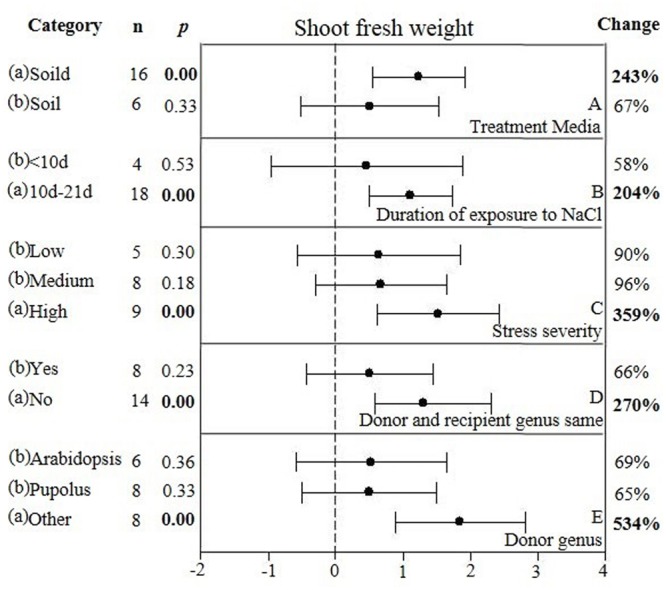
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CBL transformation modifies shoot fresh weight. (A) Treatment media, (B) Duration of exposure to NaCl, (C) Stress severity, (D) Donor and recipient genus same, and (E) Donor genus. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Table 1.
Heterogeneity statistics for the 11 summary effect sizes under NaCl stress.
| Summary effect size | Qt | p-hetero | I2 (%) | TC-induced change (%) |
|---|---|---|---|---|
| Seed germination | 22.774 | 0.199 | 21 | 77 |
| Shoot Na+ | 3.987 | 0.996 | 0.0 | –4 |
| Shoot K+ | 0.549 | 1.000 | 0.0 | 13 |
| Shoot K+/Na+ ratio | 1.179 | 0.978 | 0.0 | –2 |
| Root length | 29.332 | 0.045 | 39 | 168 |
| Shoot fresh weight | 6.67 | 0.979 | 0.0 | 70 |
| Chlorophyll | 5.024 | 0.930 | 0.0 | 114 |
| Proline | 2.337 | 0.997 | 0.0 | 22 |
| MDA | 7.795 | 0.454 | 0.0 | –43 |
| CAT | 2.054 | 0.957 | 0.0 | 57 |
| SOD | 0.301 | 1.000 | 0.0 | 25 |
Qt, total heterogeneity; p-hetero, probability that Qt was due entirely to sampling error and not to variation among true effects; I2, percentage of heterogeneity due to variation among true effects; changes to summary effects caused by foreign CIPK genes, transformed from ln R to raw percentages. For TC-induced changes, bold text signifies change was significant (p ≤ 0), with positive values indicating TC-induced promotion and negative values TC-induced inhibition.
Table 2.
Heterogeneity statistics for the six summary effect sizes under NaCl stress.
| Summary effect size | Qt | p-hetero | I2 (%) | TC-induced change (%) |
|---|---|---|---|---|
| Survival test | 9 | 0.342 | 0.0 | 74 |
| Seed germination | 21.528 | 0.308 | 11.743 | 78 |
| Shoot Na+ | 1.311 | 0.971 | 0.0 | –3 |
| Root length | 4.508 | 0.999 | 0.0 | 18 |
| Shoot fresh weight | 37.433 | 0.015 | 43.899 | 173 |
| Chlorophyll | 0.583 | 0.999 | 0.0 | 37 |
Qt, total heterogeneity; p-hetero, probability that Qt was due entirely to sampling error and not to variation among true effects; I2, percentage of heterogeneity due to variation among true effects; changes to summary effects caused by foreign CBL genes, transformed from ln R to raw percentages. For TC-induced changes, bold text signifies change was significant (p ≤ 0), with positive values indicating TC-induced promotion and negative values TC-induced inhibition.
The meta-analysis indicated that six plant parameters were significantly impacted by foreign CIPK overexpression in plants subjected to NaCl stress (Figure 1A: p ≤ 0.05). Among the six parameters, root length exhibited the largest increase, with an approximately 2.7× higher increase in transgenic (TC) plants, relative to (NC) plants, subjected to NaCl conditions. Chlorophyll levels also increased dramatically by 114% in TC plants, relative to their NC counterparts. Shoot fresh weight and seed germination were also markedly higher in TC plants. Overexpression of CIPK genes resulted in a significant reduction of the MDA content in leaves. Shoot Na+ and shoot K+/Na+ were reduced by 2 to 4%, although this was not considered significant since the 95% CI overlapped with zero. In contrast to NaCl stress conditions, the overexpression of CIPK genes did not significantly impact (p ≥ 0.05) any of the plant parameters in transgenic plants subjected to non-stress conditions (Figure 1B).
Only two summary effects (TC/NC response ratios) were significantly impacted in transgenic plants overexpressing a CBL gene when they were exposed to NaCl stress (Figure 2A: p ≤ 0.05). Shoot fresh weight exhibited a 173% increase in TC plants, relative to NC plants, subjected to NaCl stress, and seed germination increased by 78% in TC plants, relative to NC plants. None of the impacts on the summary effects were significant under non-stressed conditions (Figure 2B).
Probability (p) values for the summary effects directly reflect the magnitude of the significance of the treatment effect, but may also indirectly convey information about the availability (sample size) on which the summary effect is based on (Borenstein et al., 2009). Large but insignificant summary effects may reflect real treatment differences for which sample sizes are simply insufficient to account for the variability present in the data set (i.e., statistical power is too low to reveal significance). For example, this may be the case for CAT and SOD in CIPK overexpressing plants subjected to NaCl stress (Figure 1A), and root length in unstressed plants (Figure 2B). Therefore, more attention should be placed on the magnitude and precision of the summary effects in a meta-analysis than on tests of their statistical significance or the p-values (Cooper, 2009; Cumming, 2012).
Publication Bias
Based on the parameters that are commonly used to test for publication bias in meta-analysis, no evidence of publication bias was found (Supplementary Tables S1, S2). The Begg and Mazumdar rank correlation test derives the rank correlation between standardized effect sizes and their standard errors (estimated from their non-parametric variances) (Begg and Mazumdar, 1994). Results indicated that most summary effects had a p > 0.05, indicating little concern for bias. Across summary effects in our study, most of the funnel plots across summary effects were symmetric and also raised no concerns about publication bias (Supplementary Tables S1, S2). Kendall tau values were also below 0.3 for all of the summary effects with p > 0.05, indicating little evidence for bias. A two-tailed significance test (Egger’s p-values) in CIPK overexpressing plants suggested the possibility of a publication bias for shoot Na+, and MDA. The two-tailed significance tests (Egger’s p-values) in CBL overexpressing plants, however, suggested little concern for publication bias.
Heterogeneity and Moderator Analysis
Heterogeneity represents true differences in treatment outcomes among studies. Moderator analyses are conducted to reveal heterogeneity and to determine which experimental variables (moderators) significantly influenced treatment outcomes and which have had little or no effect. A P-hetero value for a Q-test < 0.1 or a positive I2 was used to indicate significant heterogeneity in the size of the summary effect. A p-hetero value for a Q-test > 0.1 indicates that the impact of the moderator on the summary effect is inconsistent among studies (Iacovelli et al., 2014). Due to the low statistical power brought about by a small number of available studies for a particular parameter or large within-study variance, the substantial real dispersion of true effects might frequently yield a p-hetero > 0.1. This situation often arises in meta-analyses of plant studies. As a result, although only two of the summary effects in CIPK overexpressing plants subjected to NaCl stress exhibited significant heterogeneity (Table 1, root length: p-hetero values less 0.10, and I2 = 39; seed germination: p-hetero values exceeded 0.10, however, I2 = 21), and two summary effects in CBL overexpressing plants subjected to NaCl stress exhibited statistically significant heterogeneity (Table 2, seed germination: p-hetero values exceeded 0.10, however, I2 = 11.743; Shoot fresh weight: p-hetero values less 0.10, and I2 = 43.899), a moderator analysis on all summary effects which was affected significant by foreign CIPK genes or CBL genes was still performed. Results of the meta-analysis indicated that six summary effects were significantly affected by CIPK transformation (Figure 1A) and two were significantly affected by CBL transformation (Figure 2A). Therefore, a moderator analysis on all eight summary effects was conducted and the results are displayed in Figures 3–7 and Supplementary Figures S1–S3. The results of the analysis revealed the influence of the moderator categories on the summary effects and indicated which summary effect was most affected by a moderator. If the influence of a moderator category has a significant impact due to overexpression, it is marked with an “a.” If the influence of a moderator category is not significant, it is marked with a “b.” If the sample size of a moderator category is too small (n < 3) to have the statistical significance, it was marked in red (Figures 3–7).
FIGURE 3.
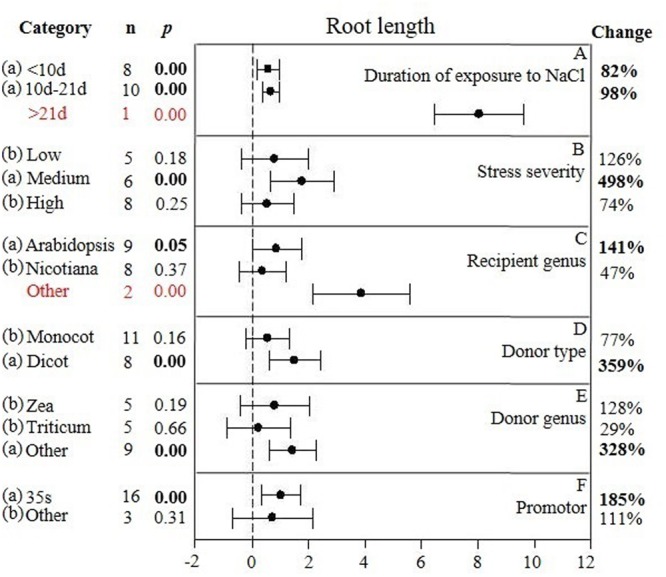
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies root length. (A) Duration of exposure to NaCl, (B) Stress severity, (C) Recipient genus, (D) Donor type, (E) Donor genus, and (F) Promotor. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
The individual impact of several different moderators on the degree to which CIPK overexpression affected root length in salt-stressed plants is presented in Figure 3. There were two of the six moderators significantly affected the TC-induced increase in root length observed in CIPK-overexpressing plants subjected to NaCl stress. Due to the small sample size (n < 3), the duration of the exposure to salt stress only had a minor effect on the TC-induced increase in root length, with the exception at the longest durations (more than 21 days) (Figure 3A). An increase in the severity of the salt stress from low to medium, or a reduction from high to medium stress levels resulted in a 4× or 7× increase in the TC-induced impact on root length, respectively (Figure 3B). When the gene recipient was Arabidopsis, it was apparent that root length was more responsive to CIPK overexpression than Nicotiana. However, even though the effect of other recipient genera appeared to be quite large, it was not statistically significant due to the small sample size (Figure 3C). The general category of donor type (monocot vs. dicot) greatly affected the magnitude of the TC-induced increase in root length when the donor was a dicot, with the impact being 4× greater than when the donor was a monocot (Figure 3D). As a gene donor genus, although Triticum had only a modest effect on the TC-induced increase in root length in plants subjected to salt stress, increasing the CIPK-overexpression effect by 29%; compared to a 128% increase in root length when the gene donor genus was Zea. The effect of Triticum and Zea as CIPK gene donors, however, was no significant impact after overexpression and was less than when the genus of the gene donor was other than these two genera (Figure 3E). The type of promoter used to overexpress CIPK had little effect on the TC-induced effect on root length in plants subjected to NaCl stress (Figure 3F).
Two of the six moderators significantly affected the TC-induced increase in seed germination observed in CIPK-overexpressing plants subjected to NaCl stress (Figure 4, Table 3, and Appendix S1). Similar to root length, the lowest (<10 days) and highest (>21 days) durations of salt stress had little impact on the TC-induced increase in seed germination (Figure 4A). The effect of an intermediate duration (10–21 days) was not included in the analysis due to insufficient sample size (n < 3). The effect of salt stress severity on TC-induced increases in seed germination was greatest at low levels of severity, followed by medium and then high levels of severity (Figure 4B). The influence of the donor/recipient combination on seed germination in CIPK-overexpressing plants subjected to salt stress was very similar, regardless if the donor/recipient were different genera or the same genera (Figure 4C). The influence of just the recipient genus on the TC-induced increase in seed germination in NaCl stressed plants was much greater when Arabidopsis was the recipient genus as compared to when Nicotiana and other genera were recipients (Figure 4D). Similar to root length, the TC-induced increases in seed germination was not affected by the general category of donor type (Figure 4E), though the more specific moderator of donor genus had a strong and statistically significant effect on the TC-induced increase in seed germination (Figure 4F). When the donor genus was Populus, the average increase was 315%, 129% when it was Triticum, and an 8% decrease, relative to non-transgenic plants when the gene donor genus was some other genus.
FIGURE 4.
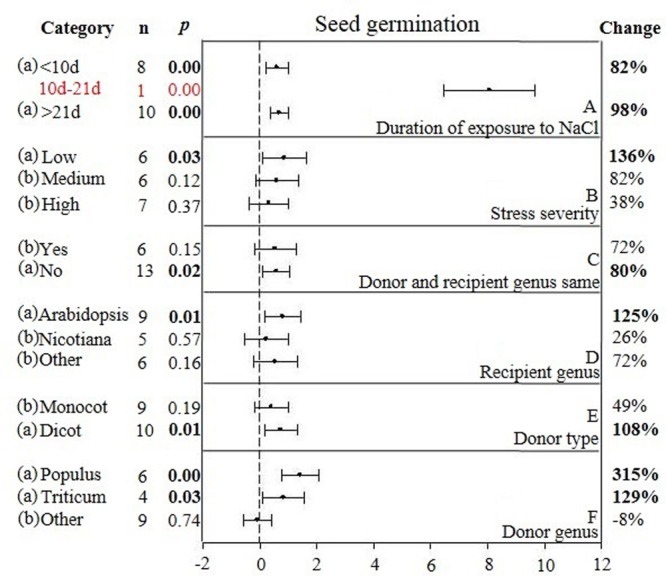
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies seed germination. (A) Duration of exposure to NaCl, (B) Stress severity, (C) Donor and recipient genus same, (D) Recipient genus, (E) Donor type, and (F) Donor genus. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Table 3.
Categorical moderators examined for the six summary effects of CIPK-overexpression that were significantly different from zero Heterogeneity p-values (p-hetero) are shown here.
| Moderator | Effect size |
|||||
|---|---|---|---|---|---|---|
| Seed germination | Root length | Shoot fresh weight | Chlorophyll | Proline | MDA | |
| Treatment media | – | – | – | 0.109 | 0.520 | 0.180 |
| Duration of exposure to NaCl | 0.000 | 0.000 | 0.627 | – | 0.545 | 0.633 |
| Stress severity | 0.598 | 0.245 | 0.815 | 0.643 | 0.691 | – |
| Recipient genus | 0.524 | 0.001 | 0.616 | 0.194 | – | 0.320 |
| Donor type | 0.423 | 0.117 | 0.470 | – | – | – |
| Donor genus | 0.000 | 0.235 | 0.396 | 0.109 | 0.740 | 0.320 |
| Donor and recipient genus same | 0.920 | – | 0.950 | 0.095 | 0.975 | 0.179 |
| Promoter | – | 0.711 | – | – | – | – |
probability that all observed (total) heterogeneity is not due solely to sampling error (expected variation) p-hetero ≤ 0.1 indicates significant heterogeneity among true effects. Dashed entries signify that insufficient data were available to perform an analysis. See supplement for associated Qm values (between study heterogeneity); n (sample size); df (degrees of freedom, levels within a moderator).
Only one of the five moderators significantly affected the TC-induced increase in chlorophyll observed in CIPK-overexpressing plants subjected to NaCl stress (Figure 5, Table 3, and Appendix S1). The effect of CIPK overexpression on chlorophyll content in salt-stressed plants was approximately twice as large (123%) when plants were grown on other media (28%) (Figure 5A). The severity of the stress severity had little differential impact on the TC-induced effect on chlorophyll content, with the exception at the low severity level, which was not considered due to a small sample size (n < 3). As with the previous summary effect, the moderator influence was the greatest when the recipient genus was Arabidopsis (125%) (Figure 5C). The TC-induced effect on chlorophyll content was greater when the donor genus was in the ‘other’ category as compared to when the donor genus was Populus (Figure 5D). The impact of CIPK-overexpression on chlorophyll content was greater when the donor/recipient combination was composed of different genera than when they were composed of the same genera (Figure 5E).
FIGURE 5.
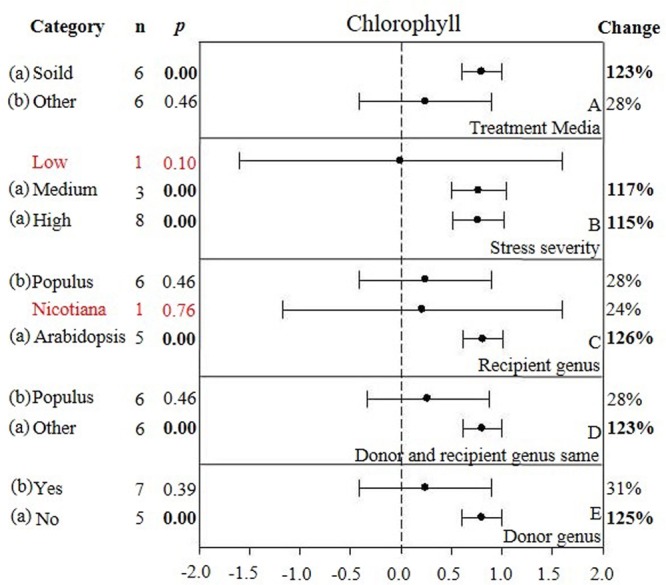
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies chlorophyll. (A) Treatment media, (B) Stress severity, (C) Recipient genus, (D) Donor and recipient genus same, and (E) Donor genus. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
There were two of the four moderators significantly affected the TC-induced increase in seed germination observed in CBL-overexpressing plants subjected to NaCl stress (Figure 6, Table 4, and Appendix S1). The level of stress severity (Figure 6A), and variations in the duration of stress exposure (Figure 6B), had differential impact on the extent to which the overexpression of CBL overexpression in transgenic plants exposed to salt stress increased seed germination. The TC-induced effect of CBL overexpression on seed germination when the donor/recipient combination was composed of different genera was only a quarter of what it was when the donor/recipient combination was composed of the same genera (51% vs. 238%, respectively) (238% vs. 51%) (Figure 6C). TC-induced enhancement of seed germination in salt stressed plants was significantly affected by the donor genus (Figure 6D). A much higher level of germination was observed when the genus of the gene donor was Arabidopsis (238%), as compared to when it was Populus (13%) or others (101%).
FIGURE 6.
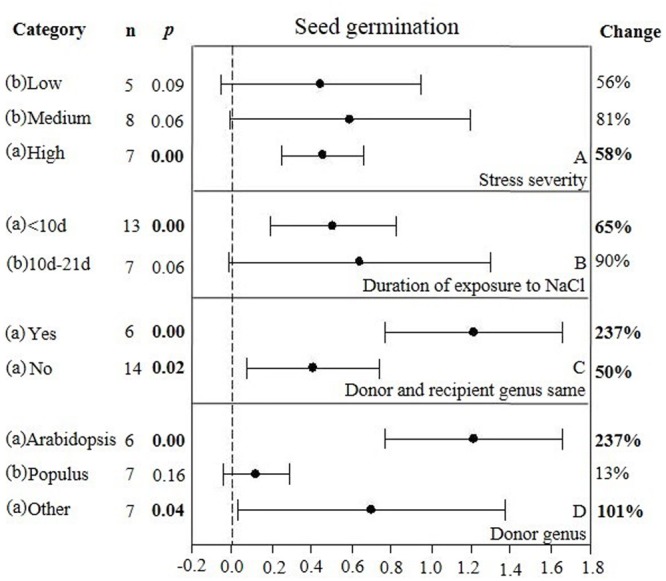
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CBL transformation modifies seed germination. (A) Stress severity, (B) Duration of exposure to NaCl, (C) Donor and recipient genus same, and (D) Donor genus. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Table 4.
Categorical moderators examined for the six summary effects of CBL- transformation that were significantly different from zero Heterogeneity p-values (p-hetero) are shown here.
| Effect size | Moderator |
||||
|---|---|---|---|---|---|
| Treatment media | Duration of exposure to NaCl | Stress severity | Donor and recipient genus same | Donor genus | |
| Seed germination | – | 0.716 | 0.915 | 0.005 | 0.000 |
| Shoot fresh weight | 0.254 | 0.412 | 0.365 | 0.190 | 0.100 |
probability that all observed (total) heterogeneity is not due solely to sampling error (expected variation) p-hetero ≤ 0.1 indicates significant heterogeneity among true effects. Dashed entries signify that insufficient data were available to perform an analysis. See supplement for associated Qm values (between study heterogeneity); n (sample size); df (degrees of freedom, levels within a moderator).
Only one of the five moderators significantly affected the TC-induced increase in fresh weight observed in CBL-overexpressing plants subjected to NaCl stress (Figure 7, Table 4, and Appendix S1). CBL overexpression had approximately 4× the effect on increased shoot fresh weight when plants were grown on solid media (236%), as compared to soil grown plants (66%) (Figure 7A). TC-induced increases in shoot fresh weight were greater when the duration of the stress was 10–21 days, as compared to when the stress was applied for shorter durations (Figure 7B). Increases in shoot fresh weight were significantly greater (358%) at the highest level of stress severity than it was at moderate and low levels of salt stress severity (90 and 96%, respectively) (Figure 7C). Whether the donor and recipient were of the same genus, had little influence on the TC-induced increase in shoot fresh weight, and of different genera, had significant influence on the TC-induced increase in shoot fresh weight in CBL-overexpressing plants subjected to NaCl stress (Figure 7D). The TC-induced increase in shoot fresh weight was significantly affected by the genus of the donor (Figure 7E). The average increase in shoot fresh weight was 69% when the donor genus was Arabidopsis, 65% when the donor genus was Populus, and 534% when the donor was a genus other than the Arabidopsis or Populus.
Discussion
The CBL-CIPK signaling network has been demonstrated to be involved in plant responses to salt stress, in which the salt-overly-sensitive (SOS) pathway is a classic. The SOS pathway is comprised of three key components: SOS1/NHX7, SOS2/CIPK24, and SOS3/CBL4 (Zhu, 2002). It has been reported that overexpression of CBL-CIPK network genes in plants could increase their ability to maintain ionic homeostasis and thus increase salt tolerance (Appendix S1). Numerous studies have been conducted in the past decade to overexpress native or ectopic CBL and CIPK genes in a variety of crop species in an attempt to develop transgenic salt-tolerant crops (Appendix S1). In the present study, a meta-analysis of published data was conducted in order to comprehensively determine the effect of overexpressing either CBL or CIPK genes on various plant parameters in transgenic plants subjected to salt stress, as well as to identify the moderators (experimental variables) that influence the impact of CBL or CIPK overexpression on those parameters.
Salt Tolerance Related Traits That Were Significantly Improved by the Overexpression of CBL-CIPK Genes in Transgenic Plants
Over-expression of CIPK gene family members significantly improves five salt-tolerance related plant parameters and decreases one parameter (Figure 1A). One of the parameters that was most significantly improved is root length, which was 168% greater in transgenic plants exposed to NaCl stress, as compared to non-transgenic plants. Another major effect was an increase in chlorophyll content, which exhibited an average increase of 114%. The higher levels of chlorophyll in transgenic plants subjected to salt stress would contribute to their ability to sustain photosynthesis and assimilation during episodes of salt stress (Smillie and Nott, 1982; Rout et al., 1997). The overexpression of CIPK genes in transgenic plants also resulted in a 77% increase in seed germination when the transgenic plants were subjected to salt stress, a reliable indicator of salt tolerance (Tahal et al., 2000; Shabala and Cuin, 2008). It is worth noting that the overexpression of CIPK gene family members had no significant effect on these plant parameters in plants growing under non-stress conditions (Figure 1B). Overexpression of CBL genes significantly improved just two plant salt-tolerance related traits in transgenic plants subjected to salt stress (Figure 2A). One of the most significant effects was a 173% improvement in the shoot fresh weight of transgenic plants, while the other major effect was a 78% increase in seed germination. No significant effect on these traits was observed when plants were grown under non-stress conditions (Figure 2B). The results of the meta-analysis clearly demonstrate that overexpression of CBL or CIPK family member genes can improve the performance of plants when they are subjected to salt stress. When plants are subjected to abiotic stress, a stress signal is generated that specifically involves Ca2+ (Rudd and Franklin-Tong, 2001; Sanders et al., 2002; Dodd et al., 2010). Previous studies have demonstrated that the interaction between CBL and CIPK requires the micromolar levels of Ca2+ (Halfter et al., 2000; Akaboshi et al., 2008). The level of Ca2+ required for the CBL-CIPK complex to be activated can vary depending on the participating CBL and CIPK members, but is still nevertheless required for activation. Ca2+ signals are elicited by many diverse stimuli, including salinity, and the CBL-CIPK system is involved in transmitting the stress perception via the Ca2+ signaling pathway (Yu et al., 2014; Zhou et al., 2014). It is plausible that this might be the reason why overexpression of the CBL and CIPK gene family members had no effect on the measured salt-tolerance related plant parameters in the absence of salt stress; since salt stress changes the Ca2+ concentration in plants and the CBL-CIPK complex requires Ca2+ to become activated.
The Influence of Experimental Variables (Moderators) on CIPK or CBL Induced Changes in Transgenic Plants Subjected to Salt Stress
The donor genus had a significant influence on the CIPK and CBL overexpression effect on seed germination. Previous studies have demonstrated that the CBL-CIPK complex acts as a negative regulator of the ABA signaling during seed germination (Kim et al., 2003; Pandey et al., 2004, 2008), however, CBL is a positive regulator of the GA pathway (Hwang et al., 2005). Therefore, we suggest, based on the meta-analysis, that CIPK and CBL differ in their regulatory function and that the regulatory mechanisms associated with these genes are genus-specific and perhaps gene family member specific. In this regard, seed germination was the greatest when the CIPK gene was obtained from Populus (315%) and the CBL gene was obtained from Arabidopsis (237%). Shoot fresh weight in transgenic plants overexpressing either CIPK or CBL genes exhibited a significant increase, relative to non-transgenic plants, when plants were grown under salt stress. The potential role of the CBL-CIPK system in inorganic nutrient sensing (Cheong et al., 2003, 2007; Xu et al., 2006; Hu et al., 2009), may explain these results. The size of the effect on shoot fresh weight, however, was affected by the donor genus in CBL transgenic plants. Therefore, we suggest that overexpression of CBL or CIPK genes can improve the shoot fresh weight of plants grown under salt stress conditions, but that the extent of the improvement is dependent on the binding capacity of CBL to stress signaling components; a CBL characteristic that is genus-specific. Further research will be required to determine how the CBLs from different genera differ in their ability to affect shoot fresh weight.
The meta-analysis indicated that gene donor/recipient combinations had a significant moderating influence on the CBL or CIPK induced increase in seed germination and chlorophyll content, respectively. When the genera of the CBL gene donor/recipient combination were different, seed germination was greater than when the genera of the CBL gene donor/recipient combination were the same. These results suggest that when studies are conducted on CBL-CIPK as a complex or a system in their response to abiotic stress (Xu et al., 2006; Pandey et al., 2008), that the impact of overexpression on stress tolerance related parameters may vary depending on the genus from which the CIPK or CBL gene is obtained; as well as the genus of the recipient plant.
Future Research
Salt stress induces many physiological changes in plants and numerous alterations in many parameters have been documented, including the compatible solutes, relative electrolyte leakage (REL), and K+ and other ionic balances (Ma et al., 2014; Song et al., 2014). Many of these parameters, however, were not measured in the available CIPK or CBL overexpression studies. Therefore, these parameters were not included in our meta-analysis due to small sample size (n < 5). Many of the classic physiological changes that occur in plants in response to salt stress were rarely examined in CBL and CIPK overexpression studies. As a result, it is unclear whether or not these parameters would even be affected in the transgenic plants, or what role they might play in salt response/tolerance.
Ca2+ has been identified as a key signaling molecule (Ditta, 2013), and the CBL-CIPK system has been identified as a calcium dependent signaling pathway. Few studies of CBL or CIPK -transformed plants, however, have included measurements of Ca2+ concentration in plant tissues. Previous studies have also shown that the CBL-CIPK network regulates an inward rectifier K+ channel, such as AKT1 (Li et al., 2006; Cheong et al., 2007), and regulates the high-affinity K+ transporter, such as AtHAK5 (Ragel et al., 2015). Again, however, K+ concentration in the root or in shoots has rarely been measured (n < 5) in CBL and CIPK-overexpressing plants. Measurement of K+ and Ca2+ changes in future studies would contribute greatly to developing a better understanding of salt tolerance in relation to the role of CBL-CIPK. Almost all of the CBL genes in the transgenic studies have been obtained from dicots. In fact, only four studies used monocots as the source of the transgene. The majority of recipient plants have also been dicots, with only two studies in one report using a monocot as a recipient. Therefore, it remains unclear whether or not CBL genes from monocotyledonous plants, when over-expressed in monocots, will improve salt tolerance as markedly as they do in dicots. A similar situation exists for the use of CIPK genes. Among 125 studies, only 8 studies overexpressed the CIPK gene in monocotyledonous plants. Therefore, it still remains unclear whether or not CIPK overexpression in monocots definitively improves salt tolerance.
Experimental evidence has suggested that CIPK genes are activated by CBL genes in a Ca2+-dependent manner (Kim et al., 2003) and that the CBL-CIPK complex post-translationally phosphorylates downstream target proteins (Xu et al., 2006). On this basis, one would predict that overexpressing both CBL and CIPK genes together in one recipient plant would be more effective in promoting salt tolerance than the overexpression of just a single one of these genes. Unfortunately, this approach was only used in two studies and, therefore, could not be used as a moderator in our meta-analysis due to the small sample size. As more studies using this approach are conducted, it would be interesting to include this approach as a moderator in future meta-analyses to better understand the effect of CIPK and CBL overexpression.
Author Contributions
YM, QC, and ZC designed the experiment. YM, QC, and HL performed the experiment. YM, JL, and YC analyzed the data. YM and ZC wrote the paper. All of the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the Postdoctoral Foundation of Jiangsu Province (1601142B); and Jiangsu Agriculture Science and Technology Innovation Fund of China [CX(12)5033] and the National Natural Sciences Foundation of China (No. 31372051, 31540052).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00856/full#supplementary-material
Details of all the paper, key word, effect size and mordoritor.
Measures used in characterizing publication bias for each effect size of CIPK transformation.
Measures used in characterizing publication bias for each effect size of CBL transformation.
Weighted summary effect sizes (ln R) and 95% confidence intervals (CIs) showing how moderator variables affect the extent to which CIPK transformation modifies shoot fresh weight. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies MDA levels. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies proline levels. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
References
- Akaboshi M., Hashimoto H., Ishida H., Saijo S., Koizumi N., Sato M., et al. (2008). The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 377 246–257. 10.1016/j.jmb.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Begg C. B., Mazumdar M. (1994). Operating characteristics of a bank correlation test for publication bias. Biometrics 50 1088–1101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- Borenstein M., Hedges L. V., Higgins J. P. T., Rothstein H. R. (2009). Introduction to Meta-Analysis. Manhattan, NY: John Wiley & Sons. [Google Scholar]
- Chaves-Sanjuan A., Sanchez-Barrena M. J., Gonzalez-Rubio J. M., Moreno M., Ragel P., Jimenez M., et al. (2014). Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. U.S.A. 111 4532–4541. 10.1073/pnas.1407610111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y. H., Kim K. N., Pandey G. K., Gupta R., Grant J. J., Luan S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15 1833–1845. 10.1105/tpc.012393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y. H., Pandey G. K., Grant J. J., Batistic O., Li L., Kim B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52 223–239. 10.1111/j.1365-313X.2007.03236.x [DOI] [PubMed] [Google Scholar]
- Cooper H. M. (2009). Research Synthesis and Meta-Analysis: A Step-by-Step Approach. Los Angeles, CA: Sage. [Google Scholar]
- Cramer G. R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming C. (2012). Understanding the New Statistics: Effect Sizes, Confidence Intervals, and Meta-Analysis. New York, NY: Routledge Taylor & Francis Group. [Google Scholar]
- Ditta A. (2013). “Salt tolerance in cereals: molecular mechanisms and applications,” in Molecular Stress Physiology of Plants ed. Rout G. (New Delhi: Springer; ) 133–154. [Google Scholar]
- Dodd A. N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61 593–620. 10.1146/annurev-arplant-070109-104628 [DOI] [PubMed] [Google Scholar]
- Gurevitch J., Hedges L. V. (1999). Statistical issues in ecological meta-analyses. Ecology 80 1142–1149. 10.1890/0012-9658(1999)080[1142:SIIEMA]2.0.CO;2 [DOI] [Google Scholar]
- Halfter U., Ishitani M., Zhu J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97 3735–3740. 10.1073/pnas.040577697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L. R., Yang X. Y., Wang L. C., Zhu L. F., Zhou T., Deng J. W., et al. (2013). Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 435 209–215. 10.1016/j.bbrc.2013.04.080 [DOI] [PubMed] [Google Scholar]
- Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratge-Faillie C., et al. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21 1116–1130. 10.1038/cr.2011.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren T., Hallgren H. B., Oberg B., Adolfsson L., Johansson K. (2013). Republished research: effect of specific exercise strategy on need for surgery in patients with subacromial impingement syndrome: randomised controlled study. Br. J. Sport Med. 47 908–908. 10.1136/bjsports-2012-e787rep [DOI] [PubMed] [Google Scholar]
- Hu D. G., Ma Q. J., Sun C. H., Sun M. H., You C. X., Hao Y. J. (2016). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 156 201–214. 10.1111/ppl.12354 [DOI] [PubMed] [Google Scholar]
- Hu H. C., Wang Y. Y., Tsay Y. F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57 264–278. 10.1111/j.1365-313X.2008.03685.x [DOI] [PubMed] [Google Scholar]
- Hwang Y. S., Bethke P. C., Cheong Y. H., Chang H. S., Zhu T., Jones R. L. (2005). A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol. 138 1347–1358. 10.1104/pp.105.062703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovelli R., Alesini D., Palazzo A., Trenta P., Santoni M., De Marchis L., et al. (2014). Targeted therapies and complete responses in first line treatment of metastatic renal cell carcinoma. A meta-analysis of published trials. Cancer Treat. Rev. 40 271–275. 10.1016/j.ctrv.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Jia W. S., Wang Y. Q., Zhang S. Q., Zhang J. H. (2002). Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 53 2201–2206. 10.1093/jxb/erf079 [DOI] [PubMed] [Google Scholar]
- Kim K. N., Cheong Y. H., Grant J. J., Pandey G. K., Luan S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15 411–423. 10.1105/tpc.006858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klümper W., Qaim M. (2014). A meta-analysis of the impacts of genetically modified crops. PLoS ONE 9:e111629 10.1371/journal.pone.0111629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Xu Q., Harter K., Gruissem W., Luan S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. U.S.A. 96 4718–4723. 10.1073/pnas.96.8.4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajeunesse M. J., Forbes M. R. (2003). Variable reporting and quantitative reviews: a comparison of three meta-analytical techniques. Ecol. Lett. 6 448–454. 10.1046/j.1461-0248.2003.00448.x [DOI] [Google Scholar]
- Lee S. C., Lan W. Z., Kim B. G., Li L. G., Cheong Y. H., Pandey G. K., et al. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. U.S.A. 104 15959–15964. 10.1073/pnas.0707912104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A., Rillig M. C. (2015). Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops – a meta-analysis. Soil Biol. Biochem. 81 147–158. 10.1016/j.soilbio.2014.11.013 [DOI] [Google Scholar]
- Li L. G., Kim B. G., Cheong Y. H., Pandey G. K., Luan S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 12625–12630. 10.1073/pnas.0605129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. P., Zhu J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945. 10.1126/science.280.5371.1943 [DOI] [PubMed] [Google Scholar]
- Ma D. M., Xu W. R., Li H. W., Jin F. X., Guo L. N., Wang J., et al. (2014). Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 251 219–231. 10.1007/s00709-013-0540-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.-C., Augé R. M., Dong C., Cheng Z.-M. (2017). Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: a meta-analysis. Plant Biotechnol. J. 15 162–173. 10.1111/pbi.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M., Caballero F., Martinez V., Rubio F. (2012). Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol. 53 423–432. 10.1093/pcp/pcr194 [DOI] [PubMed] [Google Scholar]
- Ohta M., Guo Y., Halfter U., Zhu J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C AB12. Proc. Natl. Acad. Sci. U.S.A. 100 11771–11776. 10.1073/pnas.2034853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G. K., Cheong Y. H., Kim K. N., Grant J. J., Li L. G., Hung W., et al. (2004). The calcium sensor calcineurin B-Like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16 1912–1924. 10.1105/tpc.021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G. K., Grant J. J., Cheong Y. H., Kim B. G., Li L. G., Luan S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1 238–248. 10.1093/mp/ssn003 [DOI] [PubMed] [Google Scholar]
- Pandey G. K., Kanwar P., Singh A., Steinhorst L., Pandey A., Yadav A. K., et al. (2015). Calcineurin B-Like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 169 780–792. 10.1104/pp.15.00623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P., Rodenas R., Garcia-Martin E., Andres Z., Villalta I., Nieves-Cordones M., et al. (2015). The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in arabidopsis roots. Plant Physiol. 169 2863–2873. 10.1104/pp.15.01401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout N. P., Tripathi S. B., Shaw B. P. (1997). Effect of salinity on chlorophyll and proline contents in three aquatic macrophytes. Biol. Plant. 40 453–458. [Google Scholar]
- Rudd J. J., Franklin-Tong V. E. (2001). Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol. 151 7–33. 10.1046/j.1469-8137.2001.00173.x [DOI] [PubMed] [Google Scholar]
- Ruiz-Lozano J. M., Porcel R., Azcon C., Aroca R. (2012). Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J. Exp. Bot. 63 4033–4044. 10.1093/jxb/ers126 [DOI] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J. F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 401–417. 10.1105/tpc.002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer R. N., Manson J. S., Irwin R. E. (2013). Effects of abiotic factors and species interactions on estimates of male plant function: a meta-analysis. Ecol. Lett. 16 399–408. 10.1111/ele.12044 [DOI] [PubMed] [Google Scholar]
- Shabala S., Cuin T. A. (2008). Potassium transport and plant salt tolerance. Physiol. Plant. 133 651–669. 10.1111/j.1399-3054.2007.01008.x [DOI] [PubMed] [Google Scholar]
- Smillie R. M., Nott R. (1982). Salt tolerance in crop plants monitored by chlorophyll fluorescence in vivo. Plant Physiol. 70 1049–1054. 10.1104/pp.70.4.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A. P., An J., Guan Z. Y., Jiang J. F., Chen F. D., Lou W. H., et al. (2014). The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 80 114–120. 10.1016/j.plaphy.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Sterne J. A. C., Egger M. (2005). “Regression methods to detect publication and other bias in meta-analysis,” in Publication bias in Meta-Analysis: Prevention, Assessment and Adjustments eds Rothstein H. R., Sutton A. J., Borenstein M. (Oxford: Blackwell Science; ) 99–110. [Google Scholar]
- Tahal R., Mills D., Heimer Y., Tal M. (2000). The relation between low K+/Na+ ratio and salt-tolerance in the wild tomato species Lycopersicon pennellii. J. Plant Physiol. 157 59–64. 10.1016/S0176-1617(00)80136-4 [DOI] [Google Scholar]
- Viechtbauer W. (2007). Publication bias in meta-analysis: prevention, assessment and adjustments. Psychometrika 72 269–271. 10.1007/s11336-006-1450-y [DOI] [Google Scholar]
- Wang X. P., Chen L. M., Liu W. X., Shen L. K., Wang F. L., Zhou Y., et al. (2016). AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis. Plant Physiol. 170 2264–2277. 10.1104/pp.15.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worchel E. R., Giauque H. E., Kivlin S. N. (2013). Fungal symbionts alter plant drought response. Microb. Ecol. 65 671–678. 10.1007/s00248-012-0151-6 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang Y. M., Xiong L. Z. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144 1416–1428. 10.1104/pp.107.101295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H. D., Chen L. Q., Wang Y., Liu L. L., He L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360. 10.1016/j.cell.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Yu Q. Y., An L. J., Li W. L. (2014). The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 33 203–214. 10.1007/s00299-013-1507-1 [DOI] [PubMed] [Google Scholar]
- Zhang H. F., Yang B., Liu W. Z., Li H. W., Wang L., Wang B. Y., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14:8 10.1186/1471-2229-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J. J., Bi Y. F., Wang L. K., Tang L. Z., Yu X., et al. (2014). Overexpression of PtSOS2 enhances salt tolerance in transgenic Poplars. Plant Mol. Biol. Rep. 32 185–197. 10.1007/s11105-013-0640-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of all the paper, key word, effect size and mordoritor.
Measures used in characterizing publication bias for each effect size of CIPK transformation.
Measures used in characterizing publication bias for each effect size of CBL transformation.
Weighted summary effect sizes (ln R) and 95% confidence intervals (CIs) showing how moderator variables affect the extent to which CIPK transformation modifies shoot fresh weight. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies MDA levels. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.
Weighted summary effect sizes (ln R) and 95% CIs showing how moderator variables affect the extent to which CIPK transformation modifies proline levels. A p ≤ 0.05 indicates that the moderator level was significantly different than zero; n stands for the number of studies.


