Abstract
Objective
Hypermethylation of human papillomavirus (HPV) and host genes has been reported in cervical cancer. However, the degree of methylation of different HPV types relative to the severity of the cervical lesions remains controversial. Studies of the degree of methylation associated with the host gene and the HPV genome to the severity of cervical lesions are rare. We examined the association of methylation status between host genes and late gene 1 (L1) regions of HPV16, 18, 52, and 58 in cervical brushings.
Methods
Cervical brushings from 147 HPV-infected patients were obtained. The samples comprised normal (n=28), cervical intraepithelial neoplasia (CIN) 1 (n=45), CIN2 (n=13), and CIN3/carcinoma in situ (n=61). The methylation status of HPV and host genes was measured using bisulfite pyrosequencing and quantitative methylation-specific polymerase chain reaction (PCR).
Results
The degree of methylation of L1 in HPV16, 18, and 52 was associated with the severity of the cervical lesion. In HPV52, C-phosphate-G (CpG) sites 6368m, 6405m, and 6443m showed significantly higher methylation in lesions ≥CIN3 (p=0.005, 0.003, and 0.026, respectively). Methylation of most HPV types except HPV52 (r<−0.1) was positively correlated with the degree of methylation of host genes including PAX1 and SOX1 (0.4≤r≤0.7). Combining HPV methylation with PAX1 methylation improved the clustering for ≥CIN2.
Conclusion
Our study showed that the degree of L1 methylation of HPV16, 18, and 52 but not 58 is associated with the severity of cervical lesions. The association between HPV methylation and host gene methylation suggests different responses of host cellular epigenetic machinery to different HPV genotypes.
Keywords: DNA Methylation, Viruses, Methylation, PAX1 Transcription Factor, Cervical Intraepithelial Neoplasia, Human Papillomavirus 16, Human Papilloma Virus
INTRODUCTION
In 2012, there were approximately 528,000 new cases of cervical cancer and 266,000 deaths caused by cervical cancer worldwide [1]. Human papillomavirus (HPV) infection is an important risk factor for cervical cancer: 99.7% of cervical cancers are associated with HPV infection. More than 100 HPV types have been identified and can be categorized into high-risk and low-risk based on their carcinogenic ability. The high-risk HPV (hrHPV) types include 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82 [2]. About 71% of cervical cancer is associated with infection with HPV16 and HPV18. HPV prevalence rates and types differ by ethnic group, and infection with HPV52 and HPV58 is more common in Asia than in other regions [3]. Infection with hrHPV is considered the major essential risk factor for cervical cancer and its precursor lesions [4]. HPV E6 and E7 are the 2 most important oncogenic proteins required for cervical carcinogenesis, and are known to activate oncogenic pathways and repress tumor suppressor pathways. Degradation of p53 by E6 and of retinoblastoma (Rb) by E7 results in dysregulation of the host cell cycle and cell growth [5]. Most HPV infection is transient, but persistent infection with hrHPV lasting more than 2 years can develop into high-grade cervical intraepithelial neoplasia (CIN) [6]. However, infection with hrHPV is necessary but not sufficient to cause cervical cancer [7]: other factors such as epigenetic alterations are also critical in cervical cancer development.
Epigenetic alterations such as DNA methylation and histone modifications can result in heritable gene regulation without changes to the genetic sequence and are recognized as important causes of cancer [8]. DNA methylation is an important mechanism of gene regulation, and the accumulation of alterations in DNA methylation has been shown to be related to disease severity. Alteration in DNA methylation is an early and frequent event during carcinogenesis. Such changes have been found in precancerous and malignant cervical tissue that involved global hypomethylation and the local hypermethylation of tumor suppressor gene (TSG) promoters [9]. The hypermethylation of promoters inactivates their associated genes, resulting in abnormalities in many biological functions. Genes affected include RASSF1A and MGMT, involved in DNA repair, CDKN2A, involved in cell-cycle control, PYCARD, involved in apoptosis, APC and SFRP1, involved in Wnt signaling [10]. This abnormal methylation of genes could serve as a marker for early diagnosis and cancer screening [11].
Over 20 genes, including PAX1, SOX1, ZNF582, PCDHA4, and PCDHA13 have been confirmed to be hypermethylated in cervical brushings diagnosed as CIN2 and more severe lesions. Studies have shown that their sensitivity for CIN2+ or CIN3+ is in the range of 69%–74% for clinical specimens, with a specificity in the range of 66%–82% [3,12,13]. Meta-analyses have shown the utility of PAX1 methylation status as a promising biomarker for cervical cancer screening. In addition, combining analysis of host gene methylation with hrHPV DNA testing appears to be a new strategy in cervical cancer screening. Better specificity for host gene methylation status could improve the accuracy of detection of advanced cervical abnormalities in HPV-positive women [14]. ZNF582 and a methylation panel tested in parallel with testing for hrHPV DNA has a greater accuracy for detection of CIN2 and more severe lesions than gene methylation alone [15,16]. Genetic or environmental factors may also contribute to the progression of cervical lesions. Epigenetic components such as DNA methylation constitute an interface of the genome with the environment, including viral infections. In addition to changes in methylation of the host genome, the HPV genome also shows methylation in cervical cancer [17].
Infection with viruses such as adenovirus, Epstein-Barr virus (EBV), hepatitis B virus (HBV), and retroviruses causes DNA methylation of the host genome. DNA methylation is a common cellular defense mechanism to silence invading foreign DNA and viral genomes. For adenovirus, methylation of viral promoters leads to long-term viral gene silencing [18]. For EBV and HBV, viral replication is inhibited by methylation of promoters in viral DNA and is reversed by demethylating agents [19,20]. For retroviruses, de novo methylation of retrovirus genomes will block viral gene expression [21]. The HPV genome itself is also a target of DNA methylation.
Although the correlation between the methylation status of the HPV genome (long control region [LCR], early genes, and late genes) and disease severity has been studied, the results are controversial. Some reports showed a positive correlation while others revealed a negative correlation [22]. Different HPV genotypes may be differently methylated. For HPV16 and HPV18, some studies have shown that LCR and late gene 1 (L1) methylation is associated with the severity of cervical lesions, while methylation of some C-phosphate-G (CpG) sites may be weakly associated with cervical disease severity [23,24]. However, there have also been reports demonstrating no correlation [22,25]. For HPV31 and 45 studies showed that L1 methylation status weakly distinguished cervical lesions of different severity, but that LCR methylation showed no correlation with disease severity [24,25]. It appears that the breadth and magnitude of methylation of CpG sites differ widely between HPV genotypes. The role of HPV methylation in cervical cancer remains controversial. Most studies have focused on HPV16 and HPV18, with other types such as 31, 45, 52, and 58 being rarely studied. Therefore, the role in cervical cancer of HPV methylation in other hrHPV types and in other ethnic groups needs to be validated. The correlation between HPV and host gene methylation is also an interesting but less studied issue.
In this study, we aimed to validate the methylation status of L1 CpG sites in HPV16, 18, 52, and 58 in cervical lesions from Asian patients. We further investigated the correlation of the methylation status of each CpG site in HPV with disease severity and the correlation between the degree of methylation of virus and host genes to highlight the epigenetic mechanisms involved in HPV infection and their clinical relevance.
MATERIALS AND METHODS
1. Clinical samples
We collected 147 cervical brushings from patients from the Gynecologic Committee of the Taiwan Cooperative Oncology Group (TCOG) cohorts [12] who were infected with HPV and who were diagnosed as normal (n=28), with CIN1 (n=45), CIN2 (n=13), and CIN3/carcinoma in situ (CIS, n=61). Cytological, histological, and clinical data for all patients were reviewed by a panel of colposcopists, cytologists, and pathologists. The patients' final diagnosis was reported as the most severe grade of abnormality identified in punch or cone biopsy specimens. Including criteria were patients aged ≥20 years and enrolled from December 2009 to November 2010. Exclusion criteria were pregnancy, chronic or acute systemic viral infections, a history of cervical neoplasia, skin or genital warts, an immune-compromised state, the presence of other cancers, or a history of surgery to the uterine cervix. The infecting HPV types included 16, 18, 52, and 58. The HPV genotype was determined by reverse line blot hybridization (the prototype Roche linear array HPV test; Roche Molecular Systems Inc., Pleasanton, CA, USA) and consensus polymerase chain reaction (PCR) of the L1 sequences. The demographic details of patients are listed in Supplementary Table 1. Informed consent was obtained from all patients, and this study was approved by the Institutional Review Boards of all participating medical centers.
2. Collection of specimens and preparation of genomic DNA
The cervical specimens were collected by cervical brushing (PAP BRUSH; Young Ou Co., Ltd., Yongin, Korea). After cells were brushed off, the brush was immediately placed into 4℃ sterile phosphate-buffered saline (PBS) for subsequent DNA extraction. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany).
3. Bisulfite modification and pyrosequencing
We used bisulfite pyrosequencing assay for the analysis of HPV gene methylation and used qMSP for the analysis of host gene methylation [12]. Bisulfite conversion was performed by EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA). The positive control is CpG Methylated Human Genomic DNA (Thermo Fisher, Waltham, MA, USA). The negative control is genomic DNA from healthy white blood cells. According to the manufacturer's protocol, the methylation level of target CpG sites ranges from 0% to 100%. We used PyroMark Assay Design 2.0 software (Qiagen GmbH) to design the primers, and PyroMark PCR kits to amplify the DNA (20 ng of bisulfite-converted DNA, 500 nmol/L of each primer, 1×PCR Master Mix in 20 µL total volume). The PCR conditions were as follows: denaturation at 95℃ for 15 minutes, 49 polymerization cycles (95℃ for 30 seconds; the appropriate annealing temperature for 40 seconds and 72℃ for 45 seconds), and a final extension at 72℃ for 5 minutes. The bisulfite pyrosequencing (BPS) was performed according to the manufacturer's recommendations for operation on a PyroMark Q24 System.
4. Statistical analysis
The Mann-Whitney nonparametric U test was used to identify differences in methylation level between 2 categories and the Kruskal-Wallis test for differences between 3 categories. All significant differences were assessed using a 2-tailed p<0.05. The correlation coefficient analysis of methylation profiling at 2 CpG sites used the Pearson correlation. Hierarchical clustering was used log values of methylation level to calculate the matrix of Euclidean distance. For the clustering analysis of the combining host gene and HPV methylation, the methylation values were transformed by quantile normalization. A receiver operating characteristic (ROC) curve was used to select the optimal threshold for distinguishing the control and comparison groups. The optimum threshold was calculated using Youden's method. All analyses and graphs were generated using the statistical package in R (version 3.1.2; R Foundation, Vienna, Austria; https://www.R-project.org).
RESULTS
1. Correlation between HPV methylation status and severity of cervical lesions
To evaluate the degree of HPV methylation related to the spectrum of cervical lesions, we examined HPV methylation in cervical brushings using BPS. Three CpG sites of the HPV16 and 6 CpG sites of the HPV18 L1 gene were examined according to previous studies [23,26]. We selected 3 CpG sites of HPV52 and 4 CpG sites of HPV58 from L1 gene corresponding to HPV16 L1 regions. The locations of these CpG sites are indicated in Supplementary Fig. 1.
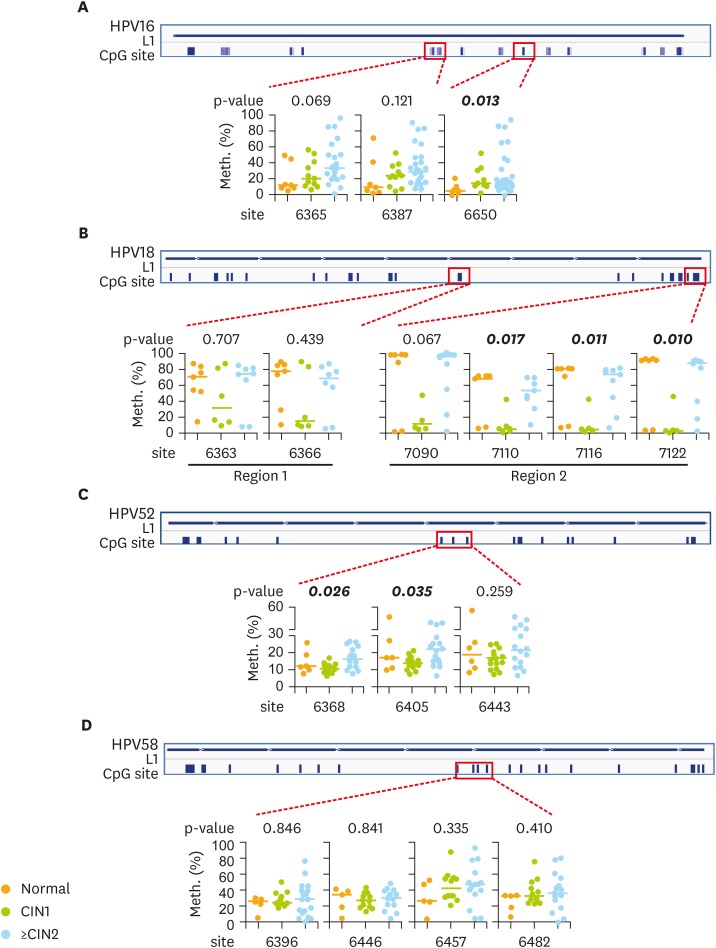
In HPV16, 1 of the 3 CpG sites, 6650m, showed significantly increased methylation with more severe disease (Fig. 1A). In HPV18, the methylation level of 3 CpG sites in region 2, 7110m, 7116m, and 7122m showed significant differences, but not those in region 1. It is interesting that the methylation level in CIN1 was significantly lower than that in both normal controls and ≥CIN2 (Fig. 1B). In HPV52, the methylation levels of the 2 CpG sites were significantly different (Fig. 1C). There was no difference in the methylation level of any CpG site in HPV58 (Fig. 1D).
Fig. 1.
The methylation level of the HPV L1 region in cervical brushings diagnosed with different levels of CIN. The methylation status detected by bisulfate-pyrosequencing in (A) HPV16, (B) HPV18, (C) HPV52, and (D) HPV58 at the indicated CpG sites, with the p-value shown above the graph.
CIN, cervical intraepithelial neoplasia; CpG, C-phosphate-G; HPV, human papillomavirus; L1, late gene 1; Meth., methylation.
In Table 1, when the data were dichotomized, the methylation of HPV16-6650m was significantly higher in ≥CIN1 than in normal controls (14.0 vs. 4.6; p=0.025). In contrast, HPV18-7122m showed significantly lower methylation in ≥CIN1 than in normal controls (43.5 vs. 91.5; p=0.034). The methylation level of HPV52-6368m and 6405m showed differences between both <CIN1 and ≥CIN2. All of these HPV52 CpG sites showed significantly higher methylation in ≥CIN3 than in other stages (Table 1). Other CpG sites showed no difference and listed on Supplementary Table 2. When using ≥CIN2 as the endpoint, CpG site 6368m had the best accuracy with the area under the ROC curve of 74%. When using ≥CIN3, CpG site 6405m conferred the best accuracy of 81% (Table 2).
Table 1. Relationships between HPV methylation level and clinical lesions.
| Variables | <CIN1 | ≥CIN1 | p-value | <CIN2 | ≥CIN2 | p-value | <CIN3 | ≥CIN3 | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV16 | |||||||||||
| 6,650 bp | 0.025 | 0.207 | 0.207 | ||||||||
| Case No. | 8 | 39 | 17 | 30 | 17 | 30 | |||||
| Median±(95% CI) | 4.6 (−5.0–32.1) | 14.0 (−16.6–32.3) | 11.4 (7.7–26.6) | 13.6 (15.8–35.5) | 11.4 (7.7–26.6) | 13.6 (15.8–35.5) | |||||
| HPV18 | |||||||||||
| 7,122 bp | 0.034 | 0.749 | 0.749 | ||||||||
| Case No. | 7 | 14 | 12 | 9 | 12 | 9 | |||||
| Median±(95% CI) | 91.5 (28.0–106.8) | 43.5 (23.8–70.1) | 26.0 (16.3–72.3) | 87.8 (39.2–93.4) | 26.0 (16.3–72.3) | 87.8 (39.2–93.4) | |||||
| HPV52 | |||||||||||
| 6,368 bp | 0.192 | 0.012 | 0.005 | ||||||||
| Case No. | 6 | 33 | 23 | 16 | 28 | 11 | |||||
| Median±(95% CI) | 11.1 (7.5–15.9) | 12.8 (12.6–16.7) | 11.3 (10.4–13.9) | 16.3 (13.9–20.5) | 11.6 (10.8–14.5) | 17.1 (14.5–21.9) | |||||
| 6,405 bp | 0.472 | 0.025 | 0.003 | ||||||||
| Case No. | 6 | 33 | 23 | 16 | 28 | 11 | |||||
| Median±(95% CI) | 15.3 (9.8–22.7) | 15.6 (16.0–22.8) | 15.0 (12.9–19.7) | 22.1 (17.3–27.8) | 14.9 (13.6–19.4) | 23.5 (18.0–31.9) | |||||
| 6,443 bp | 0.425 | 0.149 | 0.026 | ||||||||
| Case No. | 6 | 33 | 23 | 16 | 28 | 11 | |||||
| Median±(95% CI) | 14.8 (9.5–23.4) | 17.9 (16.9–24.8) | 17.0 (13.9–22.2) | 21.6 (17.0–29.5) | 15.9 (14.3–21.5) | 22.5 (17.6–34.1) | |||||
All p-values calculated by Mann-Whitney U test.
CI, confidence interval; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
Table 2. Performance of HPV52 methylation in cervical lesions.
| Location of CpG site (bp) | <CIN2 vs. ≥CIN2 | <CIN3 vs. ≥CIN3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | AUC (95% CI) | p-value | Sensitivity | Specificity | AUC (95% CI) | p-value | |
| 6,368 | 68.8 | 87.0 | 0.74 (0.57–0.91) | 0.012 | 81.8 | 82.1 | 0.79 (0.64–0.95) | 0.005 |
| 6,405 | 62.5 | 82.6 | 0.71 (0.54–0.89) | 0.013 | 72.7 | 78.6 | 0.81 (0.67–0.95) | 0.003 |
| 6,443 | 62.5 | 69.6 | 0.64 (0.45–0.83) | 0.154 | 81.8 | 67.9 | 0.73 (0.56–0.91) | 0.025 |
AUC, area under the curve of receiver operating characteristic; CI, confidence interval; CIN, cervical intraepithelial neoplasia; CpG, C-phosphate-G; HPV, human papillomavirus.
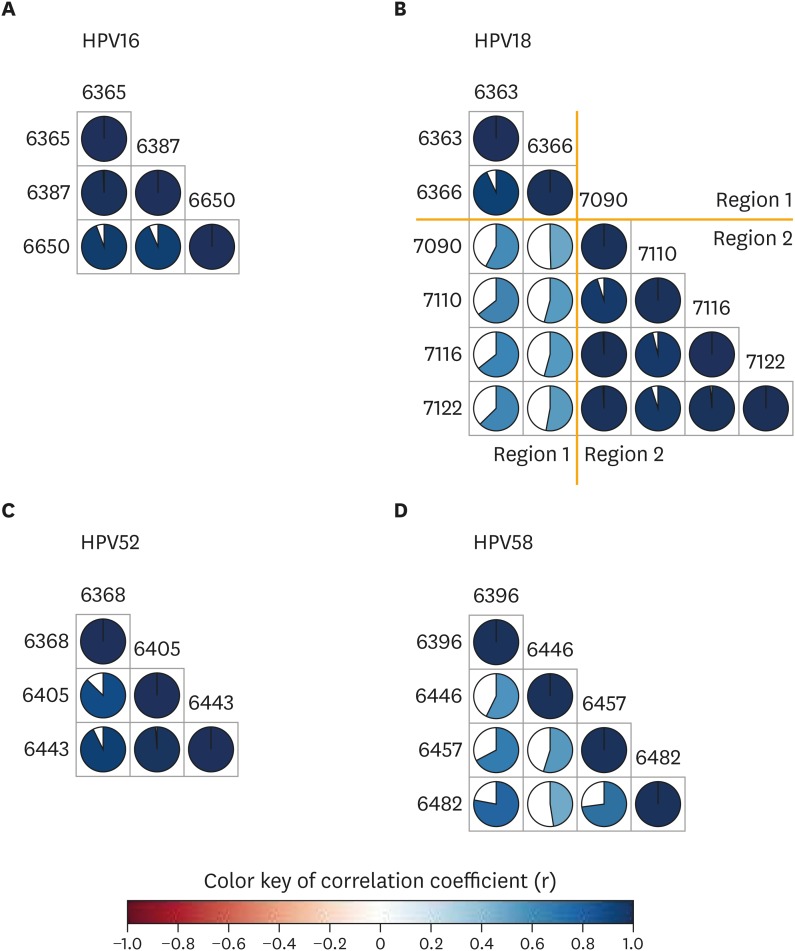
2. Boundary of HPV genome methylation
It is well known that adjacent CpG sites have similar methylation status, which suggests a methylation boundary in epigenetic modification [27]. Because no previous report identifies an HPV genome methylation boundary, we tested the correlation of the methylation levels at each CpG site. Our results demonstrated that all CpGs situated close together in HPV16 and HPV52 had highly correlated methylation status (r>0.9). In HPV18, the CpGs within regions 1 or 2 showed highly correlated methylation status (r>0.9; r>0.9), but the correlation of methylation between regions 1 and 2 was low (r=0.5), suggesting a possible methylation boundary in L1 at about 500 bps (Fig. 2).
Fig. 2.
The correlation between any 2 CpG methylation on HPV L1 region. Each circle indicates the correlation coefficient of methylation between any 2 CpG sites. (A) HPV16. (B) HPV18. (C) HPV52. (D) HPV58. Blue indicates a positive correlation and red indicates a negative correlation.
CpG, C-phosphate-G; HPV, human papillomavirus; L1, late gene 1.
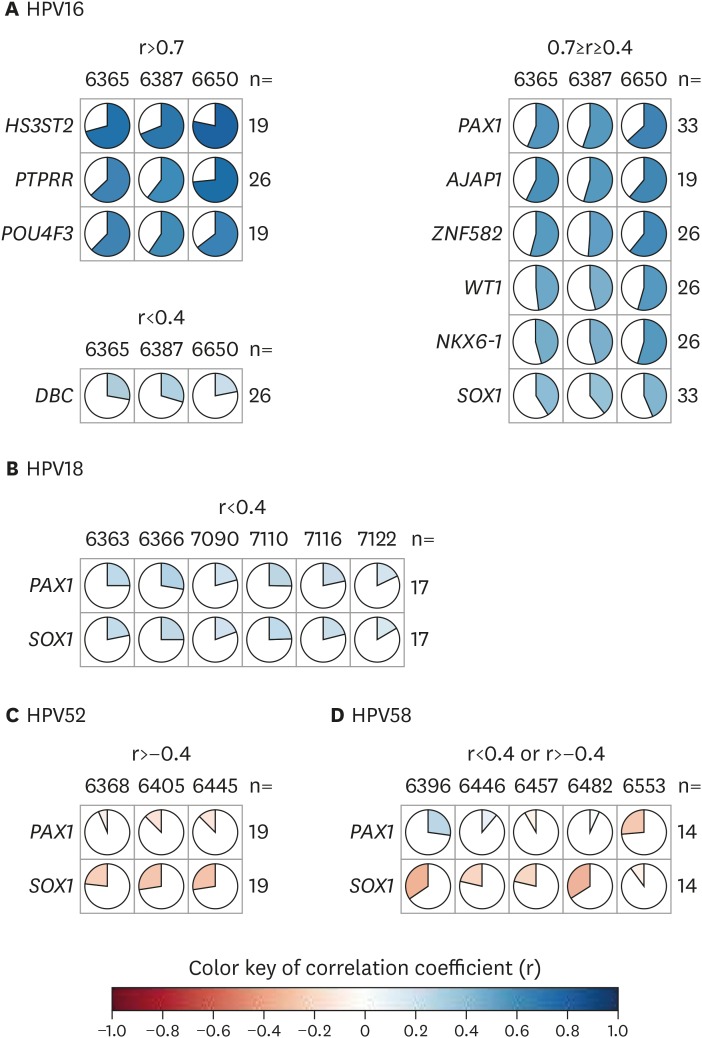
3. Correlation of HPV and candidate host gene methylation
Although the HPV genome may be methylated by the host cellular defense mechanisms, HPV may also influence the host cellular methylation mechanism to alter host genome methylation. The association between HPV genome methylation and host gene methylation is interesting but has not been tested previously. In an earlier study, we identified a correlation between HPV methylation and methylation of host genes [3,28]. Our results demonstrated that HPV16 methylation was positively correlated with host gene methylation, especially that of HS3ST2, PTPRR, and POU4F3 (r>0.7), while HPV18 methylation was weakly correlated with methylation of PAX1 and SOX1 (r<0.4). HPV52 and HPV58 methylation status showed a weak negative correlation with host gene methylation (Fig. 3).
Fig. 3.
The correlation of methylation levels between HPV and host genes. (A) In HPV16, the correlation coefficients are classified into 3 groups: >0.7, 0.7–0.4, and <0.4. Others correlation showed in HPV18 (B), HPV52 (C), and HPV58 (D). Blue and red indicate positive and negative correlation, respectively.
HPV, human papillomavirus; n, case number.
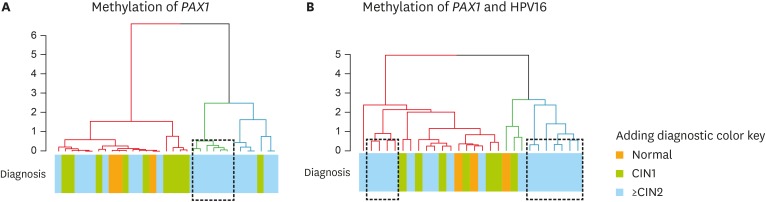
4. Combination of host and viral gene methylation improves cervical lesion screening
Although host gene methylation has been proposed as a biomarker for cervical cancer detection with high specificity, its moderate sensitivity requires improvement [12,16]. One appealing idea is to combine analysis of host and viral gene methylation to improve the sensitivity. Methylation of PAX1 is one of the most promising markers for cervical cancer screening. We tested the possibility of improving the sensitivity of PAX1 for detecting ≥CIN2 lesions. Although the analysis was limited by the number of patients for whom both PAX1 and HPV type-specific methylation data were available, we used clustering analysis to test whether the inclusion of HPV methylation status could improve the sensitivity of PAX1 methylation status for the classification of disease. PAX1 alone classified a single ≥CIN2 group, while PAX1 and HPV methylation in combination classified 2 ≥CIN2 groups, suggesting a potential benefit of adding HPV methylation status to the analysis (Fig. 4).
Fig. 4.
The dendrogram of hierarchical clustering displaying the merged groups for cervical diagnosis using DNA methylation. The dashed lines indicate the 3 merged groups. The clustering was calculated using DNA methylation values from (A) methylation of PAX1 and (B) methylation of PAX1 and HPV16.
CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
DISCUSSION
The present study demonstrated that infection with different HPV genotypes leads to different HPV methylation status and that this will correlate with cervical lesion severity. Moreover, different HPV genotypes showed different correlations between HPV methylation and host gene methylation. All these data prompted us to hypothesize that different HPV genotypes may have different effects on the host epigenetic machinery, and that the host epigenetic machinery may have different responses to different HPV genotypes. HPV16 is the most common oncogenic genotype in cervical lesions. Infection with HPV16 may trigger the epigenetic defense machinery to methylate the HPV genome, but hijacking of this defense by HPV16 results in alteration of the methylation of the host genome. This could cause the observed correlation between HPV16 genome methylation and host gene methylation ranging from normal tissue to CIS. However, HPV52, although presumably an oncogenic type, may not strongly induce the host epigenetic machinery so that both viral and host genes show low levels of methylation in the late precancerous stages (Supplementary Fig. 2). This speculation warrants further investigation, which may improve our understanding of the role of epigenetics in HPV oncogenesis.
Some studies have shown that HPV infection interferes with host epigenetic regulation. HPV infection upregulated the expression of DNMT1, DNMT3A, and DNMT3B [29,30]. The mechanism of this appeared to be degradation of p53 by the E6 protein of HPV, which resulted in specificity protein 1 (Sp1) dissociating from the p53/Sp1 complex. Then, phosphorylation of Rb by E7 of HPV resulted in E2F dissociating from the Rb/E2F complex. The released Sp1 and E2F bound to the DNMT promoters and upregulated expression of DNMTs [29,30,31]. Previous studies showed that the E6 protein of HPV16 is capable of binding to p53, which may release more Sp1 and activate more DNMTs and alter the degree of DNA methylation. Taken together, we speculate that infection with different HPV types may result in differences in the methylation status of both the host and virus genomes. This may also affect the carcinogenic activity of different HPV types.
Increasing integration of HPV DNA into the host genome correlates with cervical cancer progression [32]. The integrated form of HPV DNA was found in 80% and 100% of HPV16- and HPV18-infected patients, respectively, but in only about 25% of HPV52-infected patients [33]. Insertion of viral DNA sequences into the host genome often triggers host defense mechanisms such as the DNA methylation machinery, which then mediates the methylation of the virus genome [30,34]. Therefore, the low integration rate of HPV52 may induce a low level of epigenetic defenses. This is consistent with our result that the methylation of L1 in HPV52 was lower than that in HPV16 and HPV18.
Detection of disease-specific gene methylation is a new strategy for cancer screening, and an optimal biomarker will improve detection of cancer at an early stage. SEPT9 methylation was identified as a biomarker of colorectal cancer over 10 years ago [35] and was recently approved for clinical use by the Food and Drug Administration (FDA). In previous studies, cervical cancer-specific methylation of genes such as PAX1, SOX1, ZNF582, and POU4F3 showed their clinical usefulness in detecting ≥CIN3 precancerous samples [3,12,36]. PAX1 methylation was recently approved by Taiwan FDA as an adjunct testing for cervical cancer screening. These methylation biomarkers showed satisfactory clinical results in patients with CIN3 and more severe lesions. However, the goal of early detection is to improve the detection of precancerous lesions before the ≥CIN3 stage by, for instance, identifying new cervical cancer methylation biomarkers, possibly by including analysis of the methylation of other genes or by HPV typing. In this study, we evaluated the feasibility of combining HPV methylation with host gene methylation to improve the detection of CIN3 lesions. Our data showed that the combination of HPV methylation and specific gene methylation improved the clinical performance of classification of precancerous lesions. This result was consistent with the previous reports that HPV16 gene methylation provided some improvement in disease discrimination when combined with methylation of EPB41L3 and LMX1 [37]. A larger patient sample is required to verify the optimal combination for precancer screening of hrHPV-infected CIN patients.
The correlation coefficient for methylation of adjacent CpG sites was high in HPV16 and HPV52, but low in 2 CpG sites more than 500 bps apart in HPV18. This result might reflect the phenomenon of methylation boundaries. Sp1 and CTCCC-binding factor (CTCF) are associated with methylation boundaries in the human genome [38], and there are 2 CTCF binding motifs on the HPV18 genome that might be relevant to this phenomenon. However, HPV18 has a breakpoint at 6,775 to 6,803 bps [39] between regions 1 and 2. If HPV18 cleaves at this position and inserts into the host genome, the distance between regions 1 and 2 will change from 500 bps to more than 7,000 bps. Taken together, these results support our finding that a lower correlation between the methylation of regions 1 and 2 in HPV18 may be the result of a methylation boundary. We searched for Sp1 and CTCF binding motifs in the HPV genome, and identified 1 Sp1 binding motif in HPV52 and 3 in HPV58, but no Sp1 binding motifs in either HPV16 or HPV18. There was 1 CTCF binding motif in HPV16, 2 in HPV18, 4 in HPV52, and 5 in HPV58. Because Sp1 and CTCF protect the surrounding DNA against methylation, a greater number of binding motifs in the HPV genome implies a higher potential to form a methylation-free boundary. The abundant CTCF and Sp1 binding motifs in the HPV58 genome may explain the low methylation of HPV58.
In conclusion, our study showed that L1 methylation of HPV16, HPV18, and HPV52 is associated with cervical lesion severity. The L1 methylation of HPV16 and HPV18 had the potential to differentiate normal from ≥CIN1 cervical lesions. The L1 methylation of HPV52 had the potential to distinguish <CIN2 from ≥CIN2 and <CIN3 from ≥CIN3 cervical lesions. In addition, the combination of host and HPV genome methylation had the potential to improve the diagnosis of cervical lesions. HPV16 and HPV18 methylation was positively correlated and HPV52 methylation was negatively correlated with that of PAX1 and SOX1. These results suggest that infection with different HPV genotypes might lead to different methylation status of the HPV genome and host genes. Increasing the number of samples and exploring wider areas of relevance are the goals of our future studies.
ACKNOWLEDGMENTS
Yaw-Wen Hsu is thankful to Graduate Institute of Life Sciences for the Ph.D. Fellowship.
Footnotes
Funding: This work was supported in part by grants from Ministry of Science and Technology (NSC 102-2628-B-038-010-MY3 and MOST 105-2628-B-038-011-MY3), Taipei Medical University (105TMU-SHH-09), and the Teh-Tzer Study Group for Human Medical Research Foundation.
Conflict of Interest: The application of these candidate genes in cancer detection has been patented. National Defense Medical Center owns right. Hung-Cheng Lai is the inventor.
- Conceptualization: H.Y.W., H.R.L., L.H.C.
- Data curation: H.Y.W., H.R.L.
- Formal analysis: H.Y.W., H.R.L.
- Funding acquisition: L.H.C.
- Investigation: H.Y.W., W.H.C.
- Methodology: H.Y.W.
- Project administration: L.H.C.
- Resources: L.C.C., L.H.C.
- Supervision: L.H.C.
- Validation: H.Y.W.
- Visualization: H.Y.W., H.R.L.
- Writing - original draft: H.Y.W., H.R.L., S.P.H., C.Y.C., L.H.C.
- Writing - review & editing: H.Y.W., H.R.L., S.P.H., L.H.C.
Supplementary Material
Demographics related to the clinical samples
Methylation level of HPV L1 region in patients and control subjects
The distribution of CpG sites in 4 HPV types. The location of CpG sites and genes shown on each HPV genome by IGV. The arrows indicate regions where methylation was detected. (A) HPV16. (B) HPV18. (C) HPV52. (D) HPV58.
The epigenetic aspects of the HPV genome. We hypothesized that different HPV genotypes may have different effects on the host epigenetic machinery. In HPV16, the host methylation machinery is triggered strongly to methylate the viral and host genome. In HPV52, the viral genome methylation is accompanied by less methylation of host gene.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Huang RL, Chang CC, Su PH, Chen YC, Liao YP, Wang HC, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One. 2012;7:e41060. doi: 10.1371/journal.pone.0041060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008;26(Suppl 10):K1–K16. doi: 10.1016/j.vaccine.2008.05.064. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura A, Ono T, Ishimoto A, Dowhanick JJ, Frizzell MA, Howley PM, et al. Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J Virol. 2000;74:3752–3760. doi: 10.1128/jvi.74.8.3752-3760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasagawa T, Takagi H, Makinoda S. Immune responses against human papillomavirus (HPV) infection and evasion of host defense in cervical cancer. J Infect Chemother. 2012;18:807–815. doi: 10.1007/s10156-012-0485-5. [DOI] [PubMed] [Google Scholar]
- 7.Franco EL, Duarte-Franco E, Ferenczy A. Cervical cancer: epidemiology, prevention and the role of human papillomavirus infection. CMAJ. 2001;164:1017–1025. [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 11.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 12.Lai HC, Ou YC, Chen TC, Huang HJ, Cheng YM, Chen CH, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a Taiwanese Gynecologic Oncology Group (TGOG) study. Cancer Med. 2014;3:1062–1074. doi: 10.1002/cam4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang KH, Lin CJ, Liu CJ, Liu DW, Huang RL, Ding DC, et al. Global methylation silencing of clustered proto-cadherin genes in cervical cancer: serving as diagnostic markers comparable to HPV. Cancer Med. 2015;4:43–55. doi: 10.1002/cam4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolaidis C, Nena E, Panagopoulou M, Balgkouranidou I, Karaglani M, Chatzaki E, et al. PAX1 methylation as an auxiliary biomarker for cervical cancer screening: a meta-analysis. Cancer Epidemiol. 2015;39:682–686. doi: 10.1016/j.canep.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Liou YL, Zhang Y, Liu Y, Cao L, Qin CZ, Zhang TL, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics. 2015;7:50. doi: 10.1186/s13148-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijsink JJ, Lendvai Á, Deregowski V, Klip HG, Verpooten G, Dehaspe L, et al. A four-gene methylation marker panel as triage test in high-risk human papillomavirus positive patients. Int J Cancer. 2012;130:1861–1869. doi: 10.1002/ijc.26326. [DOI] [PubMed] [Google Scholar]
- 17.Saavedra KP, Brebi PM, Roa JC. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin Epigenetics. 2012;4:13. doi: 10.1186/1868-7083-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doerfler W. DNA methylation: a defense mechanism against the expression of foreign DNA? In: Berghaus G, Brinkmann B, Rittner C, Staak M, editors. DNA — technology and its forensic application. Berlin: Springer-Verlag Berlin Heidelberg; 1991. pp. 50–58. [Google Scholar]
- 19.Jung EJ, Lee YM, Lee BL, Chang MS, Kim WH. Lytic induction and apoptosis of Epstein-Barr virus-associated gastric cancer cell line with epigenetic modifiers and ganciclovir. Cancer Lett. 2007;247:77–83. doi: 10.1016/j.canlet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, Li Y, Mu S, Zhang J, Yan Z. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J Med Virol. 2009;81:1177–1183. doi: 10.1002/jmv.21525. [DOI] [PubMed] [Google Scholar]
- 21.Jähner D, Stuhlmann H, Stewart CL, Harbers K, Löhler J, Simon I, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 22.Patel DA, Rozek LS, Colacino JA, Van Zomeren-Dohm A, Ruffin MT, Unger ER, et al. Patterns of cellular and HPV 16 methylation as biomarkers for cervical neoplasia. J Virol Methods. 2012;184:84–92. doi: 10.1016/j.jviromet.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wentzensen N, Sun C, Ghosh A, Kinney W, Mirabello L, Wacholder S, et al. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. J Natl Cancer Inst. 2012;104:1738–1749. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marongiu L, Godi A, Parry JV, Beddows S. Human papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer. 2014;14:384. doi: 10.1186/1471-2407-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasiljević N, Scibior-Bentkowska D, Brentnall A, Cuzick J, Lorincz A. A comparison of methylation levels in HPV18, HPV31 and HPV33 genomes reveals similar associations with cervical precancers. J Clin Virol. 2014;59:161–166. doi: 10.1016/j.jcv.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123:161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 29.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 30.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–3632. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 31.Au Yeung CL, Tsang WP, Tsang TY, Co NN, Yau PL, Kwok TT. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 32.Arias-Pulido H, Peyton CL, Joste NE, Vargas H, Wheeler CM. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J Clin Microbiol. 2006;44:1755–1762. doi: 10.1128/JCM.44.5.1755-1762.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102:54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 34.Bryant D, Onions T, Raybould R, Jones S, Tristram A, Hibbitts S, et al. Increased methylation of human papillomavirus type 16 DNA correlates with viral integration in vulval intraepithelial neoplasia. J Clin Virol. 2014;61:393–399. doi: 10.1016/j.jcv.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Song L, Li Y. SEPT9: a specific circulating biomarker for colorectal cancer. Adv Clin Chem. 2015;72:171–204. doi: 10.1016/bs.acc.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Pun PB, Liao YP, Su PH, Wang HC, Chen YC, Hsu YW, et al. Triage of high-risk human papillomavirus-positive women by methylated POU4F3 . Clin Epigenetics. 2015;7:85. doi: 10.1186/s13148-015-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louvanto K, Franco EL, Ramanakumar AV, Vasiljević N, Scibior-Bentkowska D, Koushik A, et al. Methylation of viral and host genes and severity of cervical lesions associated with human papillomavirus type 16. Int J Cancer. 2015;136:E638–E645. doi: 10.1002/ijc.29196. [DOI] [PubMed] [Google Scholar]
- 38.Butcher DT, Mancini-DiNardo DN, Archer TK, Rodenhiser DI. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int J Cancer. 2004;111:669–678. doi: 10.1002/ijc.20324. [DOI] [PubMed] [Google Scholar]
- 39.Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographics related to the clinical samples
Methylation level of HPV L1 region in patients and control subjects
The distribution of CpG sites in 4 HPV types. The location of CpG sites and genes shown on each HPV genome by IGV. The arrows indicate regions where methylation was detected. (A) HPV16. (B) HPV18. (C) HPV52. (D) HPV58.
The epigenetic aspects of the HPV genome. We hypothesized that different HPV genotypes may have different effects on the host epigenetic machinery. In HPV16, the host methylation machinery is triggered strongly to methylate the viral and host genome. In HPV52, the viral genome methylation is accompanied by less methylation of host gene.