Abstract
Checkpoint kinase 1 (Chk1) is an evolutionarily conserved serine/threonine kinase that plays an important role in G2/M checkpoint signaling. Here, we evaluate the radiosensitizing effects of a novel selective Chk1 inhibitor MK-8776, comparing its efficacy with a first-generation Chk1 inhibitor UCN-01, and attempt to elucidate the mechanism of radiosensitization. In a clonogenic survival assay, MK-8776 demonstrated a more pronounced radiosensitizing effect than UCN-01, with lower cytotoxicity. Importantly, radiosensitization by MK-8776 can be achieved at doses as low as 2.5 Gy, which is a clinically applicable irradiation dose. MK-8776, but not UCN-01, exacerbated mitotic catastrophe (MC) and centrosome abnormalities, without affecting repair kinetics of DNA double strand breaks. Furthermore, live-cell imaging revealed that MK-8776 significantly abrogated the radiation-induced G2/M checkpoint, prolonged the mitotic phase, and enhanced aberrant mitosis. This suggests that Chk1 inhibition by MK-8776 activates a spindle assembly checkpoint and increases mitotic defects in irradiated EMT6 cells. In conclusion, we have shown that, at minimally toxic concentrations, MK-8776 enhances radiation-induced cell death through the enhancement of aberrant mitosis and MC, without affecting DNA damage repair.
Introduction
Checkpoint kinase 1 (Chk1) is an evolutionarily conserved serine/threonine kinase that has several important roles in cell cycle regulation. Chk1 is particularly important for the initiation of a G2/M checkpoint in response to genotoxic stress. When cells are exposed to genotoxic stresses, such as ionizing radiation (IR) or chemotherapeutic agents, Chk1 is phosphorylated and activated by ATM- and Rad3-related (ATR) kinase [1]. When activated, Chk1 phosphorylates and inhibits Cdc25 phosphatases, thereby attenuating cyclin-dependent kinase 1 (Cdk1)/cyclin B1 activity and preventing entry into mitosis [1]. In addition, depletion of Chk1 is reported to induce chromosome misalignment, lagging chromosomes, and cytokinesis failure [2]. These findings suggest that Chk1 also has a critical role in the organization of mitotic chromosomes.
In several types of human tumor cells, overexpression of Chk1 confers resistance to IR and chemotherapeutic agents such as bleomycin and cytarabin [3], [4]. Down-regulation of Chk1 during exposure to antimetabolites, such as hydroxyurea and cytarabine, increases cell death, independent of p53 or Chk2 [5]. With Chk1 inhibition alone, cells can remain viable due to up-regulation of compensatory mechanisms [6], but are made more susceptible to extrinsic DNA damage [7]. These properties have generated interest in the possible use of Chk1 inhibitors in combination therapy. 7-Hydroxystaurosporine (UCN-01), one of the first-generation Chk1 inhibitors, has been found to enhance the cytotoxicity of ionizing radiation (IR) and anticancer drugs in various tumor cell lines [8], [9]. However, clinical trials have not progressed due to the erratic pharmacokinetic properties of the compound and broad-spectrum inhibition of off-target kinases that leads to excess toxicity [10]. To overcome these issues, more specific Chk1 inhibitors have been developed and are currently under evaluation [6]. Recently, a compound named MK-8776 (also known as SCH900776) has been developed as a novel Chk1 inhibitor [11], [12]. MK-8776 is a pyrazolo[1,5-a]pyrimidine derivative that was first identified via a functional screen assay and shown to potently inhibit Chk1 [13]. The compound also exhibits chemosensitizition both in vitro and in vivo when combined with DNA-damaging agents, such as gemcitabine or hydroxyurea [14]. A recent phase I clinical study has shown promising clinical efficacy for MK-8776 treatment in patients with advanced solid tumors [15]. MK-8776 is now being tested in combination with cytarabine in a phase II clinical trial with patients that have relapsed acute myeloid leukemia [15]. These studies indicate that MK-8776 enhances cellular susceptibility to chemotherapeutic agents and has the potential to be a clinically viable chemosensitizing agent with fewer side effects.
Previously, various Chk1 inhibitors, including UCN-01, GNE-783, A-690002, and A-641397, have been shown to induce chemo- and radio-sensitization [16], [17], [18]. Several potential mechanisms for sensitization by Chk1 inhibition have been proposed, including inhibition of repair systems for DNA double strand breaks (DSBs), spindle assembly checkpoint (SAC) activation, promotion of premature mitosis, and mitotic catastrophe (MC) [17], [19], [20], [21]. However, it remains unclear how Chk1 inhibition triggers sensitization in tumor cells. In addition, most prior data investigating possible mechanisms for sensitization by Chk1 inhibition were collected when examining combinations of high-irradiation doses and/or low-specificity Chk1 inhibitors, such as UCN-01 [19], [21]. Therefore, clinically relevant irradiation doses and non-toxic concentrations of Chk1 inhibitors should be examined to precisely evaluate the mechanisms by which these drugs sensitize tumor cells during genotoxic stress. In this study, we examined whether sub-lethal concentrations (90% survival concentration when the Chk1 inhibitor was used alone) of MK-8776 or UCN-01 influence cellular clonogenicity, MC, and DNA repair kinetics in murine breast cancer EMT6 cells or human cervical carcinoma HeLa cells exposed to 2.5–10 Gy of X-irradiation. Additionally, the incidence rates of centrosome abnormalities and aberrant mitosis were examined using immunohistochemistry and live-cell imaging techniques.
Materials and Methods
Materials
UCN-01 and MK-8776 were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Cayman Chemical (Ann Arbor, MI, USA), respectively. The nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) was obtained from Thermo Fisher Scientific (Waltham, MA, USA). The following antibodies were used for immunocytochemistry: anti-γ-tubulin (Sigma-Aldrich), anti-p53-binding protein 1 (53BP1; Abcam; Cambridge, UK), anti-γ-H2AX (Cell Signaling Technology, Beverly, MA, USA), Alexa Fluor® 488 anti-rabbit IgG, and anti-mouse IgG (Thermo Fisher Scientific). Plat-E cells and pMXs-IP plasmid were generously gifted by Dr. Toshio Kitamura (University of Tokyo, Tokyo, Japan) [22], [23]. H2B-mCherry and pEF6.mCherry-TSG101 plasmids were gifted by Robert Benezra and Quan Lu (Addgene plasmid #20972 and #38318), respectively [24], [25].
Cell Culture and X-irradiation
Murine breast cancer EMT6 cells were obtained from ATCC (CRL-2755) and maintained in RPMI1640 medium (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS; Biosera; Nuaille, France) at 37°C in a humidified atmosphere of 5% CO2. Human cervical carcinoma HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific) supplemented with 10% (v/v) FBS at 37°C in a humidified atmosphere of 5% CO2. X-irradiation was performed using an X-Rad iR-225 (Precision X-Ray; North Branford, CT, USA) at 200 kVp, 15 mA with a 1.0-mm aluminum filter.
Plasmids and Stable Transfection
To generate an expression plasmid encoding human histone H2B fused to mCherry driven by EF1a promoter, histone H2B-mCherry cDNA was excised from a H2B-mCherry plasmid using KpnI/XbaI and sub-cloned into pEF6.mCherry-TSG101 plasmid (pEF6/H2B-mCherry). To generate a retroviral vector encoding human α-tubulin fused to EYFP at its N-terminus, EYFP-human α-tubulin cDNA was excised from pEYFP-Tub (Clontech; Mountain View, CA, USA) using BamHI/NotI and sub-cloned into a pMXs-IP plasmid (pMXs/EYFP-Tub). The pMXs/EYFP-Tub plasmid was transfected into Plat-E cells and virus-containing supernatants resulting from transfected cells were filtered through a 0.45-mm cellulose acetate filter and stored at −80°C. EMT6 cells that stably expressed H2B-mCherry and EYFP-Tub (EMT6/H2B-R/Tub-G) were generated using the following procedure. After the pEF6/H2B-mCherry plasmid was introduced into EMT6 cells by lipofection, cells were treated with G418 (600 μg/ml; Enzo Life Sciences; Farmingdale, NY, USA). Surviving clones with high red fluorescence were selected as EMT6/H2B-R cells. EMT6/H2B-R cells were then infected with the pMXs/EYFP-Tub retroviral vector, followed by puromycin (2 μg/ml; InvivoGen; San Diego, CA, USA) selection. Surviving clones with high red/green fluorescence were selected and stored as EMT6/H2B-R/Tub-G cells. EMT6/H2B-R/Tub-G cells were maintained in RPMI1640/10% FBS containing G418 (300 μg/ml) and puromycin (1 μg/ml).
Clonogenic Survival Assay
Cells were collected and seeded into 60-mm dishes at densities of 100–10,000 cells/dish and were allowed to adhere for 6 h. Cells were X-irradiated, followed by drug treatment. EMT6 or HeLa cells were cultured for either 6 or 14 days before methanol fixation and staining with Giemsa solution (Wako; Osaka, Japan). Colonies containing >50 cells were scored as surviving cells. Surviving fractions were calculated, correcting for the plating efficiency of cells, with or without X-irradiation.
Immunostaining
Analyses of mitotic catastrophe (MC) and centrosome numbers were performed as previously described [26], [27]. For the 53BP1/γ-H2AX foci formation assay, cells were seeded on glass coverslips coated with collagen (Cellmatrix Type I-C; Nitta Gelatin; Osaka, Japan) and cultured overnight. Cells were fixed using 3.7% paraformaldehyde/PBS for 20 min at 4°C and permeablized with PBS containing 0.5% Triton X-100 for 5 min at 4°C. Cells were incubated with PBS containing 6% (v/v) goat serum (Chemicon International; Temecula, CA, USA) for 1 h at room temperature (RT). Next, cells were incubated with anti-53BP1 antibody (1:2000 dilution) or anti-γ-H2AX antibody (1:1000 dilution) in 3% (v/v) goat serum/PBS overnight at 4°C. They were next incubated with Alexa Fluor® 488 anti-rabbit IgG away from light (1:2000 dilution) in 3% (v/v) goat serum/PBS for 1.5 h, followed by DAPI staining. Coverslips were mounted with ProLong® Gold Antifade Mountant reagent (Thermo Fisher Scientific). Fluorescent microscopic analysis was performed using an Olympus BX61 microscope (Olympus; Tokyo, Japan) using reflected light fluorescence. At least 100 cells were analyzed and the average number of the foci per cell was determined.
Cell Cycle Analysis
Cells were exposed to X-rays, and then treated with Chk1 inhibitors. After incubation, the cells were collected and washed with ice-cold PBS. Cells were fixed with ice-cold 70% (v/v) ethanol and kept at −20°C for 12 h. RNA was hydrolyzed with 100 μg/ml RNase A (NIPPON GENE, Tokyo, Japan) at 37°C for 30 min. Cells were stained with propidium iodide (Sigma-Aldrich) for 20 min. The DNA content of the cells was measured using an EPICS XL flow cytometer (Beckman Coulter; Fullerton, CA, USA).
Live-Cell Imaging
EMT6/H2B-R/Tub-G cells were seeded on a 35-mm glass-bottom dish (Matsunami Glass; Osaka, Japan) and cultured overnight. Following X-irradiation at 2.5 Gy, cells were treated with Chk1 inhibitors. After incubation for 3 h, cells were monitored using a time-lapse microscope system (LCV110, Olympus) equipped with a Retiga Exi camera (QImaging; Tokyo, Japan). Differential interference contrast (DIC) and fluorescence images were acquired every 8 min for 21 h. Images were analyzed using the MetaMorph Imaging System (Universal Imaging; CA, USA). Cell fate analysis was performed using 20 selected cells per condition. The mitotic progression of each individual cell was evaluated manually by analyzing the appearance of red-fluorescent nuclei and green-fluorescent microtubules at each time point. Based on these data, we calculated the duration from the start of observations to the onset of first mitosis, as well as the duration of the mitotic phase.
Statistical Analysis
All results are expressed as means ± S.D. from at least three experiments. Comparisons between groups were performed using a Student's t-test. The minimum level of significance was set at P < .05.
Results
Effect of Chk1 Inhibitors on Cellular Radiosensitivity
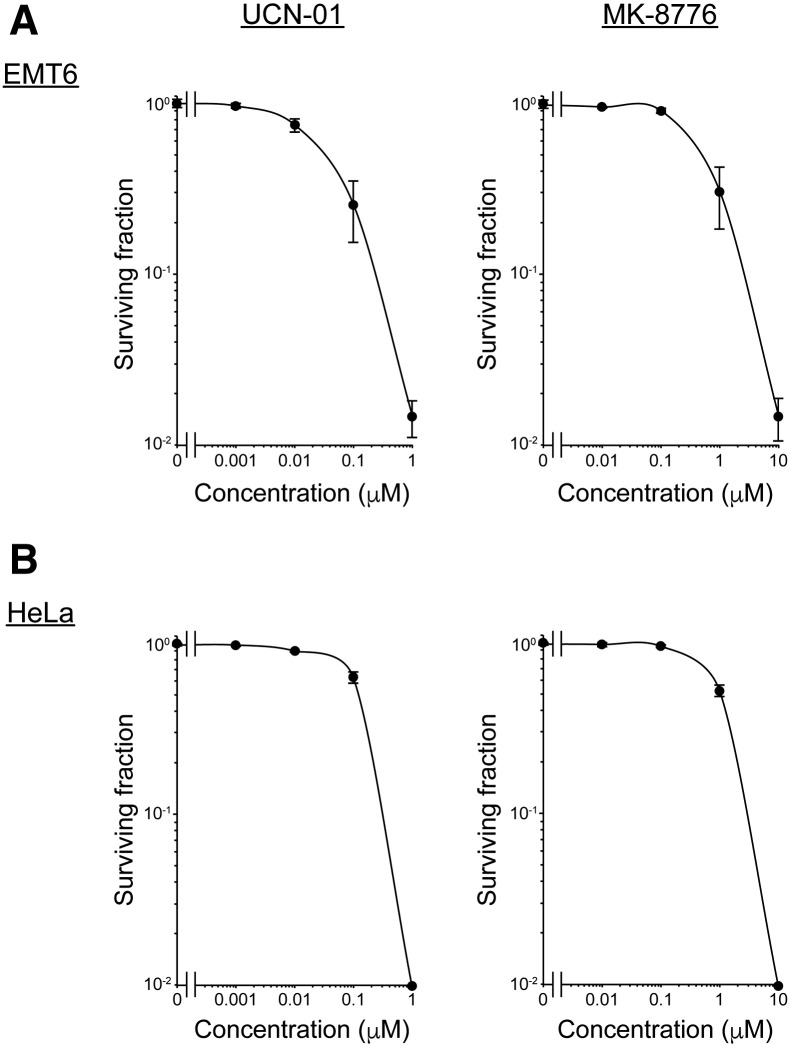
We first tested the cytotoxicity of two Chk1 inhibitors, UCN-01 and MK-8776, on two tumor cell lines, EMT6 or HeLa. Cells were treated with various concentrations of each inhibitor and cell viability was assessed using a clonogenic survival assay. As shown in Supplementary Figure S1, both Chk1 inhibitors reduced cell viability in a dose-dependent manner. UCN-01 was found to be more toxic than MK-8776 in both cell lines [IC50 = 32 nM (EMT6) and 169 nM (HeLa) for UCN-01 and 491 nM (EMT6) and 1.17 μM (HeLa) for MK-8776]. The minimally toxic concentrations (approximately 10% reduction in clonogenic survival) for each drug (5 nM UCN-01 and 200 nM MK-8776 for EMT6 cells; 10 nM UCN-01 and 500 nM MK-8776 for HeLa cells) were used to further investigate the radiosensitivity of the two cell lines.
Supplementary Figure S1.
Cytotoxicity of the two Chk1 inhibitors. Cytotoxicity of the two Chk1 inhibitors was examined using a clonogenic assay. (A) EMT6 cells were treated with UCN-01 or MK-8776 at the indicated concentrations for 7 days. (B) HeLa cells were treated with UCN-01 or MK-8776 at the indicated concentrations for 14 days. The surviving fractions with or without drug treatment were calculated with a correction for the plating efficiency of cells. Data are expressed as means ± S.D. from three experiments.
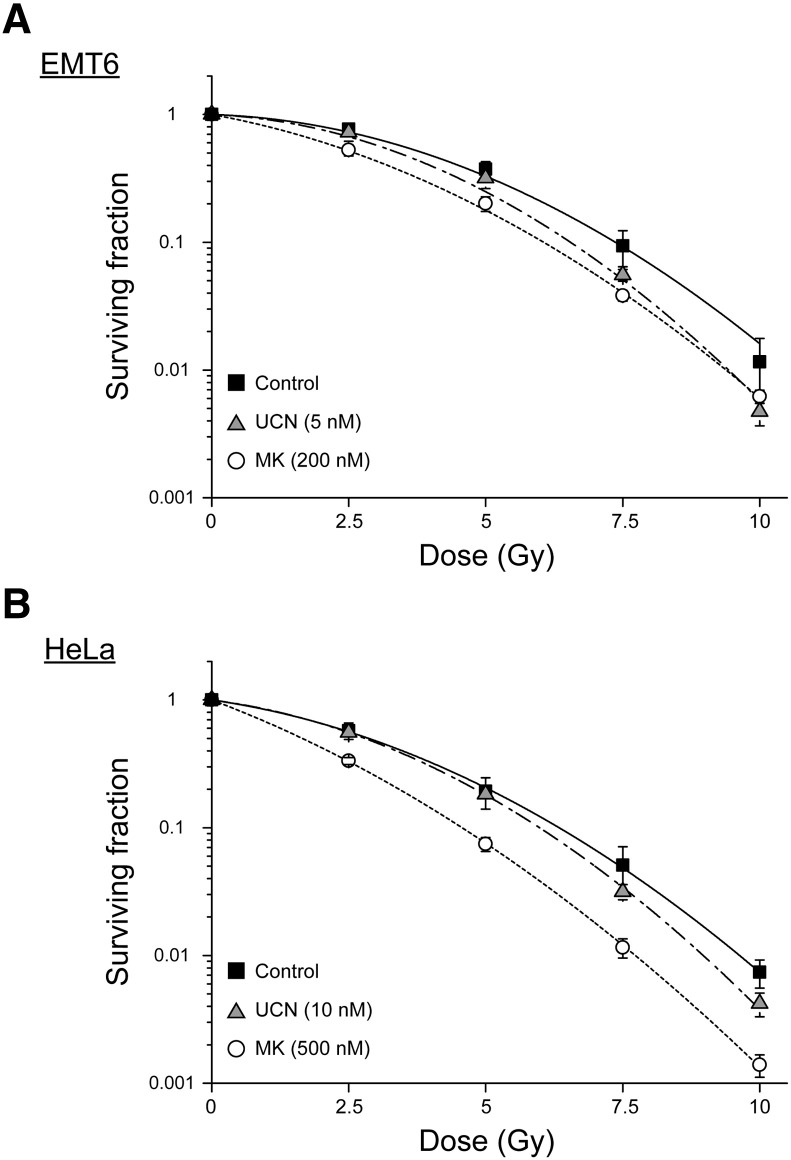
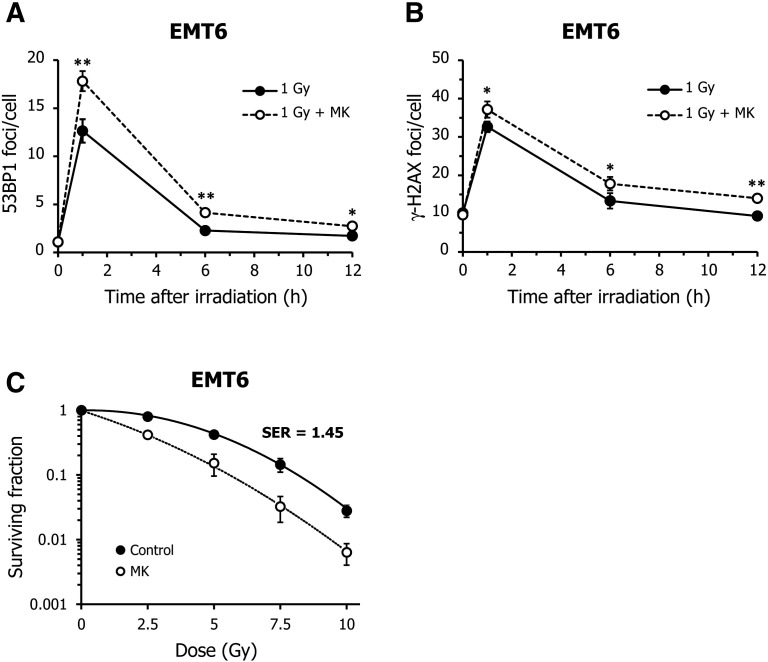
We next evaluated the effect of UCN-01 and MK-8776 on cell viability after exposure to IR using a clonogenic survival assay. As shown in Figure 1, IR decreased the viability of both tumor cell lines in a dose-dependent manner. While UCN-01 treatment enhanced radiation-induced clonogenic cell death at dosages of 7.5 Gy or higher, MK-8776 exhibited a more substantial radiosensitizing effect in both cell lines, even at 2.5 Gy. The sensitizer enhancement ratio (SER), judged using the 10% lethal dose for UCN-01 and MK-8776, was found to be 1.13 and 1.22 in EMT6 cells, and 1.07 and 1.39 in HeLa cells, respectively. Radiobiological variables were calculated, which are summarized in Table 1. These indicate that despite both Chk1 inhibitors enhancing radiosensitivity in EMT6 and HeLa cells, MK-8776 showed a more pronounced radiosensitizing effect, with lower cytotoxicity than UCN-01.
Figure 1.
Effect of Chk1 inhibitors on cellular radiosensitivity. Cellular radiosensitivity was examined using a clonogenic survival assay. (A) After X-irradiation, EMT6 cells were treated with either 5 nM UCN-01 (▲) or 200 nM MK-8776 (○) and incubated for 7 days. (B) After X-irradiation, HeLa cells were treated with either 10 nM UCN-01 (▲) or 500 nM MK-8776 (○) and incubated for 14 days. The surviving fraction was calculated using a correction for plating efficiency without X-irradiation. Data are expressed as means ± S.D. from three experiments.
Table 1.
Summary of Radiological Variables Calculated from Survival Curves
| Cell | Drug | α (Gy−1) | β (Gy−2) | α/β (Gy) | SF2.5 | D10 (Gy) | SER (D10) |
|---|---|---|---|---|---|---|---|
| EMT6 | Control | 0.03 | 0.038 | 0.84 | 0.77 | 7.37 | |
| UCN-01 | 0.04 | 0.047 | 0.88 | 0.67 | 6.55 | 1.13 | |
| MK-8776 | 0.18 | 0.033 | 5.41 | 0.53 | 6.06 | 1.22 | |
| HeLa | Control | 0.14 | 0.035 | 4.08 | 0.58 | 6.35 | |
| UCN-01 | 0.13 | 0.043 | 2.96 | 0.56 | 5.96 | 1.07 | |
| MK-8776 | 0.37 | 0.029 | 12.79 | 0.33 | 4.57 | 1.39 |
The data were fitted using a linear-quadratic model, surviving fraction (SF) = exp.(−αD−βD2). The 10% lethal dose (D10) was calculated from α and β values. The sensitizer enhancement ratio (SER) was calculated from each D10 value. SF2.5, surviving fraction at 2.5 Gy.
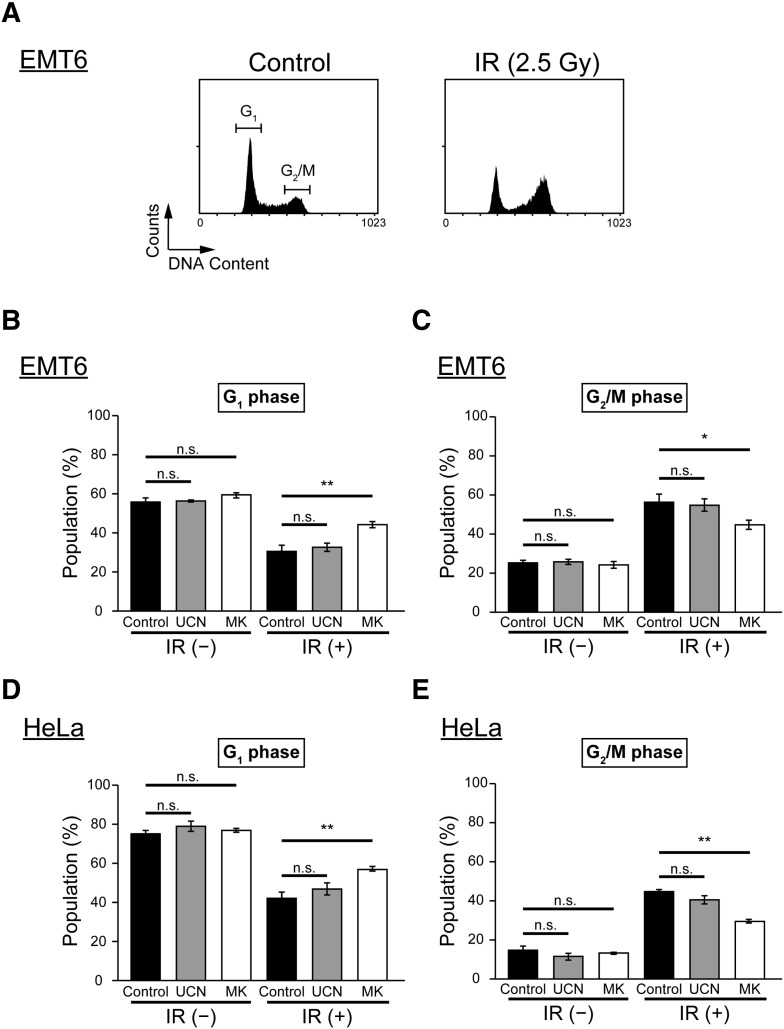
Effect of Chk1 Inhibitors on Cell Cycle Distribution After X-Irradiation
As Chk1 is one of the key proteins that regulate the cell cycle, especially G2/M checkpoint activation [28], we analyzed the distribution of cells in different cycle phases after exposure to IR and Chk1 inhibitor treatment. We used an irradiation dose of 2.5 Gy because a radiosensitizing effect was observed at this level with MK-8776, but not UCN-01. Cellular DNA content was measured using flow cytometry, and the proportions of cells in G1 or G2/M phase were determined (Figure 2A). In both cell lines, IR caused an increase in the proportion of cells in the G2/M phase and a decrease in the proportion in G1 phase. This indicates that there was radiation-induced G2/M checkpoint activation (Figure 2, B–E). UCN-01 treatment did not significantly alter cell cycle distribution in EMT6 cells, with or without irradiation (Figure 2, B and C). Conversely, MK-8776 treatment resulted in an increase in the proportion of cells in G1 phase and a decrease in cells that were in G2/M phase after exposure to IR. In addition, a similar effect on cell cycle distribution was observed in HeLa cells after treatment with Chk1 inhibitors (Figure 2, D and E). These results imply that radiation-induced G2/M arrest was mitigated by MK-8776 under our experimental settings.
Figure 2.
Effect of Chk1 inhibitors on cell cycle distribution after X-irradiation.
Cell cycle distribution was examined using flow cytometry and propidium iodide (PI) staining. (A) Flow cytometric profiles of DNA content in EMT6 cells without any treatment (left) and EMT6 cells at 4.5 h post-irradiation (right). (B and C) EMT6 cells were irradiated at 2.5 Gy and treated with vehicle (black), 5 nM UCN-01 (gray), or 200 nM MK-8776 (white) for 4.5 h. (D and E) HeLa cells were X-irradiated at 2.5 Gy and treated with vehicle (black), 10 nM UCN-01 (gray), or 500 nM MK-8776 (white) for 8 h. Data are expressed as means ± S.D. from three experiments. * P < .05, ** P < .01 vs. Chk1 inhibitors (−) (Student's t-test). n.s., not significant.
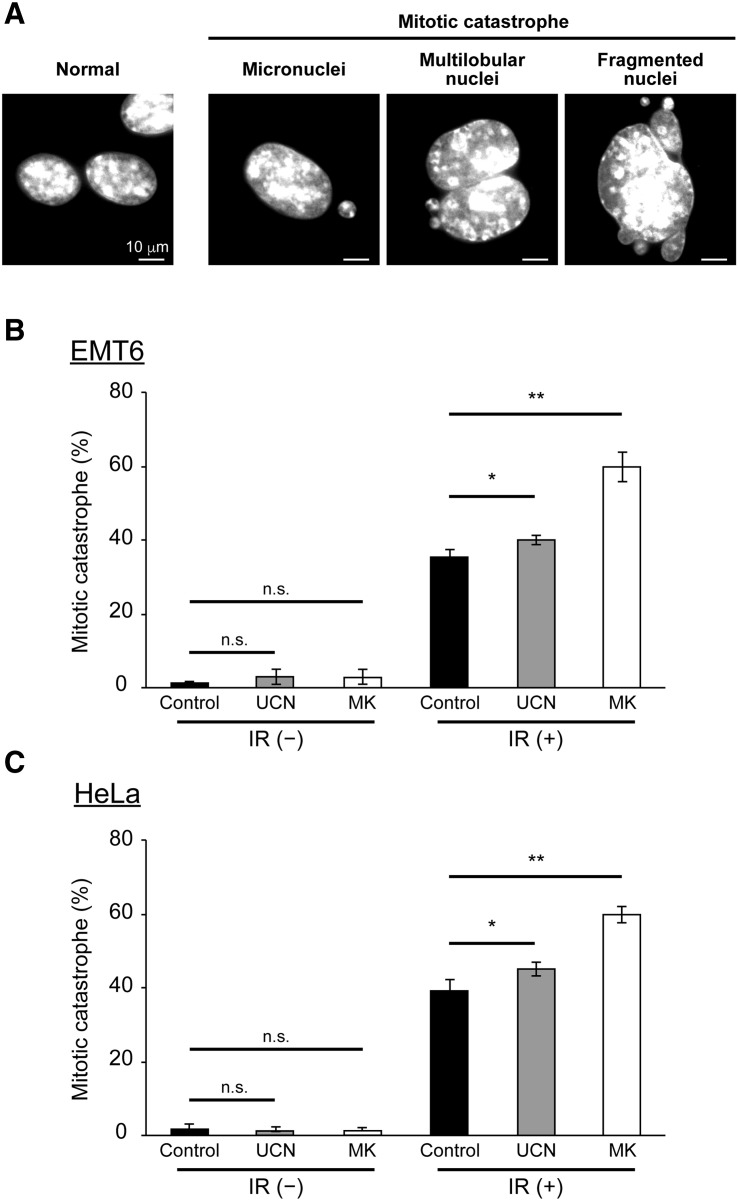
Effect of Chk1 Inhibitors on the Induction of Mitotic Catastrophe After X-Irradiation
Mitotic catastrophe (MC) is a form of cell death associated with aberrant mitosis. It is caused by uncoordinated or improper mitotic phase progression. It is considered the primary cell death mechanism following exposure to IR [29], [30], [31]. Therefore, we examined the effects of UCN-01 and MK-8776 treatment on MC induction after exposure to IR. Cells that displayed signs of aberrant nuclei, such as micronuclei, multi-lobular nuclei, or fragmented nuclei, were scored as cells undergoing MC (Figure 3A). After IR, observed MC increased from 1% to 36% in EMT6 cells (Figure 3B, black). Compared to IR alone, MC was slightly increased after IR in combination with UCN-01 (Figure 3B, gray). Interestingly, MC was significantly elevated after IR in combination with MK-8776 (Figure 3B, white). Furthermore, a similar pattern of MC induction was observed in HeLa cells with each Chk1 inhibitor (Figure 3C). This suggests that although both Chk1 inhibitors exacerbate radiation-induced MC, it is affected more by MK-8776 than UCN-01.
Figure 3.
Effect of Chk1 inhibitors on induction of mitotic catastrophe after X-irradiation.
The effect of Chk1 inhibitors on aberrant nuclei formation was examined by microscopic analysis. After cells were X-irradiated at 2.5 Gy and incubated with or without the Chk1 inhibitors for 24 h, cell nuclei were stained with DAPI. (A) Representative images of EMT6 cells possessing features of MC, such as micronuclei, multilobular nuclei, and fragmented nuclei. The scale bar represents 10 μm. (B and C) Quantitative analysis of the effect of the two Chk1 inhibitors on the induction of MC. (B) EMT6 cells were X-irradiated at 2.5 Gy and treated with vehicle (black), 5 nM UCN-01 (gray), or 200 nM MK-8776 (white) for 24 h. (C) HeLa cells were X-irradiated at 2.5 Gy and treated with vehicle (black), 10 nM UCN-01 (gray), or 500 nM MK-8776 (white) for 24 h. At least 100 cells were analyzed and the fraction of cells with aberrant mitotic nuclei was determined. Data are expressed as means ± S.D. from three experiments. * P < .05, ** P < .01 vs. Chk1 inhibitors (−) (Student's t-test). n.s., not significant.
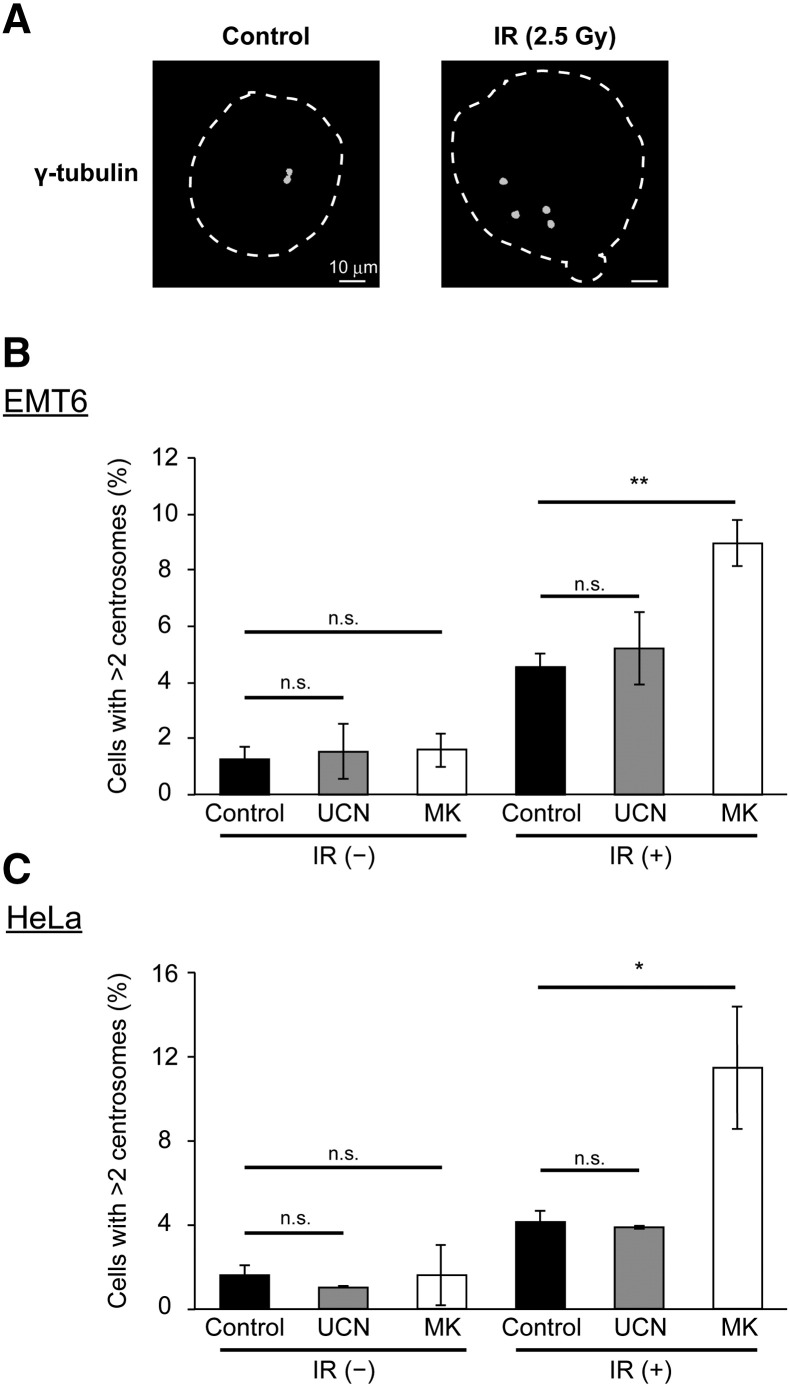
Effect of Chk1 Inhibitors on Radiation-Induced Centrosome Abnormality
Whereas normal cells contain one or two centrosomes, the centrosome duplication system is often disturbed in cells that are exposed to IR. This leads to the production of cells containing an irregular number of centrosomes [32]. Supernumerary centrosomes often result in aberrant mitosis, generating daughter cells that display aneuploidy. This is often associated with MC [32]. We therefore hypothesized that Chk1 inhibition may increase radiation-induced centrosome abnormalities and exacerbate MC. To test this hypothesis, we performed a microscopic analysis of cellular centrosomes using γ-tubulin staining to determine the proportion of cells with supernumerary centrosomes. As shown in Figure 4, A and B, X-irradiation at 2.5 Gy promoted the formation of supernumerary centrosomes in EMT6 cells. While UCN-01 did not significantly alter the centrosome status after exposure to IR, MK-8776 significantly increased the proportion of cells that possessed supernumerary centrosomes. Similarly, MK-8776, but not UCN-01, enhanced supernumerary centrosome formation after exposure to IR in HeLa cells (Figure 4C). These results demonstrate that MK-8776, but not UCN-01, increases the number of observed radiation-induced centrosome abnormalities. This suggests that the increase in observed centrosome abnormalities by MK-8776 may contribute to the increased rates of radiation-induced MC.
Figure 4.
Effect of Chk1 inhibitors on radiation-induced centrosome abnormalities.
After treatment, centrosome abnormalities were determined by immunostaining with γ-tubulin antibody. (A) Representative images of centrosomes in EMT6 cells with or without X-irradiation. The dashed line represents the nuclear outline. The scale bar represents 10 μm. (B and C) Quantitative analysis of the effect of the two Chk1 inhibitors on centrosome abnormalities. (B) EMT6 cells were X-irradiated at 2.5 Gy and treated with vehicle (white), 5 nM UCN-01 (gray), or 200 nM MK-8776 (white) for 24 h. (C) HeLa cells were X-irradiated at 2.5 Gy and treated with vehicle (black), 10 nM UCN-01 (gray), or 500 nM MK-8776 (white) for 24 h. At least 100 cells were analyzed and the percentage of cells containing more than two centrosomes was determined. Data are expressed as means ± S.D. from three experiments. * P < .05, ** P < .01 vs. Chk1 inhibitors (−) (Student's t-test). n.s., not significant.
Effect of Chk1 Inhibitors on Radiation-Induced DSBs
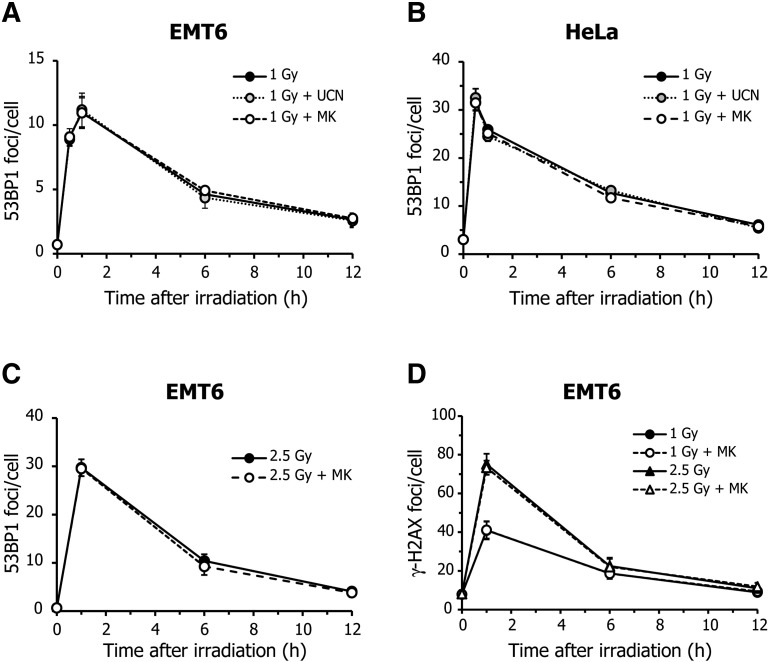
Although Chk1 is required for cell cycle regulation, it is also involved in the DNA homologous repair pathway [33]. Therefore, it may be possible that MK-8776 enhances radiation-induced MC and centrosome abnormalities by promoting entry into mitosis, even if a cell contains unrepaired DNA damage. To examine whether MK-8776 enhanced the radiosensitivity of tumor cells by inhibiting double-strand break (DSB) repair, we measured cellular DSB levels using a 53BP1 foci formation assay. When EMT6 cells were exposed to X-rays at 1 Gy, the mean number of 53BP1 foci per cell peaked 1 h after irradiation and then gradually decreased over time (Figure 5A). Neither UCN-01 nor MK-8776 influenced the kinetics of 53BP1 focal formation after irradiation. Similarly, these Chk1 inhibitors had no impact on the kinetics of radiation-induced 53BP1 focal formation in HeLa cells (Figure 5B). In addition, MK-8776 did not alter the kinetics of 53BP1 focal formation induced by X-rays at 2.5 Gy (Figure 3C). Furthermore, we also found that MK-8776 unaffected the kinetics of γ-H2AX focal formation, another DSB marker, after irradiation at 1 and 2.5 Gy (Figure 5D). These findings indicate that DSB formation and repair are likely not involved in the MC induction and radiosensitizing effects demonstrated by these Chk1 inhibitors.
Figure 5.
Effect of Chk1 inhibitors on radiation-induced double-strand breaks.
(A and B) Cells were X-irradiated at 1 Gy and incubated with vehicle (black), 5 nM UCN-01 (gray), or 200 nM MK-8776 (white) for the indicated times. The formation of double-strand breaks (DSBs) after X-irradiation was determined using a 53BP1 foci formation assay. Each graph represents the kinetics of 53BP1 focal formation after irradiation in EMT6 (A) or HeLa cells (B), in the presence or absence of Chk1 inhibitors. (C) EMT6 cells were X-irradiated at 2.5 Gy and incubated with vehicle (black) or MK-8776 (white) for the indicated times. The number of 53BP1 foci was evaluated. (D) EMT6 cells were X-irradiated at 1 or 2.5 Gy and incubated with vehicle (black) or MK-8776 (white) for the indicated times. The number of γ-H2AX foci was evaluated. At least 100 cells were analyzed and the average number of 53BP1 or γ-H2AX foci per cell was scored. Data are expressed as means ± S.D. from three experiments.
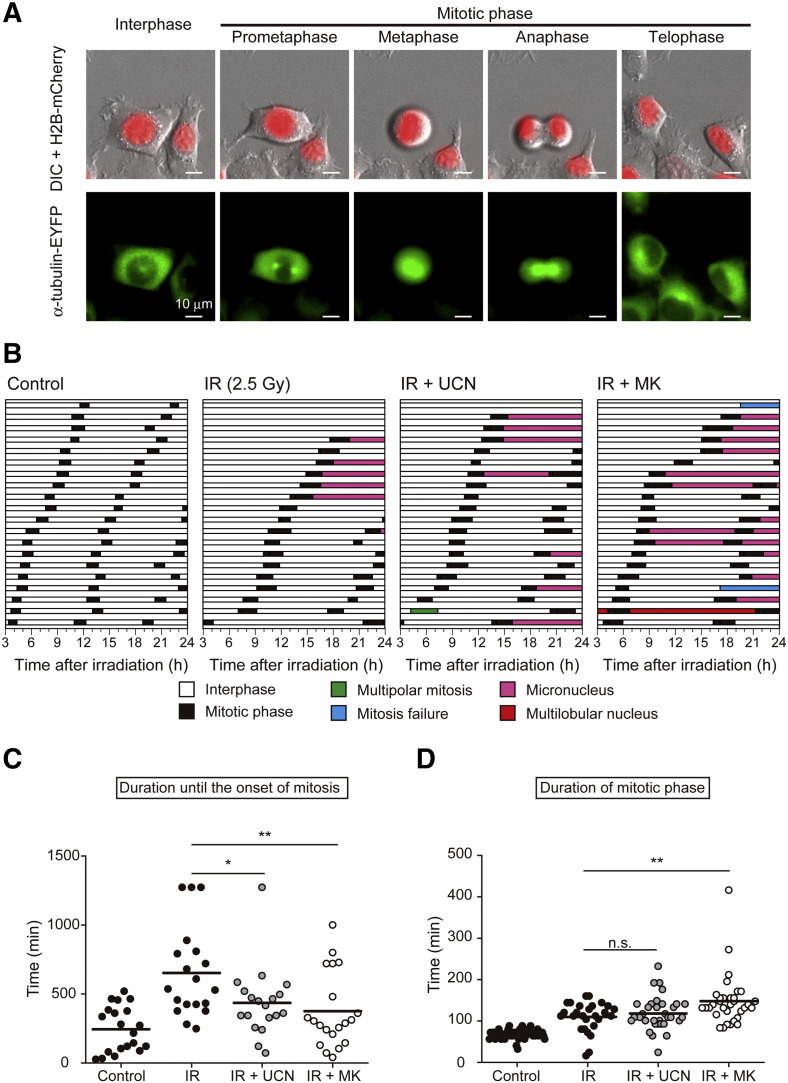
Effect of Chk1 Inhibitors on Mitotic Progression After X-Irradiation
To further analyze the mechanism by which Chk1 inhibitors trigger enhanced aberrant mitosis and centrosome abnormalities after irradiation, we next examined the effects of the two Chk1 inhibitors on mitotic progression by utilizing live-cell imaging. To achieve this, we generated EMT6 cells that stably expressed an mCherry-tagged histone H2B and an EYFP-tagged α-tubulin (EMT6/H2B-R/Tub-G cells). This cell line was used to visualize nuclei and microtubules dynamics, and to track the fate of individual cells using time-lapse microscopy. To analyze and quantify mitotic progression in EMT6/H2B-R/Tub-G cells, each cell was classified as being in interphase or the mitotic phase (consisting of prometaphase, metaphase, anaphase, and telophase) (Figure 6A). As shown in Figure 6B, control cells exhibited consistent cycles of mitotic progression and the average duration of interphase and the mitotic phase were 460 min and 68 min, respectively (Supplementary Table S1). When cells were exposed to X-rays at 2.5 Gy, mitotic cells only emerged after a prolonged interphase, indicating IR-induced G2/M arrest. In addition, IR resulted in the formation of micro-nucleated cells in 30% of cells (6/20). UCN-01 and MK-8776 shortened the duration of the prolonged interphase after irradiation and increased the frequency of mitosis. While co-treatment with UCN-01 and X-rays led to aberrant mitosis in 40% of cells (8/20), MK-8776 caused aberrant mitosis in 75% of cells (15/20). Furthermore, while micronuclei formation was the only mitotic defect observed in cells exposed to IR alone, co-treatment with the two Chk1 inhibitors resulted in other types of mitotic defects, including multi-polar mitosis and multi-lobular nuclei. This was especially apparent in MK-8776-treated cells. As shown in Figure 6C, quantitative analysis revealed that UCN-01 and MK-8776 both significantly shortened the period from the start of observations to the onset of first mitosis. This implies that both Chk1 inhibitors mitigate radiation-induced cell cycle arrest. Moreover, when we evaluated the duration of the mitotic phase, it was clearly increased by IR, likely due to radiation-induced SAC activation (Figure 6D and Supplementary Table S1). Whereas UCN-01 did not significantly affect the duration of the mitotic phase after irradiation, it was significantly extended by MK-8776. This indicates that MK-8776 treatment causes prolonged M-phase arrest. These results suggest that, while IR alone can induce damage-induced cell cycle arrest and small mitotic defects, the addition of Chk1 inhibitors, especially MK-8776, further disturbs cell cycle control and results in substantial mitotic defects.
Figure 6.
Effect of Chk1 inhibitors on mitotic progression after X-irradiation.
The effect of the two Chk1 inhibitors on cell fate after X-irradiation was evaluated using live-cell imaging. EMT6 cells stably expressing histone H2B fused to mCherry and α-tubulin fused to EYFP (EMT6/H2B-R/Tub-G cells) were X-irradiated at 2.5 Gy and incubated with or without Chk1 inhibitors for 24 h. Live-cell imaging was conducted from 3 to 24-h post-irradiation. (A) Representative images of EMT6/H2B-R/Tub-G cells undergoing mitosis. Upper panels represent the merged images of histone H2B fused to mCherry (red) and differential interference contrast (DIC; gray). Lower panels represent the fluorescent images of α-tubulin fused to EYFP (green). The scale bar represents 10 μm. (B) The fate of individual cells after treatment was evaluated by analyzing the appearance of red-fluorescent nuclei and green-fluorescent microtubules at each time point. Each horizontal bar represents one cell (n = 20). White, interphase; black, mitotic phase; magenta, micronucleus; red, multilobular nucleus; green, multipolar mitosis; blue, mitosis failure. (C) The scatter plot indicates the duration from the start of observations to the onset of the first mitotic event. (D) The scatter plot shows the duration of the mitotic phase. The bars represent mean values. * P < .05, ** P < .01 vs. radiation alone (Student's t-test). n.s., not significant.
Discussion
In the present study, we demonstrated that the novel Chk1 inhibitor MK-8776 greatly enhanced the cellular radiosensitivity of tumor cells at clinically relevant irradiation doses (Figure 1). Furthermore, we found that MK-8776 was much more effective in sensitizing tumor cells to radiation than UCN-01 when compared at minimally toxic concentrations. In line with our results, several groups have previously demonstrated that new classes of Chk1 inhibitors including MK-8776 radiosensitize solid tumor cells [20], [34], [35]. Interestingly, the SER values for MK-8776 in this study are similar to those obtained in the other studies using novel Chk1 inhibitors [20], [35], [36]. This implies that, regardless of the compound used, specific inhibition of Chk1 enhances cellular radiosensitivity to the same extent. Furthermore, Bridges et al. have recently reported that MK-8776 radiosensitizes human tumor cells without causing any appreciable cytotoxicity [20]. Their work indicates that the radiosensitizing effect shown by MK-8776 is restricted to p53-defective cells. As many types of cancer show high incidences of p53 mutation and loss of function, this property of MK-8776 makes it an excellent candidate for use as a radiosensitizing agent in cancer therapy. Collectively, these lines of evidence suggest that MK-8776 exerts a strong radiosensitizing effect at least in vitro, and may have clinical potential in combination with radiotherapy.
Our study clearly demonstrates that MK-8776 strongly induces a reduction in radiation-induced G2/M arrest (Figure 2), an enhancement in radiation-induced MC (Figure 3), and centrosome abnormality (Figure 4). Comparatively, UCN-01 only marginally affects these criteria. These data indicate that at low-toxic concentrations of UCN-01 (90% survival dose when used singularly) have a limited capacity to bypass a radiation-induced G2/M checkpoint and trigger subsequent events. Equivalent low-toxic concentrations of MK-8776 efficiently abrogate a radiation-induced G2/M checkpoint that is followed by a significant increase in aberrant mitosis. Various DNA-damaging agents, including IR, trigger MC and centrosome amplification through inappropriate mitosis progression [32]. Our data demonstrates that Chk1 inhibition by MK-8776 enhances these two events, resulting in increased radiation-induced clonogenic cell death. In contrast to our data, inhibition of Chk1 by RNA-mediated interference or treatment with UCN-01 is reported to suppress radiation-induced centrosome amplification in both A549 [37] and DT40 cells [38]. While we used a relatively low dose of X-rays (2.5 Gy) in our study, resulting in a small increase in centrosome amplification (approximately 4% of total cells), a high dose of X-rays (10 Gy) was employed in these other studies, leading to the larger increase in amplification (approximately 30% to 40% of total cells). Therefore, the magnitude of the initial damage due to IR may influence the cellular response. This consequently results in the different observed outcomes in centrosome amplification after IR.
In our study, Chk1 inhibition had no measurable impact on DSB repair kinetics after exposure to IR (Figure 5). This suggests that Chk1 inhibition does not influence the radiation-induced DSB levels and repair kinetics. However, there is conflicting evidence concerning the effects that Chk1 inhibitors have on DSB and repair. Supporting our results, Tao et al. demonstrated that another Chk1 inhibitor, Chir-124, does not affect DNA damage repair kinetics in X-irradiated HCT116 cells [26]. In contrast to this, Bridges et al. reported that MK-8776 inhibited the first phase of DSB repair within an hour of irradiation [20]. This discrepancy might be explained by the timing of Chk1 inhibitor treatment. In our experiments, MK-8776 was applied immediately after irradiation, whereas Bridges et al. exposed cells to MK-8776 prior to irradiation. Since Chk1 has been reported to phosphorylate BRCA2 to facilitate its function in homologous recombination DNA repair [39] and phosphorylated BRCA2 is recruited to DSB sites within 30 sec of DNA damage induction [40], it may be possible that pre-treatment with Chk1 inhibitor inhibits this process. This would mitigate BRCA2-dependent homologous recombination DNA repair after irradiation. In fact, when MK-8776 treatment was started at 1 h prior to irradiation, it resulted in the increase of the number of 53BP1 and γ-H2AX foci after irradiation (Supplementary Figure S2, A and B). We also found that pre-treatment of MK-8776 induced higher radiosensitizing effect than its treatment at post-IR (Supplementary Figure S2C). These lines of evidence support our assumption on the effect of Chk1 inhibition on DSB repair.
Supplementary Figure S2.
Effect of pre-treatment of MK-8776 on radiation-induced double-strand breaks and cellular radiosensitivity. (A and B) After EMT6 cells were treated with vehicle (black) or MK-8776 (white) for 1 h, they were X-irradiated at 1 Gy and incubated for the indicated times in the presence or absence of drug. At least 100 cells were analyzed and the average number of 53BP1 (A) or γ-H2AX (B) foci per cell was scored. Data are expressed as means ± S.D. from three experiments. * P < .05, ** P < .01 vs. radiation alone (Student's t-test). (C) After EMT6 cells were treated with vehicle (black) or MK-8776 (white) for 1 h, they were X-irradiated and incubated with or without drug for 18 h. Subsequently, media was replaced and the cells were cultured for 7 days in the absence of drug. Cellular radiosensitivity was determined by clonogenic survival assay. The surviving fraction was calculated using a correction for plating efficiency without X-irradiation. Data are expressed as means ± S.D. from three experiments.
As our results show that MK-8776 enhances MC in EMT6 and HeLa cells without affecting DSBs repair kinetics, it was hypothesized that radiosensitization by MK-8776 associates with an aberrant and premature onset of mitosis induced by Chk1 inhibition. Therefore, time-lapse analysis using live-cell imaging was adopted to continuously track the fate of individual cells. This demonstrated that IR alone delayed the onset of mitosis, indicating the activation of interphase checkpoints. The addition of MK-8776 canceled this radiation-induced checkpoint activation and significantly extended the mitotic period. This was followed by an increase in the proportion of cells with micronuclei and mitotic failure (Figure 6, B–D). These observations indicate that MK-8776 abrogates the radiation-induced interphase checkpoints and disturbs normal cell cycle progression in the mitotic phase (Figure 6). Previously, Chk1 was reported to localize to centrosomes and to be involved in the inhibition of CDC25B-dependent activation of Cdk1 during metaphase [41]. This implies that centrosome-associated Chk1 shields centrosomal Cdk1 from unscheduled mitotic entry by regulating the formation of mitotic spindles [41]. Furthermore, it has been reported that attenuation of Chk1 function induces mitotic defects, including binucleation, abnormal localization of Aurora B, the increase of lagging and/or nondisjunction chromosomes [2], and SAC activation through polo-like kinase 1 (PLK1) [42]. The extension of mitosis is reported to be due to SAC activation in Jurkat T cells treated with taxol or nocodazole, and SAC activation is considered to play a part in the induction of MC [43]. From these studies, the radiosensitization of tumor cells by MK-8776 likely involves the abrogation of a centrosomal Chk1-dependent defense system for unscheduled mitotic entry and the impairment of mitotic progression through SAC activation.
Conclusions
In conclusion, we have shown that, at minimally toxic concentrations, MK-8776 exerts a radiosensitizing effect without affecting DSB repair activity. Our data suggest that the induction of aberrant mitosis and consequential mitotic cell death is the main cause of the radiosensitization effect demonstrated by MK-8776. Importantly, radiosensitization by MK-8776 can be achieved at doses as low as 2.5 Gy, a clinically relevant irradiation dose. In a recent phase I clinical study, it was reported that MK-8776 was well-tolerated and showed evidence of clinical efficacy in patients with advanced solid tumors [15]. This demonstrates the safety of MK-8776 in a clinical setting. This evidence suggests that MK-8776 is a promising radiosensitizing candidate and support the future clinical assessment of MK-8776 in combination with radiotherapy.
The following are the supplementary data related to this article.
Duration of interphase and mitotic phase determined by live-cell imaging.
Acknowledgments
We would like to thank OPEN FACILITY (Hokkaido University Sousei Hall) for allowing us to use the LCV110 for time-lapse imaging. We would also like to thank Editage (www.editage.jp) for English language editing. This work was supported, in part, by the Japanese Society for the Promotion of Science KAKENHI (Grant number 26461875 [TY]) and by the SGH FOUNDATION [TY]. The sponsors had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; nor the decision to submit the manuscript for publication.
Contributor Information
Tohru Yamamori, Email: yamamorit@vetmed.hokudai.ac.jp.
Osamu Inanami, Email: inanami@vetmed.hokudai.ac.jp.
References
- 1.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 2.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci U S A. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargiela-Iparraguirre J, Prado-Marchal L, Fernandez-Fuente M, Gutierrez-Gonzalez A, Moreno-Rubio J, Munoz-Fernandez M, Sereno M, Sanchez-Prieto R, Perona R, Sanchez-Perez I. CHK1 expression in Gastric Cancer is modulated by p53 and RB1/E2F1: implications in chemo/radiotherapy response. Sci Rep. 2016;6:21519. doi: 10.1038/srep21519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David L, Fernandez-Vidal A, Bertoli S, Grgurevic S, Lepage B, Deshaies D, Prade N, Cartel M, Larrue C, Sarry JE. CHK1 as a therapeutic target to bypass chemoresistance in AML. Sci Signal. 2016;9:ra90. doi: 10.1126/scisignal.aac9704. [DOI] [PubMed] [Google Scholar]
- 5.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–139. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 6.Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Interv. 2011;11:133–140. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O'Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20:7453–7463. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O'Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88:956–965. doi: 10.1093/jnci/88.14.956. [ http://dx.doi.org/jnci.oxfordjournals.org/content/88/14/956.long] [DOI] [PubMed] [Google Scholar]
- 9.Bunch RT, Eastman A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin Cancer Res. 1996;2:791–797. [ http://dx.doi.org/clincancerres.aacrjournals.org/content/2/5/791.long] [PubMed] [Google Scholar]
- 10.Fuse E, Kuwabara T, Sparreboom A, Sausville EA, Figg WD. Review of UCN-01 development: a lesson in the importance of clinical pharmacology. J Clin Pharmacol. 2005;45:394–403. doi: 10.1177/0091270005274549. [DOI] [PubMed] [Google Scholar]
- 11.Dwyer MP, Paruch K, Labroli M, Alvarez C, Keertikar KM, Poker C, Rossman R, Fischmann TO, Duca JS, Madison V. Discovery of pyrazolo[1,5-a]pyrimidine-based CHK1 inhibitors: a template-based approach--part 1. Bioorg Med Chem Lett. 2011;21:467–470. doi: 10.1016/j.bmcl.2010.10.113. [DOI] [PubMed] [Google Scholar]
- 12.Labroli M, Paruch K, Dwyer MP, Alvarez C, Keertikar K, Poker C, Rossman R, Duca JS, Fischmann TO, Madison V. Discovery of pyrazolo[1,5-a]pyrimidine-based CHK1 inhibitors: a template-based approach--part 2. Bioorg Med Chem Lett. 2011;21:471–474. doi: 10.1016/j.bmcl.2010.10.114. [DOI] [PubMed] [Google Scholar]
- 13.Guzi TJ, Paruch K, Dwyer MP, Labroli M, Shanahan F, Davis N, Taricani L, Wiswell D, Seghezzi W, Penaflor E. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol Cancer Ther. 2011;10:591–602. doi: 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- 14.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Ther. 2012;11:427–438. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daud AI, Ashworth MT, Strosberg J, Goldman JW, Mendelson D, Springett G, Venook AP, Loechner S, Rosen LS, Shanahan F. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J Clin Oncol. 2015;33:1060–1066. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, Xiao Z, Gu WZ, Xue J, Bui MH, Kovar P, Li G, Wang G, Tao ZF, Tong Y. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–2794. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 17.Del Nagro CJ, Choi J, Xiao Y, Rangell L, Mohan S, Pandita A, Zha J, Jackson PK, O'Brien T. Chk1 inhibition in p53-deficient cell lines drives rapid chromosome fragmentation followed by caspase-independent cell death. Cell Cycle. 2014;13:303–314. doi: 10.4161/cc.27055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Y, Ramiscal J, Kowanetz K, Del Nagro C, Malek S, Evangelista M, Blackwood E, Jackson PK, O'Brien T. Identification of preferred chemotherapeutics for combining with a CHK1 inhibitor. Mol Cancer Ther. 2013;12:2285–2295. doi: 10.1158/1535-7163.MCT-13-0404. [DOI] [PubMed] [Google Scholar]
- 19.On KF, Chen Y, Ma HT, Chow JP, Poon RY. Determinants of mitotic catastrophe on abrogation of the G2 DNA damage checkpoint by UCN-01. Mol Cancer Ther. 2011;10:784–794. doi: 10.1158/1535-7163.MCT-10-0809. [DOI] [PubMed] [Google Scholar]
- 20.Bridges KA, Chen X, Liu H, Rock C, Buchholz TA, Shumway SD, Skinner HD, Meyn RE. MK-8776, a novel chk1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Oncotarget. 2016;7:71660–71672. doi: 10.18632/oncotarget.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Chow JP, Poon RY. Inhibition of Eg5 acts synergistically with checkpoint abrogation in promoting mitotic catastrophe. Mol Cancer Res. 2012;10:626–635. doi: 10.1158/1541-7786.MCR-11-0491. [DOI] [PubMed] [Google Scholar]
- 22.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 24.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamori T, Ike S, Bo T, Sasagawa T, Sakai Y, Suzuki M, Yamamoto K, Nagane M, Yasui H, Inanami O. Inhibition of the mitochondrial fission protein dynamin-related protein 1 (Drp1) impairs mitochondrial fission and mitotic catastrophe after x-irradiation. Mol Biol Cell. 2015;26:4607–4617. doi: 10.1091/mbc.E15-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Yamamori T, Yasui H, Inanami O. Effect of MPS1 Inhibition on Genotoxic Stress Responses in Murine Tumour Cells. Anticancer Res. 2016;36:2783–2792. [doi: ar.iiarjornals.org/content/36/6/2783.long] [PubMed] [Google Scholar]
- 28.Patil M, Pabla N, Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci. 2013;70:4009–4021. doi: 10.1007/s00018-013-1307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 30.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31:363–372. doi: 10.1007/s13277-010-0042-8. [DOI] [PubMed] [Google Scholar]
- 31.Lauber K, Ernst A, Orth M, Herrmann M, Belka C. Dying cell clearance and its impact on the outcome of tumor radiotherapy. Front Oncol. 2012;2:116. doi: 10.3389/fonc.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodson H, Wheatley SP, Morrison CG. Involvement of centrosome amplification in radiation-induced mitotic catastrophe. Cell Cycle. 2007;6:364–370. doi: 10.4161/cc.6.3.3834. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 34.Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J, Zaidi S, Powell NG, Meier P, Collins I. Targeted radiosensitization by the Chk1 inhibitor SAR-020106. Int J Radiat Oncol Biol Phys. 2013;85:1110–1118. doi: 10.1016/j.ijrobp.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, Arumugarajah S, Hylander-Gans L, Morosini D, Simeone DM. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelke CG, Parsels LA, Qian Y, Zhang Q, Karnak D, Robertson JR, Tanska DM, Wei D, Davis MA, Parsels JD. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clin Cancer Res. 2013;19:4412–4421. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Löffler H, Fechter A, Liu FY, Poppelreuther S, Krämer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32:2963–2972. doi: 10.1038/onc.2012.310. [DOI] [PubMed] [Google Scholar]
- 38.Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008;27:3977–3985. doi: 10.1038/onc.2008.17. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Shi J, Bian C, Yu X. Poly(ADP-Ribose) Mediates the BRCA2-Dependent Early DNA Damage Response. Cell Rep. 2015;13:678–689. doi: 10.1016/j.celrep.2015.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 42.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci U S A. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R, Simon HU. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Duration of interphase and mitotic phase determined by live-cell imaging.