Abstract
Somatic mutations of pre-mRNA splicing factors recur among patients with myelodysplastic syndromes (MDS) and related malignancies. Although these MDS-relevant mutations alter splicing of a subset of transcripts, the mechanisms by which these single amino acid substitutions change gene expression remain controversial. New structures of spliceosome intermediates and associated protein complexes shed light on the molecular interactions mediated by “hotspots” of the SF3B1 and U2AF1 pre-mRNA splicing factors. The frequently mutated SF3B1 residues contact the pre-mRNA splice site. Based on structural-homology with other spliceosome subunits and recent findings of altered RNA binding by mutant U2AF1 proteins, we suggest that affected U2AF1 residues also contact pre-mRNA. Altered pre-mRNA recognition emerges as a molecular theme among MDS-relevant mutations of pre-mRNA splicing factors.
Keywords: SF3B1, U2AF1, pre-mRNA splicing, myelodysplastic syndrome, MDS, cancer
Recurrent Somatic Mutations of Pre-mRNA Splicing Factors in Cancers
Prevalent, acquired mutations of pre-mRNA splicing factors in cancers were discovered initially from whole-exome sequencing of myelodysplastic syndromes (MDS) [1–4], chronic lymphocytic leukemias (CLL) [5, 6], and related disorders (Box 1). Primarily, four splicing factor genes are mutated among these myeloid neoplasms: SF3B1, U2AF1, SRSF2, and ZRSR2 (Figure 1, Key Figure). The recurrent SF3B1, U2AF1, and SRSF2 mutations alter amino acids at specific “hotspots” (mapped on SF3B1 and U2AF1 protein domains in Figure 1A; the SRSF2 hotspot is P95). An exception is the null as well as missense mutations found distributed throughout ZRSR2 (also called URP), which is associated with the “minor” U12-type spliceosome rather than the “major” U2-type spliceosome (Box 2) [7]. A single splicing factor often dominates the mutational spectrum of a distinct cancer subtype. For example, as expanded below, SF3B1 mutations occur in the majority of refractory anemia with ringed sideroblasts (RARS) [8]. On the other hand, SRSF2 mutations occur in more than a third of chronic myelomonocytic leukemias (CMML) [9]. Somatic mutations of certain splicing factors also recur among a variety of solid tumors – primarily, SF3B1 mutations in uveal melanoma [10, 11], pancreatic ductal adenocarcinoma [12, 13], or breast cancer [14, 15] and U2AF1 mutations in pancreatic ductal adenocarcinoma [13] or lung adenocarcinoma [16, 17].
Box 1. Myelodysplastic Syndromes and Related Myeloid Neoplasms.
Myeloid neoplasms are clonal diseases of stem cells in the bone marrow or peripheral blood that normally provide blood cells through the process of hematopoiesis (reviewed in [94]). The World Health Organization (WHO) classifies myeloid neoplasms and acute leukemias into distinct types and subtypes based on a combination of clinical features, morphology, immunophenotyping, cytogenetics, and molecular genetics [95]. Acute myeloid leukemia (AML) is a severe type of myeloid neoplasms characterized by increased malignant myeloid blasts and reduced mature blood cells. Myelodysplastic syndromes (MDS), historically called “pre-leukemias” based on progression to AML in ~30% of patients [96], are heterogeneous bone marrow disorders with the common feature of peripheral blood cytopenias. A distinct group called MDS/MPN displays both dysplastic features of MDS and proliferative features of myeloproliferative neoplasms (MPN), which overproduce blood cells. Chronic myelomonocytic leukemia (CMML) is one subtype of MDS/MPN. Peri-nuclear rings of iron granules, called ring sideroblasts (RS) [97], mark other MDS and MDS/MPN subtypes. Although the term refractory anemia (RA) is used herein to match prior literature, the WHO recently re-classified RA as MDS or MDS/MPN subtypes [98]. Other categories of myeloid neoplasms include B-cell lymphomas, of which hairy cell leukemia variant (HCL-v) is a provisional entity, as well as myeloid/lymphoid neoplasms with eosinophilia and genetic rearrangements, blastic plasmacytoid dendritic cell neoplasm, and acute leukemias of ambiguous lineage. Chronic lymphocytic leukemia (CLL), in which SF3B1 is frequently mutated, is classified as a lymphoid (rather than myeloid) neoplasm.
MDS and related myeloid neoplasms typically are acquired through genetic and epigenetic changes. Accordingly, the greatest risk factor for MDS is age. The incidence rate of MDS in the United States of is approximately 3 cases per 100,000 population per year and increases 10-fold among patients above 70 years of age [99]. The BCR-ABL1 fusion kinase is a well-known genetic defect of CML that has served as the basis for successful therapies [100]. As discussed herein, high-throughput DNA sequencing revealed pre-mRNA splicing as the most frequent target of acquired mutations in MDS [18, 23]. Other recurrently mutated genes, such as TET2, ASXL1, DNMT3a and RUNX1, affect epigenetic regulation, signaling, or DNA damage response. Currently, allogeneic stem cell transplant is the only curative therapy for MDS, yet its severity precludes use to treat elderly patients, who also are the most commonly sick. It is hoped that the field’s increasing knowledge of the underlying molecular pathways will guide future therapies for MDS.
Figure 1.
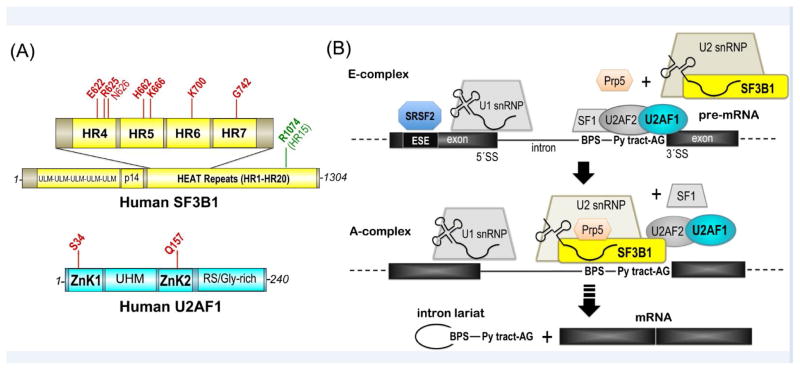
SF3B1 and U2AF1 domains and functions in pre-mRNA splicing. (A) SF3B1 (top, yellow) and U2AF1 (bottom, cyan) domains and positions of mutational “hotspots” (red) [115, 116]. N626 only recently was classified as statistically significant hotspot across tumors [116] and is shown in lighter font. Mutation of R1074 (green) confers pladienolide-resistance. ULM, U2AF ligand motif; p14, p14-binding site; HR, HEAT repeat motif; ZnK, zinc knuckle motif; UHM, U2AF homology motif; RS/Gly-rich, arginine-serine-rich repeats with poly-glycine stretch. (B) SF3B1 and U2AF1 roles in the early stages of spliceosome assembly at the 3′ splice site. snRNP, small nuclear ribonucleoprotein particle; BPS, branch point sequence; ESE, exonic splicing enhancer; Py, polypyrimidine; AG, adenosine-guanosine consensus sequence preceding the 3′ splice site (SS).
Box 2. The Spliceosome, a Molecular Machine for pre-mRNA Splicing.
The spliceosome accomplishes the feat of intron removal from the primary transcripts of eukaryotic cells. Small nuclear (sn)RNAs comprise the catalytic core, whereas multiple protein subunits chaperone assembly on the target pre-mRNA sites and ultimately scaffold the active site (reviewed in [101]). The major class of spliceosomes is composed of the U1, U2, U4, U5 and U6 snRNAs (packaged into the core ribonucleoproteins, snRNP), which assemble on consensus pre-mRNA splice site signals including a “GU” following the 5′ splice site and the BPS, Py tract, and “AG” dinucleotide preceding the 3′ splice site. A minor class of spliceosomes, composed of U11, U12, U4atac, U6atac, and a common U5 snRNP, recognizes distinct splice site signals in less than 1% of known introns (reviewed in [102]).
The recurrently mutated SF3B1, U2AF1, and SRSF2 splicing factors assist assembly of the major spliceosome. The SF3B1 and U2AF1 functions in pre-mRNA splicing are detailed in the text. SRSF2 is member of the arginine-serine-rich (RS) splicing factor family, which typically recognizes exonic splicing enhancer sequences with an RNA recognition motif (RRM) and promotes snRNA-pre-mRNA annealing [103, 104] and protein-protein interactions [105, 106] with an RS-domain. On the other hand, ZRSR2 is important for the minor spliceosome [7], for which it may displace off-target U2AF-interactions [107].
The active spliceosome assembles through a series of discrete, ATP-dependent conformational transitions that are separable by native gel electrophoresis [108] and also have been illuminated by single molecule fluorescence (reviewed in [109]). The “E”-complex of the U1 snRNP and U2AF–SF1 complex at the 5′ and 3′ splice sites is followed by the ATP-dependent “A”-complex, in which the SF3B-containing U2 snRNP stably associates. Subsequently, the U4/U5/U6 snRNP joins the “B”-complex, which is activated following ATP-dependent loss of the U4 snRNP (BACT). The BPS attacks the 5′ splice site to form a branched intron lariat in the “C”-complex, which then is activated to the “C*”-complex. A second transesterification reaction of the 5′ splice site and the downstream 3′ splice site forms the final spliced products in the “P”-complex.
Historical crystal structures have been determined for key spliceosome components, such as Prp8–Brr2 [110, 111] and the U1 snRNP [112–114]. Recent advances, including the availability of direct electron detectors, have facilitated seminal cryoEM structures of spliceosome complexes, including the BACT- [43, 44], C- [70, 71], C*- [72] and P-complexes [73, 74] – leaving only the E- and A- complexes for future structural elucidation.
Several genetic attributes hint that the hotspot mutations of splicing factors confer new functions rather than inhibit existing ones. In all cases, the allelic burdens of the mutant splicing factor alleles (~50%) are consistent with heterozygous mutations (e.g. [18]). Indeed, wild-type (WT) copies of SF3B1, SRSF2 and U2AF1 splicing factors are required for cell proliferation (e.g. [19–21] among others), although multiple single cells carrying only SF3B1 mutations have been detected in single-cell sequences from CLL patient samples [22]. Likewise, simultaneous mutations in multiple splicing factors are absent from patient samples (e.g. [23]). In support of new functional attributes conferred by the mutant splicing factors, most of the altered splice sites in the presence of the hotspot splicing factor mutations differ from affected splice sites following knockdown of that splicing factor [3, 24–27].
We refer the reader to several excellent reviews covering the potential means for progression from pre-mRNA splicing factor mutations to cancer development (e.g. [28, 29]). Here, we focus on recent ribonucleoprotein structures and RNA interactions that increase our molecular-level understanding of the consequences of SF3B1 and U2AF1 mutations in myeloid neoplasms and cancers.
SF3B1 and U2AF1 Splicing Factors Function in Spliceosome Assembly
The MDS-relevant pre-mRNA splicing factors act during the early stages of spliceosome assembly (Figure 1B, Box 2), which proceeds through a discrete series of separable complexes termed “E-“, “A-“, “B-”, “C-“, post-splicing complexes and their activated intermediates. The SF3B1 protein (also called SF3b155 or SAP155) is one of at least seven subunits that comprise the SF3b component of the U2 snRNP [30]. U2AF1 (also called U2AF35) forms a ternary complex along with U2AF2 (also called U2AF65) [31] and Splicing Factor-1 (SF1) [32, 33]. In the earliest “E-complex” of the spliceosome, U2AF1 site-specifically crosslinks with a consensus AG at the 3′ splice site junction and facilitates in vitro splicing of splice sites preceded by “weak” polypyrimidine (Py) tracts that diverge from a U-rich consensus [34–36]. In turn, U2AF2 recognizes “strong” U-rich Py tract site signals that precede the majority of 3′ splice sites [37, 38]. U2AF2 initially binds SF1, which is the first protein to recognize a branch point sequence (BPS) of the pre-mRNA (reviewed in [39]). Subsequently at the transition to the “A-complex”, U2AF2 associates with a “U2AF Ligand Motif” region of SF3B1 (reviewed in [40]), displacing SF1. The U2 snRNA anneals with the BPS [41], which ultimately offers the nucleophile for the first step of the splicing reaction. The U2AF heterodimer is thought to act in conjunction with the SF3B1 subunit to stabilize U2 snRNP association with the 3′ splice site, considering that SF3B1 crosslinks surround the BPS–U2 snRNA duplex [42]. Spliceosome assembly further depends on the ATP-dependent action of DEAD/H-box RNP unwindases, including PRP5 as well as others in the transition from the “E-“ to “A-“ complexes.
Despite extensive biochemical study, only recently has light been shed on the precise locations and interactions mediated by the MDS-relevant residues of SF3B1 and U2AF1. Breakthrough cryo-electron microscopy (cryoEM) structures of spliceosome intermediates isolated from yeast include an SF3B1-containing complex that is likely to represent a BACT-stage spliceosome [43, 44]. Although current spliceosome structures lack the U2AF1 subunit per se, two subunits (Cwc2 and Cwc24) share structural similarity to the mutated U2AF1 domains. Altogether, the spliceosome structures show that the MDS-relevant SF3B1 hotspots contact the pre-mRNA and also suggest that the mutated U2AF1 residues contact the 3′ splice site. Crystal structures of the human SF3b particle [45] and separately, of fission yeast U2AF1 bound to a U2AF2 fragment [46] further detail the hotspot locations. Below, we discuss the molecular-mechanisms of MDS-relevant mutations and new insights gained from these SF3B1 and U2AF1 structures.
Cancer-relevant SF3B1 mutations promote cryptic 3′ splice sites via alternative BPS
SF3B1 is the most frequently mutated splicing factor among a broad range of myeloid neoplasms and solid tumors. The somatic mutations of the SF3B1 gene are particularly prevalent among patients with RARS (50–75% frequencies) [2, 8], uveal melanoma (15–20% frequencies) [10, 11, 47], and CLL (~15% frequency) [5, 6]. SF3B1 mutations also recur among pancreatic or breast cancers (2–4% frequencies) [12–15, 48]. Major hotspots for cancer-relevant SF3B1 mutations are located on the fifth to eighth sequence repeats of the protein, which structures map to the fourth to seventh α-helical “HEAT” repeats (HR) (Figure 1A). A K700E substitution is the most frequent SF3B1 mutation among hematologic malignancies as well as pancreatic and breast cancers, whereas mutations of R625 (and in some cases K666) predominate in uveal melanomas. Several groups have demonstrated that SF3B1 mutations in cancers and hematologic malignancies are associated with altered selection of 3′ splice sites for a subset of transcripts [47, 49–52], including splicing-induced down-regulation of an iron transporter that may increase iron accumulation in RARS [53].
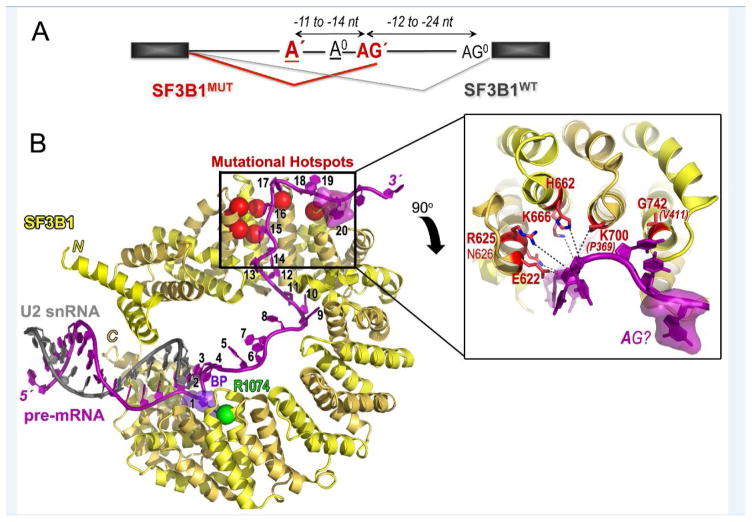
Two recent studies further investigate the molecular mechanisms for altered 3′ splice site choice in the presence of mutant SF3B1 (SF3B1MUT) (Figure 2A) [24, 25]. The studies converge on a key set of common conclusions across a range of sample contexts and SF3B1 mutations, including K700E/R625/K666 and other mutations in CLL samples, a few solid tumors, and pancreatic cancer cell lines [25] or K625/K666 mutations in uveal melanoma patient samples and minigene models in cell lines [24]. Both aberrant and normal 3′ splice sites show a consensus “AG” splice site signal preceding the intron-exon junction. SF3B1MUT stimulates use of cryptic 3′ splice sites (AG′) clustered in a −12 to −24 nucleotide window upstream of the normal splice site (AG0). Importantly, an A-containing sequence that resembles a BPS splice site signal (BPS′) is located −11 to −14 nucleotides upstream of each aberrant AG′. Indeed, the predicted complementarity of this pre-mRNA region for annealing with the U2 snRNA, and hence “strength” was greater for BPS′ than for a normal BPS (BPS0). Mutagenesis and experimental mapping confirmed that these upstream adenosines are used as the branch sites during intron lariat excision from the SF3B1MUT-sensitive splice sites. Diverse proposals have been made to explain the switch to BPS′ and AG′ in the presence of mutant SF3B1, including that mutations could enhance SF3B1MUT interactions with pre-mRNA sequences flanking BPS′, decrease SF3B1MUT interactions with BPS0, and/or trigger a conformational change in SF3B1MUT such that the “stronger” complementarity of U2 snRNA for BPS′ in turn outweighs BPS0 [24, 25]. Recent structural information raises a striking variation of these hypotheses, namely that hotspot SF3B1 mutations could influence the conformation of the bound pre-mRNA splice site.
Figure 2.
SF3B1 hotspot mutations alter branchpoint choice and cluster at a pre-mRNA “kink”. (A) Schematic representation of the effect of SF3B1MUT on pre-mRNA splicing. SF3B1MUT (red) promotes use of an upstream aberrant 3′ splice site junction (AG′) and BPS (A′), which differs from the normal 3′ splice site (AG0) and BPS0 (A0) of SF3B1WT (black). Exons, black rectangles; introns, horizontal lines; spliced junctions connected. (B) Structure of Saccharomyces cerevisiae SF3B1 (alternating HEAT repeats are yellow and gold) in the BACT spliceosome (PDB ID 5GM6) bound to pre-mRNA (magenta), which in turn anneals with U2 snRNA (gray). Pre-mRNA nucleotides are numbered from 1 at the branchpoint adenosine (violet) to 20 at the sole putative purine nucleotide, which may correspond to −2A of the conserved AG dinucleotide preceding the intron-exon junction. Mutational hotspots (red) are represented as spheres on the left and inset as ball-and-stick diagrams on the right. Dashed lines indicate putative hotspot–nucleotide interactions, although the overall 3.5 Å resolution limits interpretation. Single letter amino acid codes and residue numbers obtained from superposition with human SF3B1 (PDB ID 5IFE) are shown for clarity. The two yeast amino acids that differ in identity from the human homologue (P369 for K700 and V411 for G742) are given in parentheses and italics. Other yeast SF3B1 residues correspond to the following human residues: E291 for E622; R294 for R625; N295 for N626; H331 for H662; K335 for K666. Yeast SF3B1 R743 matches human R1074 (green), for which mutation confers pladienolide-resistance.
Spatial locations point to roles for SF3B1 hotspots in 3′ splice site conformation
The 3.5 Å resolution structure of yeast SF3B1 in the BACT spliceosome shows an α-helical HEAT domain encircling 23 nucleotides of the pre-mRNA strand [43] (Figure 2B). As mentioned above, the observed number of HR in SF3B1 [43, 45] is less than predicted based on primary sequence analysis [54]. Instead, the 3.1 Å resolution human SF3B1 structure shows that the HEAT domain is led and capped by single α-helices [45] rather than the two antiparallel α-helices that formally compose an HR (reviewed in [55]). Deep within the curved SF3B1 structure, the BPS–U2 snRNA duplex is gripped in a vise comprising an N-terminal HEAT repeat (HR1) in the minor groove and several C-terminal HEAT repeats (HR14-HR19) lining the pre-mRNA phosphodiester backbone. Notably, a key residue for which mutation to histidine confers resistance to the cell-cycle inhibitor pladienolide and its derivatives (R1074 in HR15) [56] contributes part of the binding pocket for an extrahelical branch site adenosine. Indeed, unassigned electron density in the SF3B1 crystal structure is likely to reflect co-purified branch site RNA [45]. To the 5′ side of the BPS, 20 nucleotides of single-stranded pre-mRNA cross the concave SF3B1 surface and terminate immediately following a kink at the hotspot residues. A typical BPS---AG separation of approximately 25 nucleotides among human transcripts [57] suggests that the intron-exon junction either corresponds to or closely follows the 5′ terminus of the SF3B1-bound pre-mRNA in the BACT spliceosome structure.
Superposition maps the mutational hotspots of the human SF3B1 structure on the yeast homologue in the BACT spliceosome (Figure 2B). The SF3B1 hotspots occur in four consecutive HR’s (HR4-HR7) near the N-terminus of the HEAT domain. These key residues are consistently located at the N-terminal tips of the second α-helices in the two-helix HR units, where several positively-charged residues complement the α-helical dipole moments for interactions with negatively-charged RNA. The side chains align in a spatially contiguous surface that appears to stabilize a kinked conformation for the 5′ nucleotides of the bound pre-mRNA. Most commonly, the recurrent mutations drastically change the shape and charge of the affected residue (e.g. K700E). As such, the hotspot amino acid substitutions are expected to disrupt local RNA interactions and dissociate the 5′ end of the pre-mRNA from the SF3B1MUT protein surface. Since the mutated RNA binding site is located approximately ten nucleotides from the preceding pre-mRNA contact on SF3B1 HR9 and also concludes with a unique purine that could correspond to the normal AG splice site signal, impaired pre-mRNA interactions at this interface would explain the up to ten nucleotides shorter BPS’---AG’ distance preferred by SF3B1MUT compared to normal BPS---AG spacings [24, 25]. As such, the RNA-bound SF3B1 structure in the BACT spliceosome suggests that hotspot mutations could alter the SF3B1MUT-bound conformation of the pre-mRNA, rather than RNA-sequence specificity or the protein conformation itself.
Cancer-relevant U2AF1 mutations promote alternative 3′ splice sites
Acquired mutations of the U2AF1 splicing factor are common among hematopoietic malignancies [1, 3, 4], particularly MDS without ring sideroblasts (~11%), secondary AML (~9%), or CMML (8–11%), as well as hairy-cell leukemia-variant (HCLv) (10%) [58]. Among solid tumors, U2AF1 is mutated in pancreatic ductal adenocarcinomas at similar frequencies as SF3B1 (~2% frequency) [13] and is the major mutated splicing factor gene found in lung adenocarcinomas (~3% frequency) [16, 17]. The hotspot mutations of U2AF1 narrowly focus on the codons for residues S34 and Q157. Indeed, S34F mutations account for 60–80% of U2AF1 mutations in MDS [1, 3] and nearly all documented U2AF1 mutations in HCLv, lung adenocarcinomas and pancreatic cancers [13, 16]. Less frequently, the S34 residue changes to a tyrosine (10–15% of U2AF1 mutations in MDS), which shares similar aromatic properties with the phenylalanine, or the Q157 residue changes to either proline or arginine (up to 20% of U2AF1 mutations in MDS) [1, 3]. The U2AF1 mutations are associated with altered splicing of approximately 5% of gene transcripts in patient samples and cell lines [59–62], primarily due to cassette exon skipping/inclusion or to a lesser extent, alternative 3′ splice site choice. As for SF3B1 mutations, the splicing events that are affected by U2AF1 mutations have low overlap with those affected by U2AF1 knockdown [21, 62], which suggested that these mutations subtly alter recognition of splice sites rather than completely disable the spliceosome. The mis-spliced genes fall into cancer-relevant pathways, including DNA damage response, epigenetic regulation, and apoptosis. Recently, the U2AF1 S34F mutation was discovered to increase use of distal cleavage and polyadenylation sites (~40% frequency among S34F-affected transcripts in a BaF3 cell line) [63]. Here we focus on the molecular mechanisms underlying altered 3′ splice site choice in the presence of mutant U2AF1, which hitherto has been the major research focus in the field, and touch on potential extensions to polyadenylation in the last section.
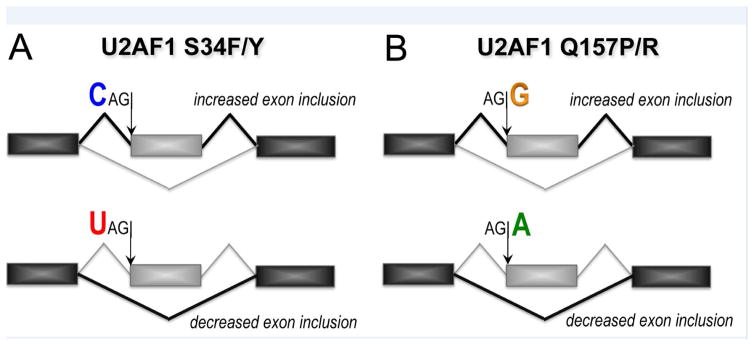
The splicing events that are altered by S34 versus Q157 mutations of U2AF1 segregate into distinct groups (Figure 3). Each of these groups shares common sequence trends surrounding the conserved “AG” consensus: S34 mutants preferentially skip splice junctions preceded by a −3U and splice those preceded by −3C/A [59–62], whereas Q157 mutants preferentially skip splice junctions followed by a +1A and splice those followed by +1G [62]. Minigenes and in vitro splicing assays of defined splice site variants recapitulate these S34F-dependent sequence trends [62]. The minimal U2AF heterodimer from fission yeast (SpU2AFMIN) and separately, nearly full length human U2AF1–U2AF2 complexes with SF1 have been prepared in sufficient quantities for quantitative RNA binding assays [21, 61, 64]. The S34F mutation reduced association of the SpU2AFMIN protein with a core 5-nucleotide RNA oligonucleotide (5′-CUAGG, AG consensus underlined, −3 and +1 nucleotides in bold) [64], although the weak binding affinity was beyond the limit of the isothermal titration calorimetry (ITC) experiment (>3-fold penalties reduce the c-value = [macromolecule in sample cell]/KD to below the limit of ~10 for a reliable fit [65]). A subtle increase in RNA binding, rather than decrease, was observed for SpU2AFMIN carrying the counterpart of the Q157R mutation to this RNA (by ~2-fold) [64]. Although surprisingly different, these mutation-induced changes in SpU2AFMIN binding the CUAGG RNA matched the sequence trends among affected splice sites; namely, the S34F mutant suffers a penalty for binding a −3U RNA, which is typically skipped in affected splice sites, whereas mutation of the Q157R counterpart prefers to bind a +1G RNA, which is typically spliced in affected splice sites. We used quantitative fluorescence anisotropy assays to compare a series of splice site RNAs with various −3 nucleotides binding to S34F or WT variants of human ternary U2AF1–U2AF2–SF1 complexes [21, 61]. Although the changes in RNA binding due to the S34F mutation were mild (2- to 7-fold), in nearly all cases these binding trends matched the observed consequences for splicing: The S34F mutation typically decreased U2AF1 binding to sites preceded by −3U and in some cases increased affinity for −3C. Altogether, these results support a hypothesis in the field that the S34 residue of U2AF1 recognizes the −3 nucleotide, whereas Q157 recognizes the +1 nucleotide of the 3′ splice site junction, and mutations alter this recognition for affected splice sites. Below we discuss support for this hypothesis gained from new structures of apo-SpU2AFMIN and structurally homologous spliceosome subunits bound to RNA.
Figure 3.
Schematic representation of the effects of U2AF1 mutations on pre-mRNA splicing. (A) Mutations of U2AF1 S34 to F or Y (S34F/Y) sense the identity of the nucleotide at the −3 position (three nucleotides preceding) the intron-exon junction, typically promoting inclusion of sites preceded by −3C (blue) and skipping sites preceded by −3U (red). (B) Mutations of U2AF1 Q157 to P or R (Q157P/R) sense the identity of the nucleotide at the +1 position immediately following the intron-exon junction, typically promoting inclusion of sites followed by +1G (yellow) and skipping sites followed by +1A (green). Exons, black or gray rectangles; spliced junctions are connected by lines.
RNA contacts in homologous structures support roles for U2AF1 hotspots in splice site recognition
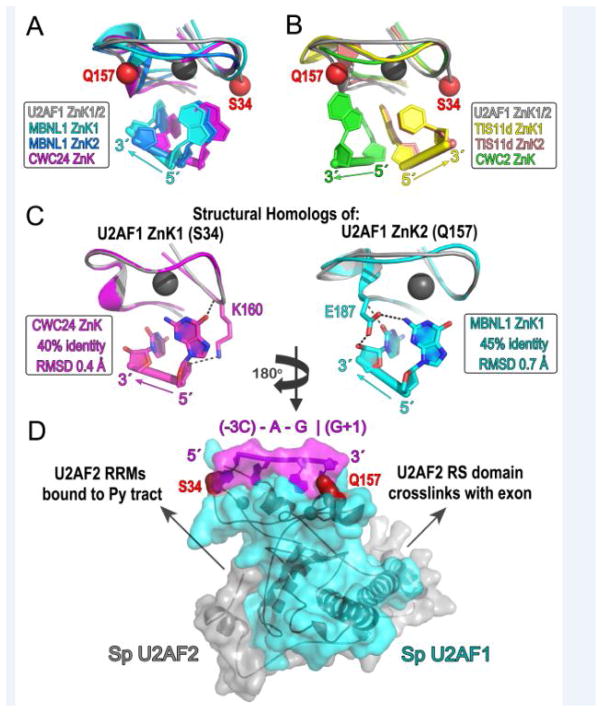
The U2AF1 hotspots are located in two CCCH-type zinc knuckles that surround the U2AF2-heterodimerization motif (UHM, [40]), namely S34 is in the N-terminal ZnK1 and Q157 in the C-terminal ZnK2 (Figure 1A). Until recently, ZnK–RNA complexes were limited to two structures: an NMR structure of TIS11d, which comprises tandem ZnK1 and ZnK2 bound to an AU-rich mRNA regulatory element [66], and a 1.7 Å resolution crystal structure of the alternative splicing regulator MBNL1, which comprises a GCU-containing RNA bound to tandem ZnK1 and ZnK2 (formally ZnK3 and ZnK4 in the intact MBNL1 protein sequence) [67]. Despite laudatory efforts [46, 62], generating a reliable model of the U2AF1–RNA complex based solely on these prior structures has met several challenges. First, the 5′-to-3′ orientations of the RNA strands relative to the bound ZnK show opposite directions between the MBNL1 and TIS11d structures (Figure 4A–B). This discrepancy was surprising considering that other RNP fold families share similar protein–RNA orientations (e.g. [55, 68, 69]). Second, the global conformations of the tandem ZnK1 and ZnK2 differ between the two structures: the MBNL1 ZnK1 and ZnK2 each bind separate RNA strands whereas the TIS11d ZnK1 and ZnK2 bind adjacent sites of a contiguous oligonucleotide. The new spliceosome structures now reveal two examples of CCCH-type ZnK’s bound to RNA: CWC24 bound to the 5′ exon of the BACT complex [43] and CWC2 bound to U6 snRNA in the BACT, C, C*, and ILS complexes [43, 44, 70–75]. Moreover, the recent SpU2AFMIN structure [46] contains the full length fission yeast U2AF1 subunit, which shares 55% sequence identity with human U2AF1 and as such provides a reliable starting point for modeling putative RNA interactions with the mutated ZnK1/ZnK2 domains.
Figure 4.
Structural comparisons support RNA contacts by U2AF1 hotspots. For clarity of viewing the affected interactions, Panels A–C are rotated two-fold about the y-axis relative to Panel D. Panel D is oriented in the typical 5′−3′ direction of the splice site consensus sequence. (A) Superpositions of MBNL1 ZnK1 (cyan) or ZnK2 (blue) (PDB ID 3D2S), CWC24 ZnK (magenta) (PDB ID 5GM6), and SpU2AF1 ZnK2 on SpU2AF1 ZnK1 (gray) (PDB ID 4YH8) by matching Cα atoms show similar RNA orientation and nucleotide binding sites. (B) Superpositions of TIS11d ZnK1 (yellow) or ZnK2 (salmon) (PDB ID 1RGO), CWC2 ZnK (green) (PDB ID 5GMK) and SpU2AF1 ZnK2 on SpU2AF1 ZnK1 (gray) (PDB ID 4YH8) show that the angle of the CWC2 RNA differs from the MBNL1 family and that the TIS11d-bound RNA runs antiparallel relative to other ZnK structures. (C) RNA-interactions of residues matching S34 (K160) or Q157 (E187) (ball-and-stick) shown for the closest structural homologs of U2AF1 ZnK1 (CWC24, magenta, left) or ZnK2 (MBNL1, cyan, right). Sequence identities of the indicated structures compared with human U2AF1 ZnK1 or ZnK2 and RMSD with matching SpU2AF1 Cα atoms are boxed. (D) Solvent-accessible surface of SpU2AF1 (cyan) (PDB ID 4YH8) bound to RNA derived from the CWC24 and MBNL1 ZnK structures. Two dinucleotides are connected by a de novo phosphate following superposition of matching Cα atoms. The nucleotide sequence was mutated to the splice site consensus (magenta label, “|” intron-exon junction). Alternative R28 and R150 rotamers were chosen to interact with the phosphodiester linkage and accommodate the bound nucleobase. Arrows indicate known pre-mRNA interactions with the N- and C-terminal domains of SpU2AF2 (gray) surrounding but absent from the bound fragment in the SpU2AF heterodimer. U2AF1 hotspots are red spheres.
The U2AF1 ZnK1 shows the greatest sequence identity with CWC24 ZnK (40% identity) whereas the U2AF1 ZnK2 resembles MBNL1 ZnK1 (45% identity) (Figure 4C). Comparison among the ZnK structures shows that the bound RNA conformation of the CWC24 ZnK closely resembles those of MBNL1 ZnK1 and ZnK2 (Figure 4A). The CWC2 ZnK–RNA also shares a similar 5′-to-3′ orientation of the RNA strand compared to the CWC24 and MBNL1 structures, although the exact trajectory of the RNA changes by approximately 30° (Figure 4B). In contrast, the orientation of the TIS11d-bound RNA strand runs antiparallel relative to other ZnK–RNA structures determined to-date (Figure 4B). Further expansion of available ZnK–RNA structures is needed to fully discern themes from outliers. Regardless, the CWC24, CWC2, MBNL1, and TIS11d structures already establish versatile means for single-stranded RNA recognition by ZnK modules, analogous to the diverse RNA interactions by members of the well-characterized RRM fold family [76].
Mutations of S34 or Q157 to phenylalanine or proline are expected to have little impact on the protein fold, considering their respective locations in either an unstructured loop or the N-terminal turn of an α-helix, which does not require backbone hydrogen bonds for proper folding. Instead, sequence-specific RNA interactions appear likely to be affected. The S34 counterpart of CWC24, as well as of its structural paralogues MBNL1 ZnK1 or ZnK2, interact with the 5′ terminal nucleotide of the bound dinucleotide. For CWC24, this residue (K160) donates hydrogen bonds from the peptide backbone to a guanine base and from the side chain to the ribose oxygen (Figure 4C, left). Although the exact consequences of the hotspot phenylalanine substitution are unpredictable, introduction of a bulky aromatic side chain certainly would alter the nucleotide position and interactions at this site. The Q157 counterparts of MBNL1 ZnK1, MBNL1 ZnK2, or CWC24 ZnK primarily interact with the 3′ terminal nucleotide of the bound dinucleotide. By analogy, the hotspot mutation of Q157 to a proline residue, which lacks the peptide N-H group, would obliterate a peptide hydrogen bond to the nucleobase. Likewise, a side chain-mediated hydrogen bond to the adjacent nucleobase would be lost.
The SpU2AFMIN structure provides a reliable scaffold for assembly of these homologous, RNA-bound structures in a composite model (Figure 4D). Together following superposition on the respective SpU2AF1 ZnK1 and ZnK2, bound dinucleotides of the CWC24 and MBNL1 structures form a contiguous RNA strand following addition of a single phosphodiester bond. Inserting an additional nucleotide in the sequence would either bulge the central nucleotide or require a conformational change that separates U2AF1 ZnK1 from ZnK2. The C-terminus of the heterodimeric SpU2AF2 is oriented towards the 5′ of the RNA strand, consistent with the C-terminal U2AF2 RRMs binding the upstream Py tract signal [77, 78]. The position of the SpU2AF2 N-terminus facing the RNA 3′ terminus is consistent with directed hydroxyl radical footprinting of the U2AF2 RS domain in the downstream exon [79]. However, footprinting of the U2AF2 N-terminus within the BPS [79, 80] suggests that the RS domain is dynamic and/or that the RNA conformation is bent. If we reverse the U2AF1-bound RNA conformation to match that of TIS11d [66], the swapped orientations of these interacting domains contradict biochemical results. Indeed, superposition of SpU2AF1 with the CWC24 and MBNL1 naturally bridges the ZnK1 and ZnK2 with a 4-nucleotide RNA, consistent with locations of the −3 nucleotide of the intron and the +1 nucleotide of the exon adjacent the S34 and Q157 residues (Figure 4D). Although alternative models have yet to be ruled out, these structural comparisons support a direct role for U2AF1 hotspots in sequence-specific read-out of splice site junctions.
Concluding Remarks
Recent structural information, together with sequence trends among affected splice sites, implicate the cancer-relevant mutational hotspots of SF3B1 and U2AF1 in pre-mRNA splice site contacts. Yet, additional evidence is needed to fully understand the mechanistic underpinnings and downstream consequences of recurrent splicing factor mutations (Outstanding Questions). Already, the RNA binding affinities of recombinant U2AF1 proteins supports the importance of the S34 and Q157 residues for recognizing the −3 and +1 nucleotides surrounding the 3′ splice site junction [21, 61, 64]. Although the recombinant SF3B1MUT protein remains capable of RNA binding at saturating concentrations [45], the RNA affinities and specificities of SF3B1 and its mutated variants remain to be evaluated for “strong” (consensus) compared to “weak” (divergent) splice sites. Regardless, a mutation-induced conformational change in the bound RNA would not necessarily alter the apparent RNA affinity and instead calls for structural characterization. Greater changes in the rate constants of RNA binding could underlie subtle apparent changes in the equilibrium RNA binding of mutant compared to WT splicing factors. Accordingly, the U2AF1 S34F mutation appears to delay the rate of spliced transcript release [81], which in turn is expected to alter splice site choice (reviewed in [82]). Notably, the P95 hotspot of SRSF2, which is frequently mutated among CMML patients, also is located at its RNA interface [83] and interferes with RNA binding when mutated [26, 84]. Such roles for hotspot residues in contacting pre-mRNA splice sites may emerge as a common theme among MDS-relevant mutations of splicing factors.
Outstanding Questions.
Do SF3B1 hotspot mutations alter the bound pre-mRNA conformation and does this structural mechanism underlie aberrant splice site choice?
Do SF3B1 hotspot mutations alter pre-mRNA binding?
Does altered RNA binding by SF3B1 hotspot mutations contribute to altered protein cofactor functions, for example of PRP5?
Do SF3b1 hotspot mutations affect auxiliary SF3b1 functions, for example nucleosome association?
Do U2AF1 or SF3B1 hotspot mutations alter the rates of pre-mRNA binding and release?
What is the structure of the U2AF1–3′ splice site RNA complex and do its S34 and Q157 hotspots recognize RNA, as predicted by RNA binding studies and sequence trends among affected splice sites?
Does altered RNA binding by U2AF1 hotspot mutations contribute to altered polyadenylation in the presence of these mutations?
What is the molecular pathway to cancer following pre-mRNA splicing factor mutations?
Yet, RNA interactions by splicing factor hotspots offer only a partial, albeit potentially complementary, explanation for multifaceted effects of hotspot mutations. Recently, the DEAD-box RNP-unwindase PRP5 (also called DDX46) was found to interact genetically and physically with the yeast SF3B1 homologue (called HSH155), and hotspot mutations altered this interaction [85, 86]. Considering SF3B1–RNA contacts, an appropriate conformation of the SF3B1–pre-mRNA complex could serve as a checkpoint for the ATPase activity of PRP5, which is required for stable association of the U2 snRNA with the BPS. Auxiliary to pre-mRNA splicing, the impacts of SF3B1 mutations on its chromatin association with and nucleosome positioning [87] have yet to be tested. For U2AF1, the S34F mutation already has been found to alter the cleavage and polyadenylation sites of a subset of affected transcripts. In particular, S34F-associated use of a distal polyadenylation site in the ATG7 transcript was sufficient to transform BaF3 or small airway cell lines, and also was observed in patient samples [63]. Although the coupling of pre-mRNA splicing to cleavage and polyadenylation and specific association of U2AF subunits with polyadenylation factors have been established ([36], reviewed in [88]) how these processes crosstalk in the downstream progression of U2AF1 mutations to malignancies remains an outstanding question. Altogether, current findings represent only a starting point for fully understanding the mechanistic effects of recurrent splicing factor mutations. Future efforts to elucidate these possibilities would guide longer-term efforts to exploit the therapeutic potential of spliceosome inhibitors [90], as recently highlighted by the spliceosome-sensitivity of MYC-driven cancers [91–93].
Trends.
Mutations of pre-mRNA splicing factors are common acquired mutations among patients with MDS and related myeloid neoplasms, and also recur in certain solid tumors.
“Hotspot” mutations of pre-mRNA splicing factors alter splicing of a subset of transcripts.
Structures of the activated B-stage spliceosome show SF3B1 hotspots contacting the pre-mRNA splice site.
Hotspot mutations of U2AF1 alter its binding affinity for splice site RNAs in a manner that correlates with changes in pre-mRNA splicing.
An apo-structure of a fission yeast homologue shows U2AF1 hotspot locations on zinc knuckle surfaces and structural homology suggests that these residues contact RNA.
Acknowledgments
Supported by grants from the NIH (GM070503 and GM117005) and the Edward P. Evans Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44(1):53–7. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makishima H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–10. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 7.Madan V, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malcovati L, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233–41. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meggendorfer M, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120(15):3080–8. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45(2):133–5. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin M, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–6. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 14.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013 doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 2015;75(10):1949–58. doi: 10.1158/0008-5472.CAN-14-2930. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672–8. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei DL, et al. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and Is Required for Cell Survival. PLoS Genet. 2016;12(10):e1006384. doi: 10.1371/journal.pgen.1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, et al. Genetic characterization of SF3B1 mutations in single chronic lymphocytic leukemia cells. Leukemia. 2013;27(11):2264–7. doi: 10.1038/leu.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsafadi S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darman RB, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13(5):1033–45. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27(5):617–30. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komeno Y, et al. SRSF2 Is Essential for Hematopoiesis, and Its Myelodysplastic Syndrome-Related Mutations Dysregulate Alternative Pre-mRNA Splicing. Mol Cell Biol. 2015;35(17):3071–82. doi: 10.1128/MCB.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue D, et al. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30(9):989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dvinge H, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–30. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will CL, et al. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21(18):4978–88. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89(3):403–12. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 33.Kramer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10(6):1503–9. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, et al. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402(6763):832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 35.Guth S, et al. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol Cell Biol. 1999;19(12):8263–71. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kralovicova J, et al. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3′ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;43(7):3747–63. doi: 10.1093/nar/gkv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, et al. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268(5214):1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 38.Shao C, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21(11):997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rymond BC. The branchpoint binding protein: in and out of the spliceosome cycle. Adv Exp Med Biol. 2010;693:123–41. [PubMed] [Google Scholar]
- 40.Loerch S, Kielkopf CL. Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise. RNA. 2016;22(12):1795–1807. doi: 10.1261/rna.057950.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker R, et al. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49(2):229–39. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 42.Gozani O, et al. A potential role for U2AF-SAP155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18(8):4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan C, et al. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016;353(6302):904–11. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 44.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353(6306):1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 45.Cretu C, et al. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol Cell. 2016;64(2):307–319. doi: 10.1016/j.molcel.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida H, et al. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 2015;29(15):1649–60. doi: 10.1101/gad.267104.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furney SJ, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3(10):1122–9. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maguire SL, et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235(4):571–80. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeBoever C, et al. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput Biol. 2015;11(3):e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gentien D, et al. A common alternative splicing signature is associated with SF3B1 mutations in malignancies from different cell lineages. Leukemia. 2014;28(6):1355–7. doi: 10.1038/leu.2014.28. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell. 2016;30(5):750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obeng EA, et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell. 2016;30(3):404–17. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolatshad H, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia. 2016;30(12):2322–2331. doi: 10.1038/leu.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, et al. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12(10):1409–14. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubinson EH, Eichman BF. Nucleic acid recognition by tandem helical repeats. Curr Opin Struct Biol. 2012;22(1):101–9. doi: 10.1016/j.sbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoi A, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278(24):4870–80. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 57.Mercer TR, et al. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25(2):290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterfall JJ, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46(1):8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Przychodzen B, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122(6):999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks AN, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One. 2014;9(1):e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okeyo-Owuor T, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29(4):909–17. doi: 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park SM, et al. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3′ End Formation. Mol Cell. 2016;62(4):479–90. doi: 10.1016/j.molcel.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida H, et al. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes & Development. 2015;29(15):1649–60. doi: 10.1101/gad.267104.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiseman T, et al. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179(1):131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 66.Hudson BP, et al. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11(3):257–64. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 67.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15(12):1343–51. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Auweter SD, et al. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34(17):4943–59. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maris C, et al. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 70.Wan R, et al. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science. 2016;353(6302):895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 71.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537(7619):197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan C, et al. Structure of a yeast step II catalytically activated spliceosome. Science. 2016 doi: 10.1126/science.aak9979. [DOI] [PubMed] [Google Scholar]
- 73.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349(6253):1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 74.Hang J, et al. Structural basis of pre-mRNA splicing. Science. 2015;349(6253):1191–8. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 75.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017 doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clery A, et al. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008 doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Mackereth CD, et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475(7356):408–11. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 78.Agrawal AA, et al. An extended U2AF65-RNA-binding domain recognizes the 3′ splice site signal. Nat Commun. 2016;7:10950. doi: 10.1038/ncomms10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kent OA, et al. Structuring of the 3′ splice site by U2AF65. J Biol Chem. 2003;278(50):50572–50577. doi: 10.1074/jbc.M307976200. [DOI] [PubMed] [Google Scholar]
- 80.Valcarcel J, et al. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273(5282):1706–9. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 81.Coulon A, et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife. 2014:3. doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moehle EA, et al. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11(4):313–9. doi: 10.4161/rna.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Daubner GM, et al. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31(1):162–74. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112(34):E4726–34. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang Q, et al. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30(24):2710–2723. doi: 10.1101/gad.291872.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carrocci TJ, et al. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkw1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kfir N, et al. SF3B1 association with chromatin determines splicing outcomes. Cell Rep. 2015;11(4):618–29. doi: 10.1016/j.celrep.2015.03.048. [DOI] [PubMed] [Google Scholar]
- 88.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA. 2011;2(4):459–70. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 89.Yang Q, et al. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol. 2011;8(5):748–53. doi: 10.4161/rna.8.5.16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Effenberger KA, et al. Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523(7558):96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 92.Hsu TY, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384–8. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hirsch CL, et al. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 2015;29(8):803–16. doi: 10.1101/gad.255109.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gangat N, et al. Myelodysplastic syndromes: Contemporary review and how we treat. Am J Hematol. 2016;91(1):76–89. doi: 10.1002/ajh.24253. [DOI] [PubMed] [Google Scholar]
- 95.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 96.Koeffler HP, Leong G. Preleukemia: one name, many meanings. Leukemia. 2017 doi: 10.1038/leu.2016.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica. 2011;96(6):789–92. doi: 10.3324/haematol.2011.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steensma DP. Good riddance to the term “refractory anemia” in myelodysplastic syndromes. Leuk Res. 2016;51:22–26. doi: 10.1016/j.leukres.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 99.Howlader N, et al. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute; 2016. [Google Scholar]
- 100.Pophali PA, Patnaik MM. The Role of New Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. Cancer J. 2016;22(1):40–50. doi: 10.1097/PPO.0000000000000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Papasaikas P, Valcarcel J. The Spliceosome: The Ultimate RNA Chaperone and Sculptor. Trends Biochem Sci. 2016;41(1):33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 102.Turunen JJ, et al. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA. 2013;4(1):61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen H, et al. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13(3):367–76. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 104.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16(3):363–73. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 105.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75(6):1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 106.Kohtz JD, et al. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368(6467):119–24. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 107.Shen H, et al. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24(21):2389–94. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ares M., Jr Analysis of splicing complexes on native gels. Cold Spring Harb Protoc. 2013;2013(10):986–9. doi: 10.1101/pdb.prot078139. [DOI] [PubMed] [Google Scholar]
- 109.DeHaven AC, et al. Lights, camera, action! Capturing the spliceosome and pre-mRNA splicing with single-molecule fluorescence microscopy. Wiley Interdiscip Rev RNA. 2016;7(5):683–701. doi: 10.1002/wrna.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mozaffari-Jovin S, et al. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341(6141):80–4. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 111.Nguyen TH, et al. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013;21(6):910–19. doi: 10.1016/j.str.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pomeranz Krummel DA, et al. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458(7237):475–80. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weber G, et al. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29(24):4172–84. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kondo Y, et al. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife. 2015:4. doi: 10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hahn CN, Scott HS. Spliceosome mutations in hematopoietic malignancies. Nat Genet. 2011;44(1):9–10. doi: 10.1038/ng.1045. [DOI] [PubMed] [Google Scholar]
- 116.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155–63. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]