Abstract
BACKGROUND & AIMS
Drug repositioning offers a shorter approval process than new drug development. We therefore searched large public datasets of drug-induced gene expression signatures to identify agents that might be effective against hepatocellular carcinoma (HCC).
METHODS
We searched public databases of mRNA expression patterns reported from HCC specimens from patients, HCC cell lines, and cells exposed to various drugs. We identified drugs that might specifically increase expression of genes that are downregulated in HCCs and reduce expression of genes upregulated in HCCs using a non-parametric, rank-based pattern-matching strategy based on the Kolmogorov–Smirnov statistic. We evaluated the anti-tumor activity of niclosamide and its ethanolamine salt (NEN) in HCC cell lines (HepG2, Huh7, Hep3B, Hep40, and PLC/PRF/5), primary human hepatocytes, and 2 mouse models of HCC. In 1 model of HCC, liver tumor development was induced by hydrodynamic delivery of a sleeping beauty transposon expressing an activated form of Ras (v12) and truncated beta catenin (N90). In another mouse model, patient-derived xenografts were established by implanting HCC cells from patients into livers of immunocompromised mice. Tumor growth was monitored by bioluminescence imaging. Tumor-bearing mice were fed a regular chow diet or a chow diet containing niclosamide or NEN. In a separate experiment using patient-derived xenografts, tumor-bearing mice were given sorafenib (the standard of care for patients with advanced HCC), NEN, or niclosamide alone; a combination of sorafenib and NEN; or a combination sorafenib and niclosamide in their drinking water, or regular water (control), and tumor growth was monitored.
RESULTS
Based on gene expression signatures, we identified 3 anthelmintics that significantly altered the expression of genes that are up- or down-regulated in HCCs. Niclosamide and NEN specifically reduced the viability of HCC cells: the agents were at least 7-fold more toxic to HCCs than primary hepatocytes. Oral administration of NEN to mice significantly slowed growth of genetically induced liver tumors and patient-derived xenografts, whereas niclosamide did not, coinciding with the observed greater bioavailability of NEN compared with niclosamide. The combination of NEN and sorafenib was more effective at slowing growth of patient-derived xenografts than either agent alone. In HepG2 cells and in patient-derived xenografts, administration of niclosamide or NEN increased expression of 20 genes downregulated in HCC and reduced expression of 29 genes upregulated in the 274-gene HCC signature. Administration of NEN to mice with patient-derived xenografts reduced expression of proteins in the Wnt–beta catenin, STAT3, AKT–mTOR, EGFR–Ras–Raf signaling pathways. Using immunoprecipitation assays, we found NEN to bind cell division cycle 37 (CDC37) protein and disrupt its interaction with heat shock protein 90 (HSP90).
Conclusions
In a bioinformatics search for agents that alter the HCC-specific gene expression pattern, we identified the anthelmintic niclosamide as a potential anti-tumor agent. Its ethanolamine salt, with greater bioavailability, was more effective than niclosamide at slowing the growth of genetically induced liver tumors and patient-derived xenografts in mice. Both agents disrupted interaction between CDC37 and HSP90 in HCC cells, with concomitant inhibition of their downstream signaling pathways. NEN might be effective for treatment of patients with HCC.
Keywords: mitochondrial uncoupler, treatment, liver cancer, big data
Introduction
Hepatocellular carcinoma (HCC), the most common adult liver malignancy, is the second most frequent cause of cancer-related deaths worldwide 1. Most patients remain asymptomatic until the disease is far advanced, when the standard of care is chemotherapy with sorafenib, the only drug approved by the FDA for the treatment of advanced HCC in the past decade. Although more than 50 molecularly targeted drugs have been in clinical trials for HCC 2, as yet, none of them have been shown to be superior to sorafenib. With recent emergence of resistance to sorafenib, there is an urgent need to identify second-line therapies for HCC patients who no longer respond to sorafenib 3.
The increasing availability of large public datasets on disease-specific and drug-induced transcriptomic signatures offers a time-efficient approach to reposition existing drugs for new indications 4–6. Compared to traditional drug discovery, drug repositioning offers a relatively short approval process and straightforward path to clinical translation 7. This approach is especially appealing for identifying alternative therapeutic options for diseases with no current satisfactory treatment 5. In oncology, recent projects funded by the National Institutes of Health to molecularly characterize cancer patient samples, cancer cell lines, and cellular drug responses, have led to the generation of huge datasets such as The Cancer Genome Atlas (TCGA) 8, the Cancer Cell Line Encyclopedia (CCLE) 9, and the Library of Integrated Network-Based Cellular Signatures (LINCS (http://www.lincsproject.org)). Effectively integrating the information across these datasets and translating them into actionable therapeutics holds promise for fatal diseases including HCC.
Here, we present a novel integrated bioinformatics approach leveraging new public gene expression profiles of HCC patient tumors, HCC cell lines, and cellular drug responses to identify drug candidates that can reverse the HCC gene expression signature and therefore have the potential to treat the disease. Three anthelmintic drugs were predicted to reverse the HCC gene expression signature, among which niclosamide was a top hit. To validate our predictions, we evaluated the anti-tumor activity of niclosamide and its ethanolamine salt (NEN) in HCC cell lines and HCC mouse models10, including a primary HCC model in which HCC is induced endogenously through hydrodynamic delivery of Sleeping Beauty transposon system with oncogene Ras (v12) and truncated β-catenin (N90) 11, and patient-derived xenograft (PDX) models 10. We further demonstrated that NEN disrupted the interaction of CDC37 and HSP90, which led to degradation of downstream protein kinases, which may in part explain its mechanism of anti-tumor action.
Materials and Methods
The detailed Materials and Methods can be found in the Supplementary Materials.
Collect and process HCC gene expression profiles
RNA-Seq profiles of HCC tumor samples were downloaded from GDAC (https://gdac.broadinstitute.org/), and their clinical data were downloaded from TCGA as of May 2014. We also searched GEO using the keyword “hepatocellular carcinoma” and manually collected seven independent datasets with at least three samples in both disease and control groups (non-tumor liver samples).
Disease gene expression signatures
Disease gene expression signatures are the differentially expressed genes computed by comparing tumors and adjacent non-tumor tissues using DESeq (version one) 12. The default parameters were used to estimate size factors and dispersions. The signatures were refined and validated using external datasets (Supplementary Materials).
Drug gene expression profiles
Each drug gene expression profile is composed of a list of genes ranked by expression change after drug treatment in cells (by comparing with vehicle control). We used two related drug gene expression databases: Connectivity Map (CMap) 13 and LINCS L1000. CMap provides genome-wide expression profiles in three non-HCC cell lines, while LINCS L1000 provides expression profiles of 978 genes (called landmark genes, which are expected to capture about 80% of the information of the human transcriptome) in two HCC cell lines. In LINCS, the expression of remaining genes was imputed. Based on previous studies 14, 15, we decided to use just the 978 landmark genes in the prediction. The process of generating the expression profiles is described in detail in Supplementary Materials.
Matching HCC gene expression and drug gene expression to predict drug hits
The method of matching disease and drug gene expression was initially proposed by Lamb et al. 13 and then expanded by others 4, 6, 16. Briefly, a non-parametric, rank-based pattern-matching strategy based on the Kolmogorov–Smirnov (KS) statistic was used to assess the enrichment of disease genes in a ranked drug gene expression list. A lower negative score indicates a reverse relationship between the disease and the drug. Significance of the score was assessed by comparing with the scores generated from random permutations, and was further corrected by the multiple hypothesis test. False Discovery Rate (FDR) < 0.05 was used to select drug hits. Each chemical perturbation was considered individually.
In addition to the KS test, other methods could be used to assess the reversal relationship 17. Moreover, parameters could be tuned to optimize the models. To evaluate the models computationally, we used drugs investigated in HCC clinical trials as a gold standard. We measured performance based on significance of enrichment of HCC drugs in the predictions. We computed enrichment scores using ssGSEA implemented in the R package GSVA. The details of model evaluation are provided in the Supplementary Materials.
Culture of HCC cell lines and primary hepatocytes
Human HCC cell lines HepG2, Huh7, Hep3B, Hep40, and PLC/PRF/5 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 μg/ml penicillin, and 100 μg/ml streptomycin. SNU-398, SNU-449, SNU-182, SNU-475 and SNU-423 were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 μg/ml penicillin and streptomycin (Invitrogen, Carlsbad, CA). Cells were cultured at 37°C in a humidified atmosphere with 5% CO2. The Huh7 cell line was a gift from Dr. Mark Kay (Stanford University, CA), and the Hep40 cell line was a gift from Dr. Xin Chen (University of California, San Francisco, CA). All other cell lines were obtained from American Type Culture Collection (Manassas, VA).
Cryopreserved primary human hepatocytes and all special media and plates needed for their culture were from Invitrogen (Durham, NC). Thawing and culture of primary hepatocytes were described previously 18.
Establishment of orthotopic HCC PDX models and drug treatment
Under approved protocol from the Institutional Review Board at Stanford University, HCC tissues were collected from HCC patients who had undergone liver resection as part of their treatment. The PDX models were established as described previously, with tumor growth being monitored by bioluminescence imaging 10. Within a week of tumor implantation onto the liver, tumor-bearing mice were randomized into two groups (n = 5 or 6) to be fed with regular chow or food containing NEN (1,500 p.p.m).
For combination treatment, within a week of tumor implantation onto the liver, tumor-bearing mice (PDX-HCC-2) were randomized into six groups to be treated with sorafenib alone (50 mg/Kg); NEN (200 mg/Kg) or niclosamide (400 mg/Kg) alone; sorafenib (50 mg/Kg) combined with NEN (200 mg/Kg) or niclosamide (400 mg/Kg); or drinking water control. Drugs were given daily by oral gavage for four weeks.
Tumor growth was monitored weekly using the Xenogen IVIS in vivo imaging system, and growth curves were plotted using average bioluminescence within either group. After four–six weeks treatment, the mice were sacrificed and the livers and xenografts were harvested. Tumor size was measured with digital calipers and tumor volume was calculated using the formula π/6 × larger diameter × [smaller diameter]2. Body weight of mice was also monitored throughout the treatment period. The niclosamide levels in non-tumor livers and xenografts were measured by LC/MS/MS analysis as previously described 19 (Integrated Analytical Solutions, Berkeley, CA).
Statistics
Bioinformatics analysis of the pre-clinical data was carried out in the R (version 3.0.2) and statistical analysis of experimental results was performed in the Prism GraphPad software (version 7). No statistical method was used to predetermine sample size for all experiments (in vitro and in vivo). The investigators were not blinded to treatment allocation for the in vivo experiments. Unpaired Student’s t-test was used to compute statistical significance, and significance levels for p-values were set at 0.05 unless specified. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s.: not significant. Data are represented as mean ± SEM.
Data and code availability
The microarray dataset has been deposited in the NCBI Gene Expression Omnibus (GEO) at accession number GSE64836. The code and data necessary to correlate tumor samples and cell lines and predict drug hits are available at Synapse (syn6173892). The code is also available at https://github.com/Bin-Chen-Lab/HCC_NEN.
Results
Niclosamide was identified as the top drug candidate that significantly reversed the HCC gene expression signature
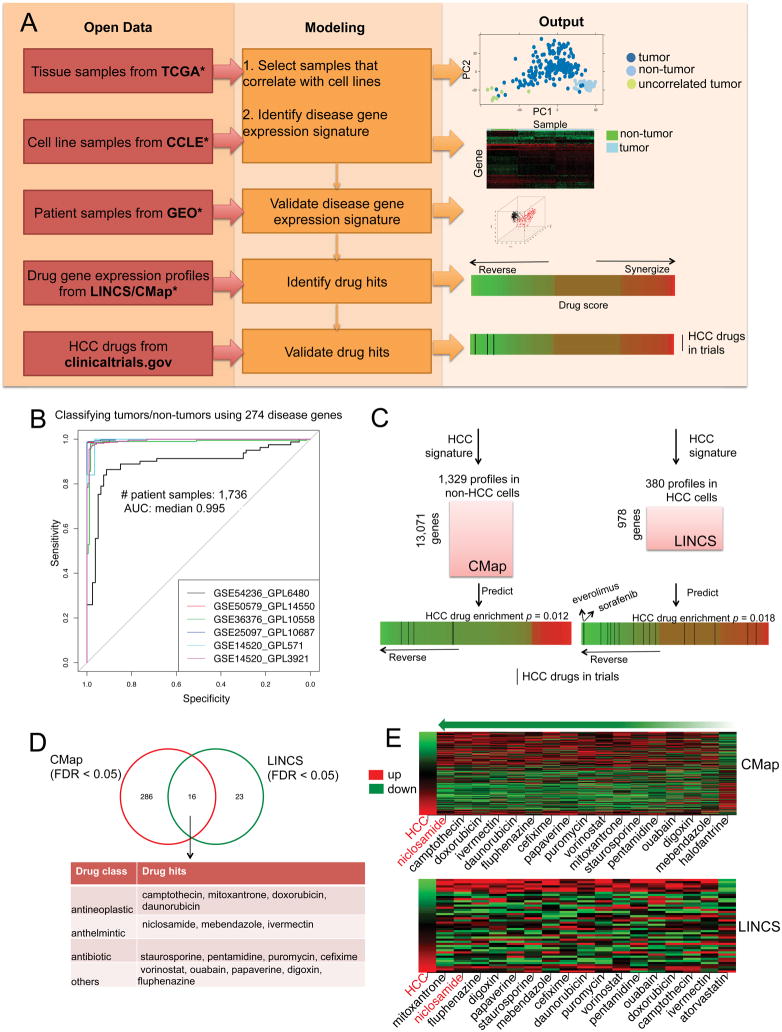
We built a rigorous drug-repositioning pipeline that leverages the new publicly available gene expression profiles (Figure 1A). We first downloaded RNA-Seq profiles of 200 HCC tumor samples and 50 adjacent non-tumor samples from TCGA. As we later use drug signatures profiled in cell lines to make drug predictions, and cell lines to validate drug hits, we chose tumor samples that correlate to cell lines. Among the 200 HCC tumor samples, we found 192 were significantly correlated with 25 HCC cell lines from CCLE by gene expression profiles (Supplemental Materials). We therefore used these 192 HCC tumor samples (and 50 adjacent non-tumor samples) to create a 274-gene expression signature (163 up- and 111 down-regulated genes, Supplemental Table 1), defined as significantly differentially expressed genes between HCC tumor and adjacent non-tumor samples. In order to evaluate the disease signature, we manually collected additional 1,736 patient samples from six studies downloaded from GEO (Supplemental Table 2). This 274-gene signature was sufficient to classify HCC tumor samples from non-tumor samples in external independent HCC datasets (Figure 1B; median AUC: 0.995), suggesting this set of genes constitutes a robust signature.
Figure 1. Integrative modeling of open data identifies repurposed drug candidates for HCC.
(A) Overview of the computational and validation pipeline. TCGA: The Cancer Genome Atlas, CCLE: Cancer Cell Line Encyclopedia, GEO: Gene Expression Omnibus, CMap: Connectivity Map, LINCS: Library of Integrated Network-based Cellular Signatures. (B) Validating the 274-disease gene signature using six external datasets. (C) Enrichment of HCC drugs in the predictions. Two drug gene expression databases CMap and LINCS were used. The significance of enrichment was computed based on the enrichment scores of permuted predictions (n = 10,000). (D) Common drug hits predicted from both CMap and LINCS. (E) Reversal relationship between the common drug hits and the disease in each drug gene expression database. The first column in the heat map represents HCC gene expression ranked by fold change and the remaining columns represent gene expression change after drug treatment in cancer cell lines. For those drugs with multiple profiles, the drug profile leading to the best score was selected for visualization. Red shows up-regulated genes and green shows down-regulated genes. The drugs are ranked based on their score at reversing disease gene expression. As a control, the most right column represents the drug that does not reverse disease gene expression. Niclosamide, the top ranked drug hit, is colored red.
The HCC gene expression signature was compared with individual drug gene expression profiles to identify drugs that likely reverse the HCC gene expression. Previous studies 4, 5, 13, 16 used the whole-genome drug expression profiles from CMap, but this original dataset did not have drugs tested in HCC cell lines. The more recent drug gene expression profile database LINCS includes two HCC cell lines (HepG2 and Huh7), which are significantly correlated to HCC tumor samples (Supplementary Figure 1), but only 978 genes were profiled (imputation of the expression of the remaining genes remains challenging) 15. To increase the predictive power, we decided to incorporate drug hits predicted from two databases. First, we collected 1,329 genome-wide expression profiles of 1,174 distinct drugs tested mainly in three cancer cell lines from CMap 13 after controlling for quality and restricting to FDA-approved drugs. We then collected 380 expression profiles of 249 distinct drugs tested in HCC cell lines from LINCS. To allow using the 978 landmark genes in LINCS, we additionally created a reduced 44-gene signature comprising 38 up- and 6-down regulated genes. This reduced HCC gene expression signature also significantly classified HCC tumor and non-tumor samples (Supplementary Figure 2).
To evaluate the model, we built a gold standard comprising 76 drugs from clinicaltrials.gov that have been investigated in HCC trials. Within the gold standard, seven and 16 drugs were included in CMap and LINCS respectively. Through measuring the enrichment of these drugs in the predictions, we systematically evaluated and optimized the model (Supplementary Text, Supplementary Table 3). The prediction from our model shows that HCC drugs are more likely to reverse HCC gene expression than random in both CMap (P = 0.012, permutation test, n = 10000) and LINCS (P = 0.018, permutation test, n = 10000) (Figure 1C). The standard of care treatment, sorafenib, was covered in the LINCS database, and our prediction indicates that sorafenib is more likely to reverse disease gene expression than other HCC drugs in LINCS (Figure 1C).
Next, we found that 302 drugs (from CMap) and 39 drugs (from LINCS) significantly reverse disease gene expression (FDR < 0.05, full list in Supplemental Table 4). Sixteen drugs were commonly predicted in CMap and LINCS. Among them were four antineoplastics, three anthelmintics, four antibiotics, and five in other drug classes (Figure 1D). Two drugs, vorinostat and doxorubicin, were previously investigated in HCC clinical trials, providing confidence to our predictions. Then we ranked the 16 drugs based on their scores in two databases. The top candidate was niclosamide (rank 1st in CMap and 2nd in LINCS), an FDA-approved drug used to treat tapeworm infection (Figure 1E). Niclosamide was reported to have anti-tumor activities in multiple cancers, including liver cancer 20, 21, 22. However, the effect of niclosamide in animal models of HCC have not been studied. Since niclosamide is our top hit from the unbiased virtual screening of two large drug libraries, and is predicted to reverse the HCC gene expression signature derived from clinical patient samples, we decided to validate its potential as an HCC drug candidate in advanced preclinical HCC models. It is also known that niclosamide is poorly soluble in water, with very limited systemic bioavailability 23, which may hamper clinical effectiveness if insufficient drug reaches the tumor site. Thus, we further evaluated NEN, an ethanolamine salt reported to have improved water solubility and systemic bioavailability over niclosamide 23.
Effect of niclosamide and NEN on a mouse primary HCC model
We initially confirmed that both niclosamide and NEN (chemical structures shown in Figure 2A) exert anti-proliferative effects in HCC cell lines, HepG2 and Huh7 (two HCC cell lines that closely correlate with HCC tumor samples) (Supplemental Figure 3A). Both compounds were also at least seven-fold more cytotoxic to HCC cells compared to primary hepatocytes obtained from donors without liver disease (Table 1, characteristics of primary hepatocytes in Supplemental Table 5). We next evaluated their anti-tumor effects in a primary HCC mouse model where the HCC was induced endogenously through hydrodynamic delivery of Sleeping Beauty transposon system with oncogene Ras (v12) and truncated β-catenin (N90). At the 2-week, 4-week, 12-week time points, mouse AFP levels were measured (Supplemental Table 6) and liver histology was studied (including H&E staining, AFP, and Ki67 immunostaining) (Supplemental Figure 4). Elevated serum AFP levels were observed within 2-weeks (after hydrodynamic injection) and histologic evidence of tumor within 4 weeks. Tumors were visible by AFP immunostaining and showed increased proliferative activity at 12 weeks compared to 4 weeks (by Ki-67 immunohistostaining).
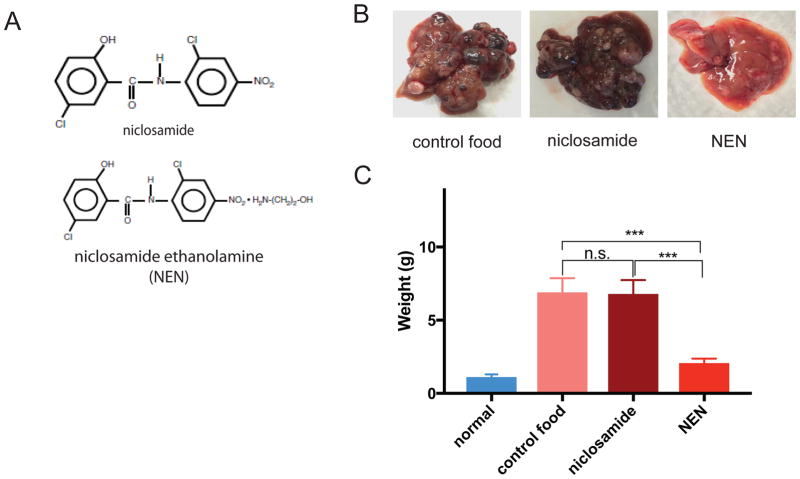
Figure 2. Effect of niclosamide and niclosamide ethanolamine (NEN) on primary HCC mouse model.
(A) Chemical structures of niclosamide and NEN. (B) Representative gross images of mouse liver with tumors after 12 weeks of treatment with regular chow, food containing niclosamide (1500 p.p.m), and food containing NEN (1500 p.p.m). (C) Weight (g) of mouse liver and tumors for normal mice (no tumor induced) and tumor-induced mice after treatment with control food, food containing niclosamide (1500 p.p.m), and food containing NEN (1500 p.p.m)(Student’s t test; ***P < 0.001, n.s., not significant; n = 6). Data are represented as mean ± SEM.
Table 1.
Effect of niclosamide and NEN on viability of HCC cell lines and primary hepatocytes.
| IC50 in HCC cell lines (μM) | IC50 in primary hepatocytes (μM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HepG2 | Huh7 | Hep3B | Hep40 | PLC/PRF/5 | SNU-398 | SNU-449 | SNU-182 | SNU-475 | SNU-423 | Hu8216 | Hu1748 | Hu1651 | |
| niclosamide | 0.85 | 0.74 | 0.20 | 0.81 | 0.77 | 0.24 | 0.67 | 0.36 | 0.40 | 0.32 | 4.31 | 3.64 | 3.58 |
| NEN | 0.46 | 0.63 | 0.17 | 0.74 | 0.53 | 0.20 | 0.53 | 0.34 | 0.35 | 0.26 | 4.07 | 3.58 | 2.66 |
Induced mice were treated with autoclaved food (control), or food containing niclosamide or NEN. NEN visibly reduced the number of tumor nodules in the livers of induced mice compared to those treated with control food, whereas niclosamide showed a much lesser effect than NEN (Figure 2B). When the whole livers (liver plus tumor nodules) were harvested and weighed at the end of the treatment period, the NEN-treated induced mice had significantly reduced liver weight compared to the weight of whole livers from both the control and niclosamide treated groups (Figure 2C). Our results suggest that NEN has superior anti-tumor effects compared to niclosamide in this primary HCC mouse model. (Figure 2B and 2C).
NEN inhibited growth of HCC PDXs
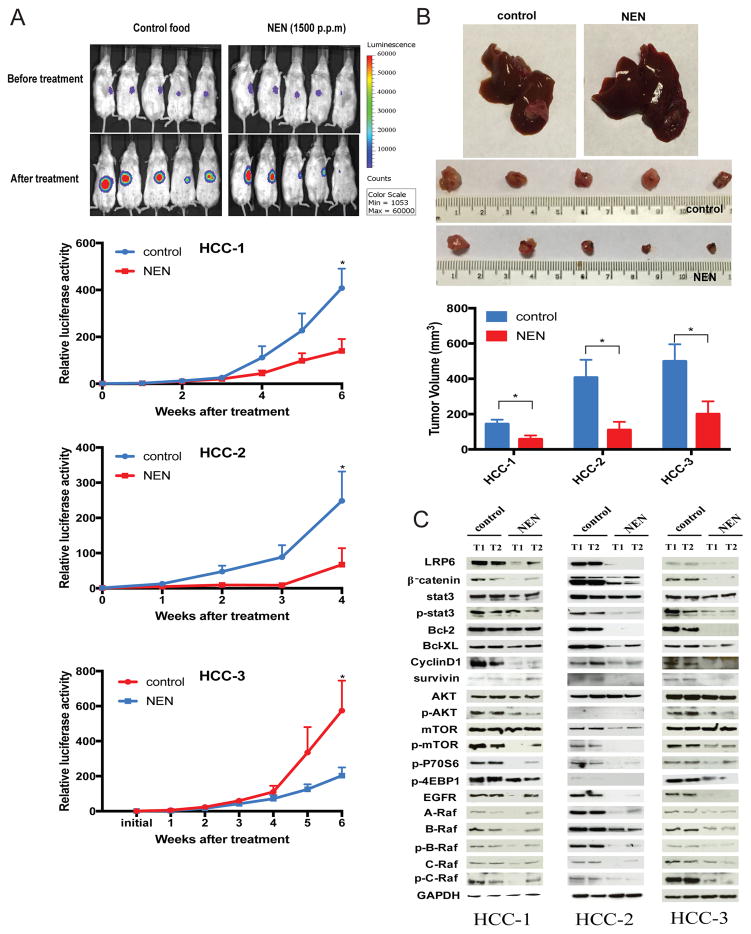
When NEN was administered orally (at 1,500 p.p.m mixed in autoclaved food) to mice bearing orthotopic PDX established from three HCC patients (patient characteristics shown in Supplemental Table 7), NEN significantly decreased PDX growth, based on bioluminescence signals (Figure 3A) and tumor volumes (Figure 3B), compared to tumor-bearing mice fed regular chow. At the end of the treatment period, no significant body weight changes were observed between treated and control groups (Supplemental Table 8). The decrease in tumor volumes induced by NEN treatment was associated with decreased expression levels of proteins involved in multiple signaling pathways, including Wnt/β-catenin, Stat3, AKT/mTOR, and EGFR/Ras/Raf (Figure 3C), similar to what we observed in HepG2 and Huh7 cells treated with NEN (Supplemental Figure 3B–E).
Figure 3. Effect of NEN on PDX models.
(A) Representative bioluminescence images of mice bearing PDX from HCC patient (HCC-1), before and after treatment with regular chow and food containing NEN (1500 p.p.m). Growth curves of tumors based on bioluminescence signals for all three PDX models (HCC-1, HCC-2, and HCC-3) throughout the treatment period are also shown (Student’s t test, *P < 0.05, n = 5 for treatment with NEN or control food). Data are represented as mean ± SEM. (B) Representative images of gross PDX tumor (from HCC-1) on mouse liver, and gross images of tumors harvested from livers at the end of the treatment period for HCC-1. Lower panel graph depicts tumor volumes of all PDX models (HCC-1, HCC-2, and HCC-3) at the end of their treatment periods (Student’s t test, *P < 0.05, n = 5 for treatment with NEN or control food). Data are represented as mean ± SEM. (C) Critical proteins involved in Wnt/β-catenin, STAT3, AKT/mTOR, and ERK/Ras/Raf pathways in three PDX models, detected by Western blotting. GAPDH was used as the loading control.
As in the primary HCC mouse model, niclosamide given at the same dose did not produce any significant effects on tumor growth (Figure 2B and 2C). At the end of the treatment period for PDX model, xenografts and non-tumor liver tissues were harvested and levels of niclosamide were detected and measured by LC/MS/MS analysis. We observed that niclosamide levels were over 15 fold higher in xenografts following administration with NEN than with niclosamide (Supplemental Figure 5A), which may explain the observed superior anti-tumor effect of NEN compared to niclosamide. In a separate experiment where NEN or niclosamide was given as a single oral dose (40 mg/Kg) to mice bearing PDX, we also observed that niclosamide levels in xenograft and non-tumor liver tissues were higher following administration with NEN than with niclosamide (Supplemental Figure 5A). Plasma levels were similarly higher after NEN was given (Supplemental Figure 5B), confirming greater bioavailability of NEN compared to niclosamide.
NEN in combination with sorafenib inhibited growth of HCC PDXs
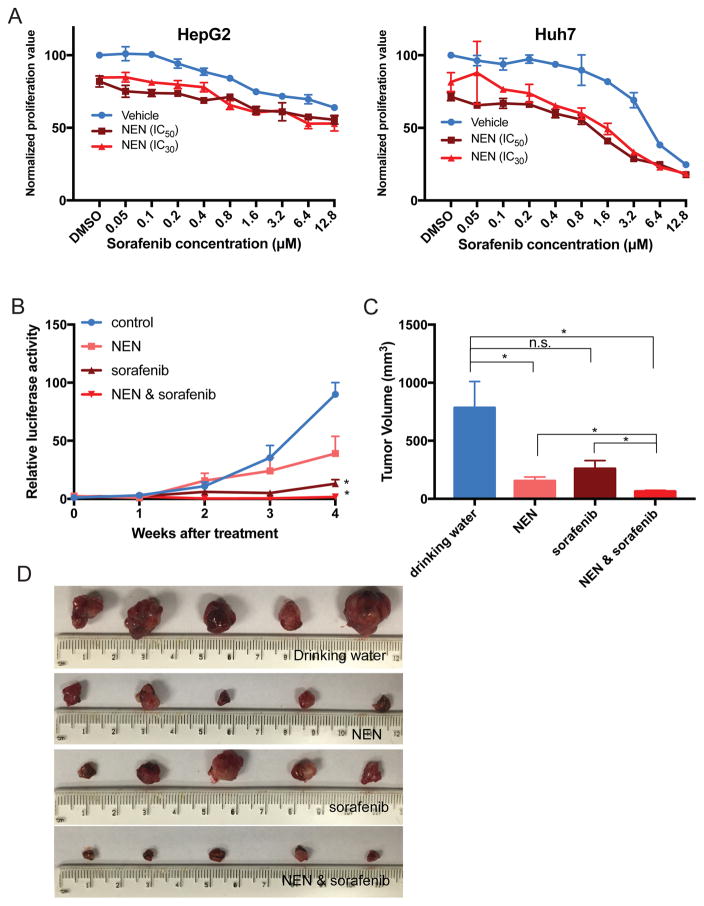
To determine whether NEN may be useful in combination therapy with sorafenib 3, we tested combination treatment of NEN with sorafenib in vitro and in vivo. In vitro, the addition of NEN (at its IC30 or IC50) to HCC cells treated over a range of sorafenib concentrations reduced their IC50 values to sorafenib (Figure 4A, Supplemental Table 9). Consistently, in our PDX models, we observed that NEN combined with sorafenib significantly decreased PDX growth based on bioluminescence signals (Figure 4B). The combination of NEN and sorafenib also caused significant reductions in tumor volumes compared to the drinking water group (P = 0.013), NEN alone (P = 0.030), or sorafenib alone (P = 0.024) (Figure 4C and 4D).
Figure 4. Effect of NEN in combination with sorafenib in PDX model.
(A) In vitro anti-proliferation assay of NEN combined with sorafenib. HepG2 and Huh7 cells were treated with a range of concentrations of sorafenib combined with NEN (at its IC30 or IC50, with DMSO as control). For HepG2, IC50 is 0.46 μM and IC30 is 0.4 μM; for Huh7, IC50 is 0.63 μM and IC30 is 0.5 μM. (B) Growth curves of tumors based on bioluminescence signals for PDX (HCC-2) throughout the combination treatment period (Student’s t test; *P < 0.05, n.s., not significant; n=5 for treatment with NEN alone, sorafenib alone, or combination). Data are represented as mean ± SEM. (C) Tumor volumes of PDX model at the end of their treatment periods (Student’s t test; *P < 0.05; n = 5 for treatment with NEN alone, sorafenib alone, or combination). Data are represented as mean ± SEM. (D) Images of tumors harvested from livers at the end of the treatment period.
In comparison, niclosamide showed similar sensitizing effects to sorafenib in both cell lines (Supplemental Figure 6A and Supplemental Table 9). However, treatment of mice bearing PDX with niclosamide alone or combined with sorafenib did not decrease tumor volume (Supplemental Figur 6, B–D). The greater bioavailability of NEN ompared to niclosamide (Supplemental Figure 4) may explain the differential activities of NEN and niclosamide in vivo. Thus, NEN is a preferred therapeutic candidate for treatment of HCC.
NEN reversed the HCC gene expression signature in vitro and in vivo
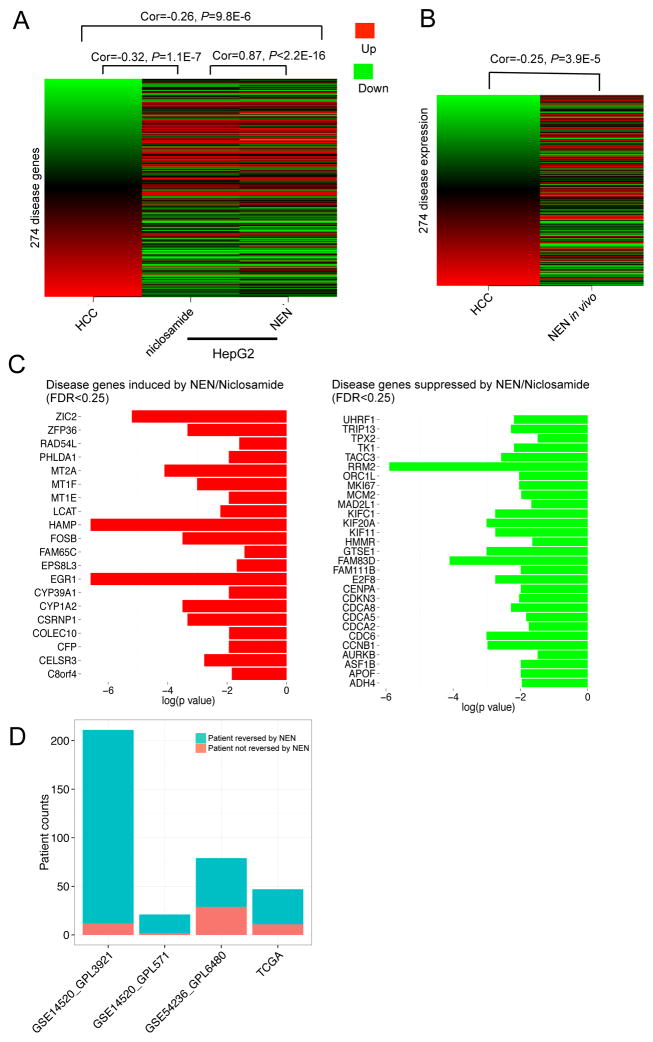
We next validated whether niclosamide and NEN can both reverse global HCC gene expression as predicted from our initial computational methods, under our experimental conditions. First, HepG2 cells were treated for 6 h with 10 μM of NEN, 10 μM of niclosamide, or DMSO as control. When compared to DMSO control, we observed that both compounds induced similar expression changes (Spearman correlation of 0.87 between the niclosamide signature and the NEN signature, P < 2.2×10−16; Figure 5A), and both compounds significantly reversed the 274-gene HCC signature (niclosamide: Spearman correlation of −0.32 between the HCC signature and the niclosamide signature, P = 1.1×10−7, KS test: P < 1×10−4; NEN: Spearman correlation of −0.26 between the HCC signature and the NEN signature, P = 9.8×10−6, KS test: P < 1×10−4) (Figure 5A). In vivo, we observed that the gene expression changes induced by NEN in the PDX model (when compared to the group given regular chow) were significantly anti-correlated with the 274-gene HCC signature (Spearman correlation of −0.25 between the HCC signature and the NEN signature, P = 3.9×10−5; KS test: P < 1×10−4; Figure 5B), again indicating that NEN can reverse the HCC gene expression signature, as predicted.
Figure 5. Reverse correlation between HCC gene expression and niclosamide/NEN-mediated expression.
HepG2 cells were harvested after 6 h treatment with 10 μM NEN, 10 μM niclosamide, or 0.1% DMSO as vehicle control. Xenograft cells were harvested after 6 weeks treatment with NEN (1,500 p.p.m). Cells were profiled using Illumina HumanHT-12 v4 Expression BeadChip. Drug-mediated gene expression changes were analyzed by comparing the profiles of a treatment group with a control group. (A) Correlation between HCC gene expression and niclosamide/NEN-mediated expression in HepG2 cells. The first column represents HCC gene expression ranked by fold change. The remaining columns represent gene expression changes of the disease genes after drug treatment. (B) Correlation between HCC gene expression and NEN-mediated expression in vivo. (C) HCC genes reversed by niclosamide and NEN. (D) Correlation between gene expression of individual HCC patients (from four public datasets) and NEN-mediated expression in vivo. A negative correlation (Spearman correlation coefficient < 0, adjusted P value < 0.01) indicates that the patient’s gene expression can be reversed by NEN.
To further understand the mechanisms of action of niclosamide and NEN in HCC, we analyzed HCC genes that were reversed by both compounds in HepG2 cells and in one PDX model. We observed that both compounds induced 20 genes and suppressed 29 genes within the 274-gene HCC signature, in vitro and in vivo (FDR< 0.25, permutation test; Figure 5C). Additional gene set enrichment analysis of these reversed genes indicated that cell cycle pathways, which are up regulated in HCC tumors (Supplemental Figure 7), are suppressed by both compounds (Supplemental Table 10). Apart from genes within the HCC gene expression signature, we also observed that both compounds inhibited respiratory electron transport in the mitochondria (Supplemental Table 11), consistent with one known function of niclosamide as a mitochondrial uncoupler, and its reported ability to reduce intracellular ATP concentrations in HepG2 cells 23.
To estimate the potential benefits of NEN to HCC patients in the clinic, we next analyzed whether NEN could reverse HCC gene expression in individual HCC patient samples. Using gene expression profiles of 358 patient tumors (compared with matched adjacent non-tumor liver) from three sources (TCGA, GSE14520, GSE54236) and the NEN-induced gene expression profile in the clinically relevant PDX model, we found that NEN was likely to reverse the 274-gene HCC signature in 317 patient profiles (88.5%; Spearman correlation < 0 and adjusted P value < 0.01; Figure 5D). Our results suggest that NEN treatment in vitro and in vivo significantly reversed the HCC gene expression signature, and is likely to reverse HCC gene expression in the majority of HCC patients, providing rationale for its use as a potential treatment option for HCC.
NEN binds to CDC37 and disrupts the CDC37/HSP90 interaction
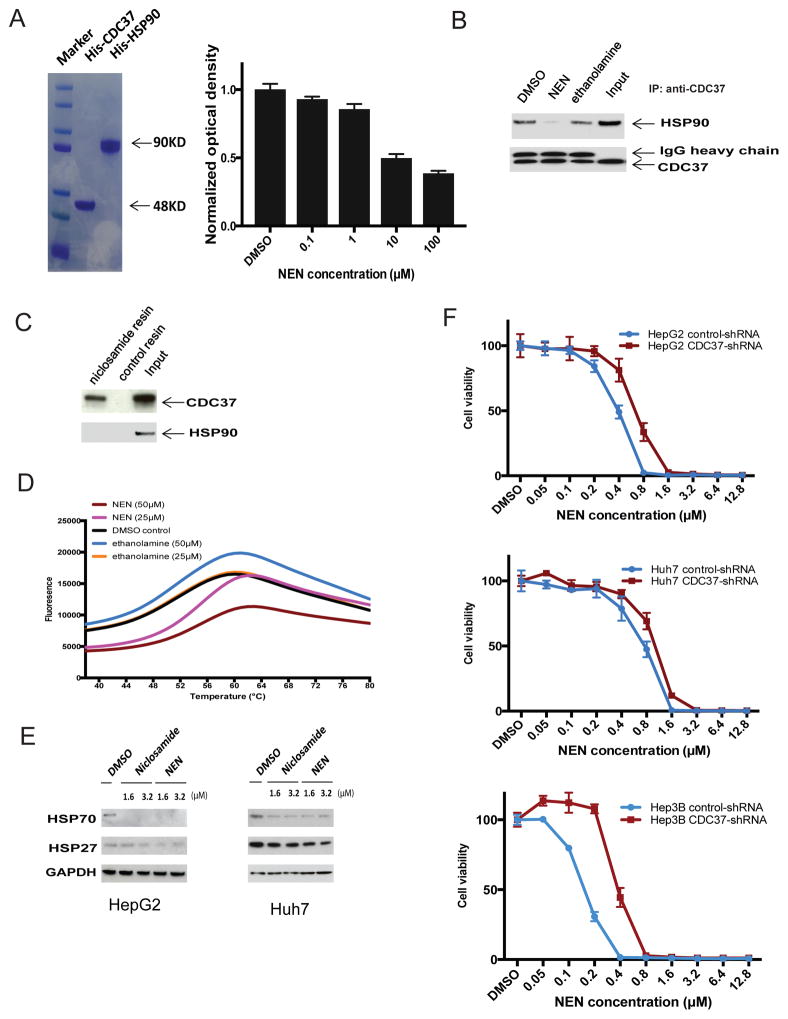
The Heat Shock Protein 90 (HSP90) and its co-chaperone CDC37 regulate the activity of multiple kinases that are inhibited by NEN (e.g., AKT 24, EGFR 25, Raf family 26, STAT3 27, LRP6 28). Using purified recombinant HSP90 and CDC37 in ELISA and co-immunoprecipitation assays, we found that NEN inhibited the HSP90/CDC37 interaction (Figure 6A and 6B). We next used a pull-down assay, in which niclosamide was conjugated to epoxy-activated sepharose beads, to determine whether NEN binds to HSP90 or CDC37. We found that niclosamide pulled down CDC37 but not HSP90 (Figure 6C). Thermal shift assay additionally suggested that NEN bound to and enhanced the thermal stability of CDC37 (Figure 6D). Furthermore, NEN did not induce the classical heat shock response of existing HSP90 inhibitors (Figure 6E), in which activation of the heat shock transcription factor HSF1 further induces expression of HSP70 and HSP27 to protect tumor cells from apoptosis (which limits their clinical use 29).
Figure 6. NEN binds to CDC37 and disrupts the HSP90/CDC37 interaction.
(A). SDS-PAGE analysis of purified His-CDC37 and His-HSP90 and effect of NEN on HSP90/CDC37 complex analyzed by ELISA assay. CDC37 recombinant protein or PBS (background) was coated onto ELISA plates, and biotin-HSP90 solution containing serial dilution of NEN was incubated after blocking with 1% BSA. Bound biotin-HSP90 was then detected by adding streptavidin-HRP and TMB substrate. The yellow end product was read at OD450. Each experiment was done in triplicate, and absorbance was calculated as relative value to the background. (B). Effect of NEN on HSP90/CDC37 complex analyzed by co-IP assay. Rabbit anti-CDC37 antibody was used for immunoprecipitation and both rabbit anti-CDC37 antibody and mouse anti-HSP90 antibody were used for immunoblotting. (C). CDC37-niclosamide interaction analyzed by pull-down assay. (D). CDC37-NEN binding analyzed by Thermal Shift Assay. Representative melting curves were shown for DMSO control, NEN, and ethanolamine. The respective average Tm value is 56.03 °C for 50 μM NEN, 54.97 °C for 25 μM NEN, 53.2 °C for 50 μM ethanolamine, 52.93 °C for 25 μM ethanolamine, and 52.3 °C for DMSO control. (E). Western blot analysis for HSP70 and HSP27 expression in NEN treated cell lines. (F). Effect of NEN on cell viability of control shRNA and CDC37 shRNA stable transfected HepG2, Huh7, and Hep3B cell lines. NEN was added at desired final concentrations and incubated for 6 days before cell viability was assessed using CellTiter-Glo Luminescent Cell Viability Assay.
CDC37 is over-expressed in 80% of HCC tissues compared to normal livers, and most HCC cell lines over-express CDC37 30. Besides, the enforced over-expression of CDC37 alone in normal cells did not promote tumorigenesis or cell proliferation 31, 32. We therefore used RNA interference to study the impact of stable CDC37 knockdown on the anti-proliferative effects of NEN in HCC cells 30. In all three HCC cell lines tested, the IC50 of NEN increased in the CDC37 knockdown counterparts compared to their mock transfected parental cell lines (from 0.35 μM to 0.61 μM for HepG2; from 0.61 μM compared to 0.93 μM for Huh7; and from 0.16 μM to 0.52 μM for Hep3B) after 6 days of treatment (Figure 6F). Cells lacking CDC37 expression were less sensitive to NEN, suggesting that its anti-tumor effects are at least in part dependent on CDC37. Our results suggest that NEN binds to CDC37 instead of HSP90, and disrupts CDC37/HSP90 interaction leading to degradation of an array of protein kinases in multiple signaling pathways critical to HCC development.
Discussion
Recent studies have mainly focused on targeting individual alterations derived from specific molecular features 33. As none of the molecularly targeted drugs for HCC have been shown to be superior to sorafenib, we sought to examine the effects of drugs on a spectrum of differentially expressed genes between tumors and non-tumors. The central hypothesis of this method is that drugs that can reverse a disease gene expression signature can be potential drug candidates for that disease. Several studies have highlighted the value of using similar computational approaches to identify new drug candidates for a variety of diseases 4–6, 34. We additionally attempted to increase predictive power by relating different models (e.g., patient tissues, cell lines) based on their molecular features, and to leverage LINCS L1000, a newer and much larger drug library. We systematically evaluated and optimized our model using a gold standard comprising drugs being used in HCC trials. Afterwards, we used the model to computationally screen FDA-approved drugs and observed an enrichment of three anthelmintic drugs that were predicted to reverse HCC gene expression.
To increase the chances of successful clinical translation, we used clinically relevant HCC cell lines, primary HCC mouse model, and orthotopic PDX models to validate our predictions. Previous studies by our group and others revealed that some cancer cell lines may not reflect the real biology of tumors, largely due to contamination, loss of origin, loss of microenvironment, tumor impurity, and tumor heterogeneity 35–37. For our validation study, we selected two HCC cell lines, HepG2 and Huh7, whose gene expression signatures are highly correlated with the gene expression of HCC tumor samples from the TCGA and GEO. Among the three anthelmintic drugs, niclosamide and mebendazole were effective at inhibiting the proliferation of HepG2 and Huh7 cells in vitro (niclosamide IC50: 0.65–1.77 μM; mebendazole IC50: 0.38–1.65 μM,), whereas ivermectin was least effective (IC50: > 10 μM) (Supplemental Figure 8). Combining this with our in silico validation using known HCC drugs as gold standards, we demonstrate the feasibility of using our computational approach to find drug candidates.
Due to the poor water solubility and limited systemic bioavailability of niclosamide 20, we decided to intensively evaluate NEN, a salt with improved water solubility, favorable systemic bioavailability, and a similar safety profile compared to niclosamide 23, 38. In vitro, we observed that NEN inhibited HCC cell proliferation at slightly lower IC50s than niclosamide, suggesting greater potency. Both compounds were also demonstrated to have preferential cytotoxicity to HCC cells compared to primary hepatocytes, suggesting a potentially favorable therapeutic window. Using a dose of NEN that was previously reported to have no adverse effect in experimental mice 23, we observed that only NEN caused significant growth retardation in a primary HCC model and all three PDX models (derived from three HCC patients), whereas niclosamide (at the same dose) had no significant effect on tumor growth. This could be explained by the higher drug levels achieved in the xenograft tissues when NEN was given, and underscores the poor bioavailability of niclosamide which may hamper its clinical efficacy. All three HCC patients (from whom the PDX were derived) had undergone surgical resection and post-surgery transarterial chemoembolization (Supplemental Table 7). Whereas HCC-3 was recurrence-free for six years post-treatment, HCC-1 and HCC-2 had tumor recurrence despite standard treatment. Of note, NEN was shown to be effective against all three PDX models, including two for which surgery and chemoembolization were not curative.
Additionally, NEN significantly reversed the HCC gene expression signature in vitro and in vivo, consistent with our computational prediction. Genes that were significantly reversed by NEN (and niclosamide) include those involved in the cell cycle, which are highly expressed and frequently altered in HCC 39. Mechanistically, we found that both niclosamide and NEN decreased expression levels of critical HCC proteins in the Wnt/β-catenin, STAT3, and AKT/mTOR pathways 40–42, consistent with previous reports 20.. We additionally observed that these compounds also disrupt the EGFR/Ras/Raf pathway, which is also fundamental to HCC development 43. Since all these protein kinases are regulated by HSP90, we studied whether NEN can inhibit HSP90 function. Unlike existing HSP90 inhibitors, which induce the undesirable anti-apoptotic heat shock response 29, NEN binds to CDC37 instead of HSP90, therefore obviating the heat shock response 44. In addition, since CDC37 is specifically over expressed in HCC 30, 45, the discovery of new CDC37 inhibitors holds promise to replace existing HSP90 inhibitors, which have not proven to be effective in HCC 46. Therapeutic targeting of CDC37 in tumor cells was shown to inhibit tumor cell proliferation by suppressing the function of most protein kinases, which are over-expressed and activated in HCC cells 10. Additionally, the recent emergence of resistance towards sorafenib was attributed to multiple signaling pathways, including Wnt/β-catenin 47, AKT/mTOR 48, and EGFR mediated signaling pathways 49. Thus, by simultaneously inhibiting these pathways, NEN may also help to overcome acquired resistance to sorafenib.
In summary, our computational modeling on open data led to the discovery of NEN as a promising clinical candidate for HCC. By combining computational approaches and in vitro and in vivo validations, we provide confidence for the utility and generalizability of our computational approach to identify potential drug candidates in other diseases.
Supplementary Material
Acknowledgments
Grants
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM079719.
We would like to thank Dr. B. Oskotsky for IT support and thank Drs. Kelly Zalocusky, Jieming Chen, Uta Grieshammer and Marina Sirota for their critical comments.
Abbreviations used in this paper
- CCLE
Cancer Cell Line Encyclopedia
- CMap
Connectivity Map
- FDR
False Discovery Rate
- GEO
Gene Expression Omnibus
- HCC
Hepatocellular carcinoma
- HSP90
Heat Shock Protein 90
- KS
Kolmogorov–Smirnov
- LINCS
Library of Integrated Network-Based Cellular Signatures
- NEN
Niclosamide Ethanolamine
- PDX
Patient-Derived Xenografts
- TCGA
The Cancer Genome Atlas
Footnotes
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Transcript Profiling
GSE77322 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ijcfcswivzujloj&acc=GSE77322)
Author contributions
B.C. conceived the study, performed bioinformatics analysis, analyzed and interpreted the data, coordinated the work and wrote the manuscript. W.W. conceived the study, performed the majority of the experiments, analyzed and interpreted the data, and wrote the manuscript. L.M. provided technical assistance with animal experiments and in vitro validation. B.Y. provided technical assistance with animal experiments and in vitro validation. R.M.G. provided assistance with histology analysis and reviewed the manuscript. M.S.C. conceived the study, wrote the manuscript and supervised the study. A.J.B. and S.S. supervised the study, edited the manuscript, and provided overall research direction.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–26. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol Res. 2013;43:147–54. doi: 10.1111/j.1872-034X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahchan NS, Dudley JT, Mazur PK, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–77. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Noort V, Scholch S, Iskar M, et al. Novel drug candidates for the treatment of metastatic colorectal cancer through global inverse gene-expression profiling. Cancer Res. 2014;74:5690–9. doi: 10.1158/0008-5472.CAN-13-3540. [DOI] [PubMed] [Google Scholar]
- 7.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W, Wu S, Wang X, et al. Novel celastrol derivatives inhibit the growth of hepatocellular carcinoma patient-derived xenografts. Oncotarget. 2014;5:5819–31. doi: 10.18632/oncotarget.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SA, Ho C, Roy R, et al. Integration of genomic analysis and in vivo transfection to identify sprouty 2 as a candidate tumor suppressor in liver cancer. Hepatology. 2008;47:1200–10. doi: 10.1002/hep.22169. [DOI] [PubMed] [Google Scholar]
- 12.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 14.Vidovic D, Koleti A, Schurer SC. Large-scale integration of small molecule-induced genome-wide transcriptional responses, Kinome-wide binding affinities and cell-growth inhibition profiles reveal global trends characterizing systems-level drug action. Front Genet. 2014;5:342. doi: 10.3389/fgene.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, Greenside P, Paik H, et al. Relating Chemical Structure to Cellular Response: An Integrative Analysis of Gene Expression, Bioactivity, and 26 Structural Data Across 11,000 Compounds. CPT Pharmacometrics Syst Pharmacol. 2015;4:576–84. doi: 10.1002/psp4.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng J, Xie Q, Kumar V, et al. Evaluation of analytical methods for connectivity map data. Pac Symp Biocomput. 2013:5–16. [PubMed] [Google Scholar]
- 18.Wei W, Chua MS, Grepper S, et al. Small molecule antagonists of Tcf4/beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int J Cancer. 2010;126:2426–36. doi: 10.1002/ijc.24810. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y-WY, Teng-Kuang, Lin Ke-Ta, Chen Wei-Cheng, Yao Hsien-Tsung, Lan Shih-Jung, Wu Yu-Shan, Hsieh Hsing-Pang, Chen Chi-Min, Chen Chiung-Tong. Pharmacokinetics of anti–SARS-CoV agent niclosamide and its analogs in rats. Journal of Food and Drug Analysis. 2006:329–333. [Google Scholar]
- 20.Li Y, Li PK, Roberts MJ, et al. Multi-targeted therapy of cancer by niclosamide: A new application for an old drug. Cancer Lett. 2014;349:8–14. doi: 10.1016/j.canlet.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomizawa M, Shinozaki F, Motoyoshi Y, et al. Niclosamide suppresses Hepatoma cell proliferation via the Wnt pathway. Onco Targets Ther. 2013;6:1685–93. doi: 10.2147/OTT.S50065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng S, Zhou L, Deng Q, et al. Niclosamide induced cell apoptosis via upregulation of ATF3 and activation of PERK in Hepatocellular carcinoma cells. BMC Gastroenterol. 2016;16:25. doi: 10.1186/s12876-016-0442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao H, Zhang Y, Zeng X, et al. Niclosamide ethanolamine-induced mild mitochondrial uncoupling improves diabetic symptoms in mice. Nat Med. 2014;20:1263–9. doi: 10.1038/nm.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basso AD, Solit DB, Chiosis G, et al. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–66. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 25.Lavictoire SJ, Parolin DA, Klimowicz AC, et al. Interaction of Hsp90 with the nascent form of the mutant epidermal growth factor receptor EGFRvIII. J Biol Chem. 2003;278:5292–9. doi: 10.1074/jbc.M209494200. [DOI] [PubMed] [Google Scholar]
- 26.Silverstein AM, Grammatikakis N, Cochran BH, et al. p50(cdc37) binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J Biol Chem. 1998;273:20090–5. doi: 10.1074/jbc.273.32.20090. [DOI] [PubMed] [Google Scholar]
- 27.Kim HL, Cassone M, Otvos L, Jr, et al. HIF-1alpha and STAT3 client proteins interacting with the cancer chaperone Hsp90: therapeutic considerations. Cancer Biol Ther. 2008;7:10–4. doi: 10.4161/cbt.7.1.5458. [DOI] [PubMed] [Google Scholar]
- 28.Ansa-Addo EA, Thaxton J, Hong F, et al. Clients and Oncogenic Roles of Molecular Chaperone gp96/grp94. Curr Top Med Chem. 2016;16:2765–78. doi: 10.2174/1568026616666160413141613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trepel J, Mollapour M, Giaccone G, et al. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Wei W, Sun CK, et al. Suppressing the CDC37 cochaperone in hepatocellular carcinoma cells inhibits cell cycle progression and cell growth. Liver Int. 2015;35:1403–15. doi: 10.1111/liv.12651. [DOI] [PubMed] [Google Scholar]
- 31.Stepanova L, Finegold M, DeMayo F, et al. The oncoprotein kinase chaperone CDC37 functions as an oncogene in mice and collaborates with both c-myc and cyclin D1 in transformation of multiple tissues. Mol Cell Biol. 2000;20:4462–73. doi: 10.1128/mcb.20.12.4462-4473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepanova L, Yang G, DeMayo F, et al. Induction of human Cdc37 in prostate cancer correlates with the ability of targeted Cdc37 expression to promote prostatic hyperplasia. Oncogene. 2000;19:2186–93. doi: 10.1038/sj.onc.1203561. [DOI] [PubMed] [Google Scholar]
- 33.Rubio-Perez C, Tamborero D, Schroeder MP, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27:382–96. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Chen MH, Yang WL, Lin KT, et al. Gene expression-based chemical genomics identifies potential therapeutic drugs in hepatocellular carcinoma. PLoS One. 2011;6:e27186. doi: 10.1371/journal.pone.0027186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–84. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
- 36.Gazdar AF, Girard L, Lockwood WW, et al. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–21. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen B, Sirota M, Fan-Minogue H, et al. Relating hepatocellular carcinoma tumor samples and cell lines using gene expression data in translational research. BMC Med Genomics. 2015;8(Suppl 2):S5. doi: 10.1186/1755-8794-8-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews P, Thyssen J, Lorke D. The biology and toxicology of molluscicides, Bayluscide. Pharmacol Ther. 1982;19:245–95. doi: 10.1016/0163-7258(82)90064-x. [DOI] [PubMed] [Google Scholar]
- 39.Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016 doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 40.Pez F, Lopez A, Kim M, et al. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59:1107–17. doi: 10.1016/j.jhep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 41.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–68. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matter MS, Decaens T, Andersen JB, et al. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855–65. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito Y, Sasaki Y, Horimoto M, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951–8. doi: 10.1002/hep.510270409. [DOI] [PubMed] [Google Scholar]
- 44.Smith JR, Clarke PA, de Billy E, et al. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene. 2009;28:157–69. doi: 10.1038/onc.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascale RM, Simile MM, Calvisi DF, et al. Role of HSP90, CDC37, and CRM1 as modulators of P16(INK4A) activity in rat liver carcinogenesis and human liver cancer. Hepatology. 2005;42:1310–9. doi: 10.1002/hep.20962. [DOI] [PubMed] [Google Scholar]
- 46.Goyal L, Wadlow RC, Blaszkowsky LS, et al. A phase I and pharmacokinetic study of ganetespib (STA-9090) in advanced hepatocellular carcinoma. Invest New Drugs. 2015;33:128–37. doi: 10.1007/s10637-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Ye X, Zhang JB, et al. PROX1 promotes hepatocellular carcinoma proliferation and sorafenib resistance by enhancing beta-catenin expression and nuclear translocation. Oncogene. 2015;34:5524–35. doi: 10.1038/onc.2015.7. [DOI] [PubMed] [Google Scholar]
- 48.Zhai B, Hu F, Jiang X, et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol Cancer Ther. 2014;13:1589–98. doi: 10.1158/1535-7163.MCT-13-1043. [DOI] [PubMed] [Google Scholar]
- 49.Blivet-Van Eggelpoel MJ, Chettouh H, Fartoux L, et al. Epidermal growth factor receptor and HER-3 restrict cell response to sorafenib in hepatocellular carcinoma cells. J Hepatol. 2012;57:108–15. doi: 10.1016/j.jhep.2012.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray dataset has been deposited in the NCBI Gene Expression Omnibus (GEO) at accession number GSE64836. The code and data necessary to correlate tumor samples and cell lines and predict drug hits are available at Synapse (syn6173892). The code is also available at https://github.com/Bin-Chen-Lab/HCC_NEN.