Abstract
Polycomb Repressive Complex 2 (PRC2) is a multiprotein complex that catalyzes the methylation of lysine 27 on histone H3 (H3K27me). This histone modification is a feature of facultative heterochromatin in many eukaryotes and maintains transcriptional repression established during early development. Understanding how PRC2 targets regions of the genome to be methylated remains poorly understood. Different cell types can show disparate patterns of H3K27me, and chromatin perturbations, such as loss of marks of constitutive heterochromatin, can cause redistribution of H3K27me, implying that DNA sequence, per se, is not sufficient to define the distribution of this mark. Emerging information supports the idea that the chromatin context – including histone modifications, DNA methylation, transcription, chromatin structure and organization within the nucleus – informs PRC2 target selection.
Introduction
Methylation of lysine 27 on histone H3 (H3K27me) by Polycomb Repressive Complex 2 (PRC2) is a hallmark of facultative heterochromatin in numerous organisms but, despite decades of research, it is not yet completely clear how this chromatin mark functions in gene repression and how it is controlled. Work in Drosophila first identified and characterized the Polycomb group (PcG) protein complexes as writers and readers of H3K27me and demonstrated that it maintains repression established early in development [1–3]. The SET domain of the PRC2 component EZH2 catalyzes H3K27 di-methylation and tri-methylation (H3K27me2/3) and requires both EED and SUZ12 to perform this function; these proteins are the core components of PRC2. H3K27me3 can be recognized by a chromodomain protein in the canonical Polycomb Repressive Complex 1 (PRC1) [4–6]. PRC1 is thought to help mediate transcriptional repression via H2AK119 monoubiquitylation (H2Aub1) and chromatin compaction [7–9].
PRC2 exists in some single celled eukaryotes, many fungi (although neither S. cerevisae nor S. pombe), plants and metazoans [10]. While plants contain a PRC1-like complex [11], true PRC1 homologs appear to be limited to metazoans [10]. The high degree of evolutionary conservation speaks to a critical role for PcG complexes; however, differences among species provide clues to general mechanisms of control and function of H3K27me. One major difference among organisms is the role of PRC1 in repression.
That H3K27me serves as a repressive mark even in organisms lacking PRC1 [12], implicates alternative repressive mechanisms downstream of H3K27me. This idea is also supported by findings showing that PRC1 does not localize to all H3K27me marked repressed targets [13] and that its enzymatic activity is not required for all repression [14]. Another intriguing difference among species is in the role of PRC2 in development. In some fungi such as Neurospora crassa and Cryptococcus neoformans, PRC2 and H3K27me are dispensable for normal growth and development [12,15]. Drosophila and higher eukaryotes are sensitive to disruption of PRC2 function, perhaps because lineage specification programs depend on the Polycomb system [16–20].
PcG proteins maintain gene silencing that is established during early development and is required for appropriate cell fate specification. The significance of H3K27me in maintaining appropriate long-term gene expression patterns is demonstrated by the range of mutations in PRC2 complex members, and its substrate (H3K27), in cancers. Both loss of function and change of function mutations in PRC2 have been reported in cancers, but a common outcome is an altered distribution of H3K27me, which perturbs differentiation [21]. Similarly, a heterozygous histone H3K27M mutation in only one of multiple genes encoding the histone H3 protein leads to glioblastoma [22]. H3K27M acts as a partially dominant negative mutation by binding to PRC2 with 22-fold higher affinity than to H3K27 [23]. This results in an overall reduction, but not complete absence, of H3K27me.
Despite the critical role of PRC2 in development and disease, some fundamental questions remain. The mechanisms controlling placement of H3K27me by PRC2, which will be the focus of this review, remain unclear. Direct targeting mechanisms have been proposed but likely only account for PRC2 recruitment in certain organisms or to certain loci. Partially defined DNA sequences called Polycomb Response Elements (PREs) recruit PRC2 in Drosophila but equivalent elements do not seem to target PRC2 in other organisms that have been examined [24]. Furthermore, even in Drosophila, PREs may not be directly required for transcriptional repression as deletion of four well-characterized strong Drosophila PREs had no effect on H3K27me-mediated silencing [25]. Sequence-specific transcription factors and noncoding RNAs can direct PRC2 to specific loci in mouse embryonic stem cells [26,27] and human cell lines [28,29], respectively, but again these do not seem to be prominent targeting strategies genome-wide. We favor a model similar to that proposed previously by others [30–33] suggesting that the local chromatin environment dictates H3K27me deposition. Our model considers the chromatin environment broadly and accounts for organisms that do not require PRC1 for PRC2 recruitment.
Features of constitutive heterochromatin prevent H3K27me deposition
In principle, the distribution of a histone mark, such as H3K27me, could be directly dictated by the sequence of associated DNA. In the case of this mark, however, this simple model is not highly compatible with recent findings. H3K27me regions greatly vary among different developmental stages, cell types, and in healthy versus diseased states of the same organism, even in cases in which the primary sequence of the genome is invariant. This raises the possibility that changes in chromatin environment may be responsible for the observed plasticity. Some candidate variables include histone modifications, DNA methylation, transcriptional state and nucleosome occupancy. While outside the scope of this review, sub-nuclear location and higher-order chromatin structure which are known to be influenced by H3K27me [34–36], may also impact placement of H3K27me. Striking examples of the plasticity of H3K27me, and indications that 'chromatin environment' is important for the distribution of the mark were provided by recent observations of redistribution of H3K27me when features of constitutive heterochromatin were disrupted [15,37–39] (see Table 1).
Table 1.
Examples of alterations to chromatin that cause redistribution of H3K27me
| Organism | Normal H3K27me distribution | Perturbation | H3K27me redistribution |
|---|---|---|---|
| A. thaliana | Promoters and gene bodies[77] | Loss of MET1 (DNA methyltransferase) |
|
| C. neoformans | Sub-telomeres | Loss of Ccc1 (chromodomain protein in PRC2 complex) |
|
| N. crassa | Sub-telomeres and interspersed genic regions |
|
|
| C. elegans | Alternating H3K27me3 and H3K36me3 domains on autosomes | Loss of MES-4 (H3K36 methyltransferase) |
|
| M. musculus (mesenchymal progenitor cells) | Promoters and gene bodies | H3.3K36M or depletion of Nsd1/2 and Setd2 (H3K36 methyltransferases) |
|
| M. musculus (neural stem cells) | Promoters and gene bodies | DNMT3a KO (DNA methyltransferase) |
|
| M. musculus (embryonic fibroblasts) | Promoters and gene bodies | DNMT1n/n (DNA methyltransferase hypomorph) |
|
The first indication that perturbation of constitutive heterochromatin impacts the distribution of H3K27me came over a decade ago in work with mouse embryonic stem cells (ESCs). Cytological observations showed that loss of H3K9me3 in pericentric heterochromatin in Suv39h double null cells was associated with new domains of H3K27me3 in patterns resembling those typical of H3K9me3 [37]. More recent studies with fungi revealed similar redistribution of H3K27me to centromeric regions. In C. neoformans, a chromodomain protein, associated with PRC2 in this organism, is required for appropriate H3K27me3 localization. When the chromodomain of this protein was mutated, H3K27me3 was lost from its normal location near the telomeres, and ectopic H3K27me3 appeared at the centromeres. The authors proposed that the chromodomain protein normally binds H3K27me3 to restrict this histone modification to its normal location, while in its absence the EED subunit of PRC2 binds to H3K9me2 at the centromere leading to methylation of H3K27 at that region [15]. Chromodomain proteins are not typically found in PRC2 complexes, but redistribution of H3K27me2/3 was also observed in N. crassa, which has a canonical PRC2 complex [12,38] and this was confirmed in another study [40]. When H3K9me3 was eliminated, H3K27me2/3 was reduced or lost at most of its normal locations and redistributed to constitutive heterochromatin, including centromeres. Moreover, elimination of HP1, normally bound to H3K9me3, caused equivalent redistribution of H3K27me2/3, without affecting H3K9me3, providing a rare example of cohabitation of these two marks on the same molecule [38]. These findings are consistent with the results with ESCs but contrast the situation in C. neoformans, where redistribution apparently depends on H3K9me. The findings with N. crassa are also consistent with experiments in mouse zygotes, where Hp1β prevents ectopic accumulation of H3K27me3 on maternal pericentric heterochromatin [39].
There are indications that another repressive mark, DNA methylation, can also, directly or indirectly, antagonize H3K27me. In a variety of cell types, loss of DNA methylation, caused by disruption of DNA methyltransferase genes or treatment with the demethylating agent 5-azacytidine, results in accumulation of H3K27me at regions previously marked by 5-methylcytosine [41–46]. These studies are consistent with data demonstrating that unmethylated CpG islands inserted at ectopic genomic locations can efficiently recruit H3K27me [47,48]. Neither H3K9me nor DNA methylation directly inhibits PRC2 activity in vitro as measured by histone methyltransferase assays [49,50] suggesting that these modifications, or proteins that recognize and bind to them, may prevent PRC2 recruitment or productive association with chromatin. Supporting this idea, DNA methylation prevents binding of factors, such as KDM2B and BEND3, that may be required for PcG recruitment [51–54]. It is noteworthy that DNA methylation is not universally important for the normal distribution of H3K27me as elimination of DNA methylation in N. crassa has no effect on the distribution of H3K27me [38] and both D. melanogaster and C. elegans lack DNA methylation [55]. The impact of perturbing DNA methylation in higher organisms may reflect recognized effects of DNA methylation on H3K9me3 in these systems [38, 56,57].
Active histone marks antagonize H3K27me
Actively transcribed regions of the genome marked by H3K4me3 and H3K36me2/3 are generally distinct from those marked by H3K27me3. H3K4me3 and H3K27me3 are mutually exclusive at HOX genes in Drosophila embryos [58] and differentiated mammalian cells, but these modifications can coexist in “bivalent” domains in ESCs [59]. In C. elegans embryos and murine mesenchymal progenitor cells, loss of H3K36 methyltransferases results in methylation of H3K27 at previously H3K36me regions [60,61]. A mutation found in human chondroblastomas, H3.3K36M, which causes global loss of H3K36me2/3, also induces gains of H3K27me3 in regions that have lost H3K36me [61]. More broadly, quantitative mass spectrometry experiments demonstrated that H3K4me3 and H3K27me3[62] or H3K36me3 and H3K27me3 [63,64] rarely co-exist on the same H3 molecule in mouse or human cell lines. A mechanistic basis for this was demonstrated by finding that PRC2 catalytic activity is inhibited by H3K4me3 and H3K36me2/3 in vitro [50].
Histone acetylation is also associated with gene expression [65]. Because acetylation of H3K27 (H3K27ac) is not compatible with methylation of this residue, histone deacetylation is required to create a chromatin context permissive for PRC2 activity [66–69]. Together these findings support the idea that active chromatin regions are refractory to PRC2 activity, preventing H3K27me3-mediated repression of transcriptionally active genes.
H2Aub1 deposited by PRC1 recruits H3K27me
Histone modifications can render a chromatin environment permissive for H3K27me. The recent observation that H2Aub1, a mark catalyzed by a RING protein in PRC1, can recruit PRC2 and cause H3K27me3 has challenged the classical view of PcG recruitment in which H3K27me3 deposited by PRC2 serves as a binding platform for chromodomain proteins in the canonical PRC1 complex [49,70,71]. Tethering a variant PRC1 complex to a bacterial artificial chromosome containing human DNA provided evidence in mouse ESCs that H2Aub1 catalyzed by the tethered protein can recruit PRC2 and H3K27me3. A complementary study demonstrated that variant PRC1 can also recruit PRC2 and H3K27me3 to pericentric heterochromatin in mouse ESCs [49]. In addition to promoting PRC2 recruitment, H2Aub1 apparently stimulates its enzymatic activity [72].
Transcription inhibits H3K27me
In addition to responding, directly or indirectly, to histone modifications and DNA methylation, PRC2 can respond directly to transcription. The complex can bind RNA and there are indications that PRC2 is sensitive to the transcriptional state of promoters. Given the role of PRC2 in transcriptional repression, it was surprising that RIP-seq studies, which detect RNAs associated with a protein of interest, suggested that PRC2 binds promiscuously to nascent RNAs at active promoters genome-wide [73,74]. This observation was reconciled by data demonstrating that RNA binding inhibits the catalytic activity of PRC2, perhaps accounting for the absence of H3K27me in regions producing RNA bound by PRC2. ChIP-seq experiments with human cell lines revealed chromatin-associated PRC2 enriched at promoters of repressed genes, the majority of which were marked by H3K27me3 [74]. Additional support for the idea that PRC2 responds to the transcriptional status of chromatin comes from studies demonstrating that chemical inhibition of RNA pol II-dependent transcription in mESCs is sufficient to recruit PRC2 and H3K27me3 to thousands of genes. Importantly, nearly all of the genes that gained H3K27me3 upon transcriptional repression are bona fide PRC2 target genes in differentiated tissues, although not in mESCs [32]. This observation was reiterated by finding that deletion of the genomic region containing the transcription start site led to loss of transcription, histone deacetylation and accumulation of H3K27me3 [75]. Taken together these data support a model in which PRC2 takes cues from the chromatin environment where it can sense transcription by interacting with nascent transcripts, which then prevent productive interaction with chromatin and modification of H3K27.
Conclusions
The findings summarized above are consistent with the idea that H3K27me catalyzed by PRC2 is directed by various inputs from the chromatin environment (Figure 1). There are at least three layers of regulation that contribute to PRC2 target selection: 1. Recruitment 2. Loading onto chromatin and 3. Regulation of catalytic activity. Some loci may utilize sequence-specific PcG targeting mechanisms using lncRNAs or transcription factor binding motifs, but many loci appear to lack any direct recruitment strategy. H3K27me targeting to these loci may rely on the ability of PRC2 to sense the chromatin environment. Once PRC2 is recruited to a given genomic location, it must productively associate with chromatin. This step likely requires the chromatin to be “accessible”, which might in part be dictated by lack of transcription and associated chromatin marks, as well as the absence of repressive epigenetic marks and proteins that recognize and bind to these modifications. Both high [76] and low nucleosome density [32] have been reported to promote H3K27me. Other factors such as chromosomal conformation and spatial location within the nucleus may also facilitate PRC2 access to chromatin. The final layer of regulation is the catalytic activity of PRC2. Signs of active transcription, in the form of RNA, H3K4me3, H3K36me2/3, and histone acetylation may prevent methylation of H3K27. While repressive marks, such as DNA methylation and H3K9me, do not directly inhibit PRC2 enzymatic activity, the effects of proteins that bind to these modifications have not been directly tested. Conversely the chromatin environment, specifically marks associated with PcG, H3K27me and H2Aub1, can stimulate PRC2 activity.
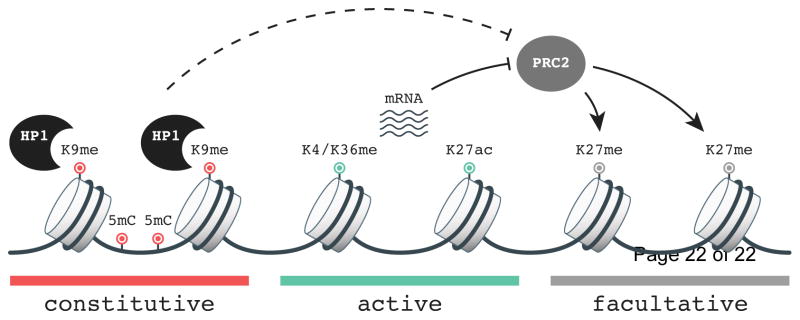
Figure 1. Model in which PRC2 responds to the chromatin environment to establish H3K27me domains.
Repressive features of constitutive heterochromatin including DNA methylation (5mC), H3K9me (K9me), and HP1 binding influence methylation of H3K27 by PRC2. Conversely, histone modifications associated with transcription such as H3K4me (K4me), H3K36me (K36me), and H3K27ac (K27ac), as well as RNA can directly inhibit PRC2 catalytic activity to prevent H3K27me at regions of active gene expression. Genomic regions that do not contain features of active or repressed chromatin may be targeted by PRC2. Other properties of the chromatin environment such as nucleosome occupancy, chromosome conformation and location within the nucleus (not pictured) may also contribute to PRC2 target selection.
The ability of PRC2 to respond to a variety of local signals in chromatin is striking. Available information suggests PRC2 does not have reciprocal effects on other chromatin modifiers. For example, PRC2 has little effect on the distribution of DNA-me or H3K9me [38,45]. Similarly, while H3K36me2/3 inhibits PRC2 activity, H3K27me does not inhibit the H3K36 methyltransferases HYBP [63] or NSD2 [50]. While the adaptability of PRC2 to different chromatin environments may preclude a unifying model for control of H3K27me, this remarkable plasticity may be crucial to its ability to maintain silencing of specific genes in different cell lineages.
Acknowledgments
We thank Kevin McNaught and Scott Stewart for comments on the manuscript and Travis Wiles for help with the figure.
Funding
This work was supported by a National Institutes of Health grant [GM093061] to EUS and an American Heart Association grant (14POST20450071) to ETW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
** Papers of outstanding interest:
* Papers of special interest:
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 4.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Development. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & Development. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 7.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb Group Proteins Ring1A/B Link Ubiquitylation of Histone H2A to Heritable Gene Silencing and X Inactivation. Developmental Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 9.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Molecular Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaver S, Casas-Mollano JA, Cerny RL, Cerutti H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics. 2010;5:301–312. doi: 10.4161/epi.5.4.11608. [DOI] [PubMed] [Google Scholar]
- 11.Molitor A, Shen W-H. The Polycomb Complex PRC1: Composition and Function in Plants. Journal of Genetics and Genomics. 2013;40:231–238. doi: 10.1016/j.jgg.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Jamieson K, Rountree MR, Lewis ZA, Stajich JE, Selker EU. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc Natl Acad Sci USA. 2013;110:6027–6032. doi: 10.1073/pnas.1303750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. PLoS Genet. 2008;4:e1000242–14. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pengelly AR, Kalb R, Finkl K, Müller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes & Development. 2015;29:1487–1492. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Dumesic PA, Homer CM, Moresco JJ, Pack LR, Shanle EK, Coyle SM, Strahl BD, Fujimori DG, Yates JR, III, Madhani HD. Product Binding Enforces the Genomic Specificity of a Yeast Polycomb Repressive Complex. Cell. 2015;160:204–218. doi: 10.1016/j.cell.2014.11.039. Demonstrates plasticity of H3K27me and how PRC2 can sense chromatin context by showing that H3K27me is lost from normal telomeric regions and redistributed to centromeric regions when a chromodomain protein that is part of the PRC2 complex in this organism is mutated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Müller J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- 17.Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 19.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. The EMBO Journal. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and Cellular Biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Current Opinion in Cell Biology. 2015;37:42–48. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nature Communications. 2016;7:1–11. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 25**.De S, Mitra A, Cheng Y, Pfeifer K, Kassis JA. Formation of a Polycomb-Domain in the Absence of Strong Polycomb Response Elements. PLoS Genet. 2016;12:e1006200–22. doi: 10.1371/journal.pgen.1006200. Challenges the view that PREs are required for recruitment of PRC2 and H3K27me mediated silencing by showing that deletion of four well-characterized strong Drosophlia PREs are not required for H3K27me or gene silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, et al. Polycomb Complex 2 Is Required for E-cadherin Repression by the Snail1 Transcription Factor. Molecular and Cellular Biology. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold P, Scholer A, Pachkov M, Balwierz PJ, Jorgensen H, Stadler MB, van Nimwegen E, Schubeler D. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Research. 2012;23:60–73. doi: 10.1101/gr.142661.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Kruijsbergen I, Hontelez S, Veenstra GJC. Recruiting polycomb to chromatin. International Journal of Biochemistry and Cell Biology. 2015;67:177–187. doi: 10.1016/j.biocel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression modifications to a complex story. Vol. 16. Nature Publishing Group; 2015. pp. 643–649. Comprehensive review covering various potential PRC1 and PRC2 chromatin targeting strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene Silencing Triggers Polycomb Repressive Complex 2 Recruitment to CpG Islands Genome Wide. Molecular Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. Shows that chemical inhibition of transcription in mESCs triggers PRC2 recruitment to CpG islands that are nucleosome depleted. [DOI] [PubMed] [Google Scholar]
- 33.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wijchers PJ, Krijger PHL, Geeven G, Zhu Y, Denker A, Verstegen MJAM, Valdes-Quezada C, Vermeulen C, Janssen M, Teunissen H, et al. Cause and Consequence of Tethering a SubTAD to Different Nuclear Compartments. Molecular Cell. 2016;61:461–473. doi: 10.1016/j.molcel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheutin T, Cavalli G. ScienceDirect Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Current Opinion in Genetics & Development. 2014;25:30–37. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 36.Entrevan M, Schuettengruber B, Cavalli G. Regulation of Genome Architecture and Function by Polycomb Proteins. Trends in Cell Biology. 2016;26:511–525. doi: 10.1016/j.tcb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Molecular Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 38**.Jamieson K, Wiles ET, McNaught KJ, Sidoli S, Leggett N, Shao Y, Garcia BA, Selker EU. Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Research. 2015;26:97–107. doi: 10.1101/gr.194555.115. Demonstrates plasticity of H3K27me2/3 by showing that disruption of H3K9me3 or HP1, but not DNA methylation, causes redistribution of H3K27me2/3 to regions of constitutive heterochromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Tardat M, Albert M, Kunzmann R, Liu Z, Kaustov L, Thierry R, Duan S, Brykczynska U, Arrowsmith CH, Peters AHFM. Cbx2 Targets PRC1 to Constitutive Heterochromatin in Mouse Zygotes in a Parent-of-Origin-Dependent Manner. Molecular Cell. 2015;58:157–171. doi: 10.1016/j.molcel.2015.02.013. Shows that the PRC1 component Cbx2 uses its chromodomain and AT-hook domain to sense the chromatin environment and that Hp1β prevents PRC1 from binding to maternal pericentric heterochromatin to restrict H3K27me3 to paternal pericentric heterochromatin. [DOI] [PubMed] [Google Scholar]
- 40.Basenko EY, Sasaki T, Ji L, Prybol CJ, Burckhardt RM, Schmitz RJ, Lewis ZA. Genome-wide redistribution of H3K27me3 is linked to genotoxic stress and defective growth. Proc Natl Acad Sci USA. 2015;112:E6339–E6348. doi: 10.1073/pnas.1511377112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. The EMBO Journal. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE. Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003062–13. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, Pasini D, Miró X, Donohoe ME, Lee JT, et al. Antagonism between DNA and H3K27 Methylation at the Imprinted Rasgrf1 Locus. PLoS Genet. 2008;4:e1000145. doi: 10.1371/journal.pgen.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagarman JA, Motley MP, Kristjansdottir K, Soloway PD. Coordinate Regulation of DNA Methylation and H3K27me3 in Mouse Embryonic Stem Cells. PLoS ONE. 2013;8:e53880–10. doi: 10.1371/journal.pone.0053880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-Dependent Nonpromoter DNA Methylation Facilitates Transcription of Neurogenic Genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells. PLoS Genet. 2010;6:e1001244–10. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Jermann P, Hoerner L, Burger L, Schubeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci USA. 2014;111:E3415–E3421. doi: 10.1073/pnas.1400672111. Shows that CpG rich DNA ectopically inserted into mESCs is sufficient to recruit H3K27me3 when the surrounding genomic region is devoid of transcription and the sequence is unmethylated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. Targeting Polycomb to Pericentric Heterochromatin in Embryonic Stem Cells Reveals a Role for H2AK119u1 in PRC2 Recruitment. CellReports. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone Methylation by PRC2 Is Inhibited by Active Chromatin Marks. Molecular Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 51.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife. 2012:1. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Molecular Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla M-E, et al. Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation. Molecular Cell. 2014;56:580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Colot V, Rossignol J-L. Eukaryotic DNA methylation as an evolutionary device. BioEssays. 1999;21:402–411. doi: 10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 56.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papp B. Histone trimethylation and the maintenance of transcriptional ONand OFF states by trxG and PcG proteins. Genes & Development. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry Ben, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 60.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and Polycomb Repressive Complex 2 Promotes Appropriate Gene Expression in C. elegans Germ Cells. CellReports. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High Throughput Characterization of Combinatorial Histone Codes. Molecular & Cellular Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 Methylation Antagonizes PRC2-mediated H3K27 Methylation. Journal of Biological Chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwammle V, Aspalter CM, Sidoli S, Jensen ON. Large Scale Analysis of Coexisting Post-translational Modifications in Histone Tails Reveals Global Fine Structure of Cross-talk. Molecular & Cellular Proteomics. 2014;13:1855–1865. doi: 10.1074/mcp.O113.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner BM. Histone acetylation and control of gene expression. J Cell Sci. 1991;99(Pt 1):13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- 66.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. The EMBO Journal. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qüesta JI, Song J, Geraldo N, An H, Dean C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science. 2016;353:485–488. doi: 10.1126/science.aaf7354. [DOI] [PubMed] [Google Scholar]
- 70.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical Recruitment of Polycomb Group Silencing Complexes. Molecular Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 72.Kalb R, Latwiel S, Baymaz HI, Jansen PWTC, Müller CW, Vermeulen M, Müller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nature Publishing Group. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 73.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nature Publishing Group. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hosogane M, Funayama R, Shirota M, Nakayama K. Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell Reports. 2016;16:696–706. doi: 10.1016/j.celrep.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 76.Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. Dense Chromatin Activates Polycomb Repressive Complex 2 to Regulate H3 Lysine 27 Methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129–10. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]