Abstract
MicroRNA (miRNA) expression varies in association with different tissue types and in diseases. Having been found in body fluids including blood and cerebrospinal fluid (CSF), miRNAs constitute potential biomarkers. CSF miRNAs have been proposed as biomarkers for neurodegenerative diseases; however, there is a lack of consensus about the best candidate miRNA biomarkers and there has been variability in results from different research centers, perhaps due to technical factors. Here, we sought to optimize technical parameters for CSF miRNA studies. We examined different RNA isolation methods and performed miRNA expression profiling with TaqMan® miRNA Arrays. More specifically, we developed a customized CSF-miRNA low-density array (TLDA) panel that contains 47 targets: miRNAs shown previously to be relevant to neurodegenerative disease, miRNAs that are abundant in CSF, data normalizers, and controls for potential blood and tissue contamination. The advantages of using this CSF-miRNA TLDA panel include specificity, sensitivity, fast processing and data analysis, and cost effectiveness. We optimized technical parameters for this assay. Further, the TLDA panel can be tailored to other specific purposes. We tested whether the profile of miRNAs in the CSF resembled miRNAs isolated from brain tissue (hippocampus or cerebellum), blood, or the choroid plexus. We found that the CSF miRNA expression profile most closely resembles that of choroid plexus tissue, underscoring the potential importance of choroid plexus-derived signaling through CSF miRNAs. In summary, the TLDA miRNA array panel will enable evaluation and discovery of CSF miRNA biomarkers and can potentially be utilized in clinical diagnosis and disease stage monitoring.
Keywords: MicroRNA, Cerebrospinal fluid, Biomarker, Neurodegenerative disease, Biofluid, Choroid plexus
Introduction
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression posttranscriptionally. MiRNAs have been implicated in neurodegenerative diseases, such as Alzheimer’s disease (AD) [1–4]. MiRNAs are expressed in virtually all known mammalian cells and can be found in body fluids including serum, cerebrospinal fluid (CSF), saliva, urine, and breast milk [5], providing a potential opportunity to monitor disease presence and severity. Studies have shown that miRNAs in serum and plasma are stable at room temperature for several days and even resistant to RNase treatment [6]. Furthermore, they are also easier to detect and quantify compared to many protein biomarkers [7]. Thus, biofluid miRNAs are good candidates for biomarkers.
Despite extensive studies, no miRNA has been firmly established as a clinically relevant biomarker. Challenges include the complexity of the pathophysiological conditions (various stages of disease and comorbid pathologies) and also probably heterogeneous human cohorts [8, 9]. This is particularly true for neurodegenerative diseases. Furthermore, preanalytical and analytical factors such as sampling methods, specimen storage, RNA isolation and detection platform, and data normalization all contribute to the mixed outcomes of the analysis [10–12].
CSF provides a special context for biomarkers. Already used as an AD biomarker detecting Aβ, tau, and other analytes, CSF can be obtained via routine lumbar puncture. This clear proteinaceous fluid is normally in direct contact with the brain after being produced in the choroid plexus, which exchanges chemicals, proteins, electrolytes, and other molecules with the blood stream. Thus, it is a critical context for searching biomarkers in association with CNS diseases.
Prior studies have analyzed CSF miRNA expression in association with CNS diseases (see for example [13–19]). These studies employed various isolation and detection methods, mostly array and deep sequencing-based profiling. Prior studies generated valuable information in searching for reliable biomarkers. However, contradictory results were also reported possibly because of biological complexity and the various study designs and methodological factors. Furthermore, array- and sequencing-based analysis generally have poorer sensitivity and higher variability for low-quantity RNA inputs, which are typical in clinic-derived CSF samples.
Here, we describe a custom-made miRNA RT-qPCR panel that can overcome some of the aforementioned shortcomings in CSF miRNA analysis. The goal was to produce a relatively economic platform that includes miRNAs known to be present in CSF and include those miRNAs that previously were implicated as possible biomarkers. We also used this panel to test whether the miRNA profile of CSF more closely resembles RNA isolated from blood, the choroid plexus, or brain tissues derived from the University of Kentucky AD Center (UK-ADC) biobank.
Materials and Methods
Human Cerebrospinal Fluid, Brain Tissues, and Blood Samples
Human CSF and tissues used in this work were obtained from the UK-ADC biobank and analyzed in compliance with University of Kentucky Institutional Review board protocol. Clinical lumbar puncture-derived CSF, blood samples, and snap-frozen brain tissues from individuals obtained at autopsy, as described previously [20, 21], were coded anonymously.
RNA Isolation from Brain Tissues and Blood
Total RNA was extracted from human tissues and blood samples using TRIzol LS reagent (Thermo Fisher Scientific) following the modified procedure described previously [3, 22, 23]. The RNA concentrations and purity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific).
Testing CSF RNA Isolation Methods
For the study of RNA isolation technical parameters, ten lumbar puncture-harvested CSF samples were mixed and spun at 3000 × g for 5 min. Aliquots of 200 μl supernatant from the mixed sample were used for each RNA isolation procedure. Four replicates were evaluated for each method. Total RNA was extracted and compared using commercially available kits: MagMAX™ mirVana™ Total RNA Isolation Kit (Thermo Fisher Scientific), miRCURY™ RNA Isolation Kit–Biofluids (Exiqon), miRNeasy Serum/Plasma Kit (QIAGEN), NucleoSpin miRNA Plasma (MACHEREYNAGEL), PureLink™ RNA Mini Kit (Thermo Fisher Scientific), and TRIzol LS reagent (Thermo Fisher Scientific). RNA isolations were carried out following manufacturers’ instructions except for TRIzol LS reagent which was done exactly as described previously [3, 22, 23]. RNA isolation experiments were performed with or without nucleic acid carriers, glycogen, and bacteriophage MS2 RNA (both from Roche). RNA was eluted or dissolved with 30 μl of nuclease-free water containing RNAsin (0.5 U/μl, Promega). After a preliminary comparison by single-tube TaqMan® miRNA assays (data not shown), three kits including miRCURY™ RNA Isolation Kit–Biofluids, miRNeasy Serum/Plasma Kit, and TRIzol LS reagent were selected for further assessments by miR-15/107 TaqMan® Low-Density Array (TLDA) analysis [24].
CSF miRNA Profiling Using TaqMan® Array Human MicroRNA A+B Cards Set v3.0
Clinical CSF samples were collected as described above from additional four controls and four AD patients and the RNA subjected to TaqMan® Array analysis using Human MicroRNA A+B Cards Set v3.0 (Life Technologies). CSF RNA was isolated using Exiqon’s miRCURY™ RNA Isolation Kit (Biofluids) following manufacturer’s protocol. Briefly, 200 μl of CSF was transferred to a fresh tube after centrifuge at 3000 g for 5 min; 60 μl of Lysis Solution BF was added to the supernatant following a 5-s vortex. After a 3-min incubation, 1 μg of bacteriophage MS2 RNA (carrier, Roche) and 3.5 μl of Cel-miR-39-3p (spike-in RNA, QIAGEN) at 1.6 × 108 copies/μl was added to the mixture. Protein Precipitation Solution BF (20 μl) was added, vortex, incubated for 1 min, and centrifuged. The clear supernatant was transferred to a fresh collection tube with addition of 270 μl isopropanol and mixed. Column binding, washing, and drying were conducted as instructed by the protocol, and RNA was eluted with 30 μl of nuclease-free water containing 0.5 U/μl RNAsin (Promega). Next, equal volume of total CSF RNA (3 μl) was reverse transcribed using the TaqMan® MicroRNA Reverse Transcription Kit (Life Technologies) with Megaplex™ Primer Pools, Human Pool A and B, respectively (Life Technologies). The resulting complementary DNA (cDNA) was preamplified using corresponding Human Pool A or B Megaplex™ PreAmp Primers (Life Technologies) before running the qPCR Array. qPCR was carried out on the ViiA™ 7 Real-Time PCR System and quantitative PCR cycle (Cq) was determined using ViiA 7 RUO software (Life Technologies) with automatic baseline and a fixed threshold set at 0.15.
TaqMan® Low-Density Array (TLDA) Analysis
Two separate sets of custom-developed TLDA cards were used in the study: miR-15/107 TLDA cards and CSF-miRNA TLDA cards. MiR-15/107 TLDA cards have been described in detail previously [24]. The CSF-miRNA TLDA card incorporated a combination of miRNAs related to CSF and neurodegenerative diseases and the miRNA selection was discussed in “Results.” Standard manufacturer’s protocol was followed for all TLDA cards. Briefly, 3 μl of CSF RNA or tissue RNA (at 10 ng/μl) was reverse transcribed using TaqMan® MicroRNA Reverse Transcription Kit (Life Technologies) with pooled specific RT primers for each panel. An aliquot of RT product was then preamplified using TaqMan® PreAmp Master Mix (Life Technologies) with pooled preamplification primers (Life Technologies). RT products with or without preamplification were served as templates. Premixed template and TaqMan® Universal PCR Master Mix (No AmpErase UNG, Life Technologies) were transferred to each port of TLDA cards and loaded to each well by spinning the cards in a centrifuge according to the manufacturer’s instructions. qPCR was performed using ViiA™ 7 Real-Time PCR System (Life Technologies).
TaqMan® Single-Tube RT-qPCR for Individual miRNAs
Individual miRNA and spike-in Cel-miR-39 were subjected to single-tube TaqMan® RT-qPCR assay (LifeTechnologies) according to the manufacturer’s instructions. Equal volume of CSF RNA (3 μl) for reverse transcription and qPCR was performed in triplicate.
Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) was followed throughout all the RT-qPCR procedure [25].
Data and Statistical Analysis
Raw PCR Cq values from custom or inventory array cards were first determined using ViiA 7 RUO software (Life Technologies) with automatic baseline and threshold set at 0.15. Data that failed QC, including those such as bad passive reference signal and failed in exponential algorithm and thresholding algorithm (amplification too early or too late, low amplification or no amplification), were treated as undetectable in the analysis. Outliers such as a data point that is much higher or much lower in Cq among replicates in the data set was also examined manually, and an amplification curve that was abnormal (i.e., flat curve which resulted in low Cq) was also treated as undetectable. DataAssist® software (Life Technologies) was employed to further analyze miRNA array data. A Cq value equal to or greater than 32 was considered as undetectable. The global mean normalization method [26] was selected and a Cq cutoff was set at 35 for inventory and miR-15/107 custom TLDA cards and 32 for CSF-miRNA TLDA cards (e.g., a Cq value equal to or more than 32 was considered as undetectable in custom CSF-miRNA TDLA arrays). Correlation coefficient (R2) of the datasets from customized CSF panel and inventory array cards was obtained using linear regression function.
Welch’s two-sample t test was used to evaluate the difference of the mean and total qPCR array Cq data between the control and AD groups. To determine the differential expression pattern between CSF and tissues, array data were evaluated using an unpaired Student’s t test. Hierarchical clustering based on the similarity of miRNA expression patterns was calculated using Pearson’s distance-Average Linkage. Benjamini–Hochberg tests were used to control false discovery rate (FDR). p value that is less than 0.05 was considered as statistically significant.
Correlation coefficient (R2) of miRNA expression pattern between CSF and each individual tissue group was obtained using linear regression function, and evaluation of correlations between CSF and various nonindependent tissues was done using confidence intervals according to the method described by Zou [27].
Results
Empirical Evaluation of CSF RNA Isolation Methods
Isolating quality RNA is a critical first step in studying CSF miRNA. The three most effective kits in our hands were as follows: Exiqon’s miRCURY™ RNA Isolation Kit–Biofluids; QIAGEN’s miRNeasy Serum/Plasma Kit; and Thermo Fisher Scientific’s TRIzol® LS Reagent. Because the content of RNA is generally very low in clinical CSF samples, NanoDrop or other methods of concentration measurements were not sensitive enough for assessing the quantity and quality of CSF RNA. Instead, we used customized miR-15/107 TaqMan® Low-Density Array (TLDA, format 24; Life Technologies) [24] to assess the quantity and quality of CSF RNA isolations. Figure 1 illustrates the procedure used in evaluating CSF RNA isolation. RNA quantity and quality were defined by the following standards: (1) low quantification cycle (Cq); (2) high ΔCq above the background; (3) low technical variability (standard deviation) among replicates.
Fig. 1.
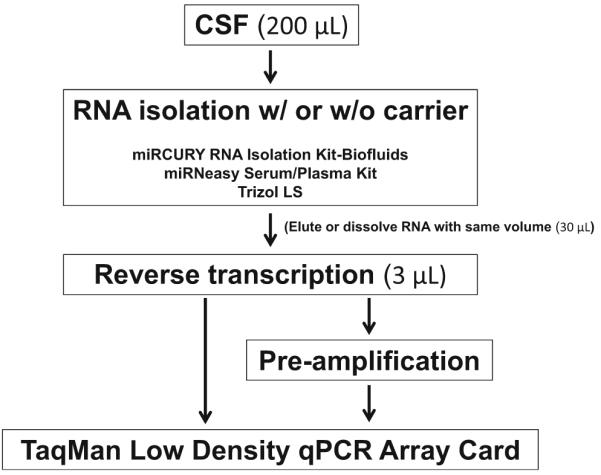
Workflow for testing CSF RNA isolation methods
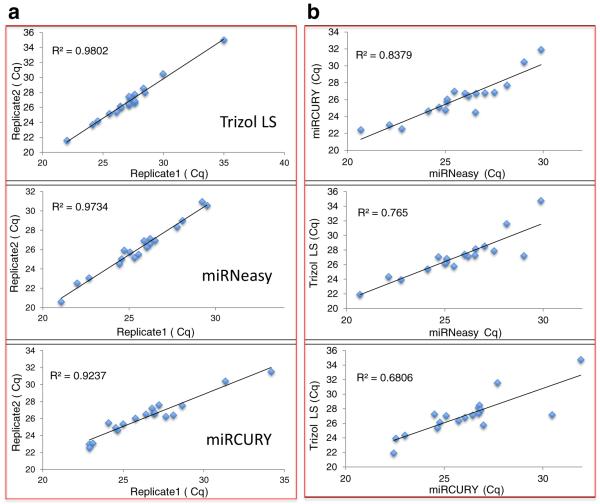
Nucleic acid carriers are additives that enhance the yield of RNA isolation, which is particularly helpful in biological samples with low-concentration RNAs. We evaluated two different carriers, glycogen and bacteriophage MS2 RNA. Both MS2 RNA and glycogen carriers enhanced the RNA yield (Suppl. Table 1), with MS2 RNA carrier slightly increasing technical background levels (Table 1). Next, we tested the consistency of each RNA isolation method. Four technical replicate isolations were carried out and TLDA array qPCRs were performed. All three tested methods gave high consistency and there was no statistically significant difference among the isolation methods tested although some trends were noted (Fig. 2a, Table 2). TRIzol isolation seemed to perform better as judged by the trends for lower background (high Cq in background), higher ΔCq above background, and better consistency among replicate isolations (smaller STDEV). When comparing qPCR Cq from different isolation methods, they generally agreed with each other, but were far from perfect (Fig. 2b). From our data, it is clearly important to use one selected method consistently throughout a group of samples. On the other hand, when validating the results, it may be necessary to use a different approach, to avoid the inherent bias or false positives/negatives generated from the same isolation and detection systems.
Table 1.
Comparison of CSF RNA isolated by various methods on miR-15/107 TLDA cards (n = 4)
| Isolation method |
Carrier | Carrier only w/ PreAmp |
CSF RNA w/o PreAmp |
CSF RNA w/ PreAmp |
ΔCq
above background |
|---|---|---|---|---|---|
| miRNeasy | MS2 | 32.77 | 32.93 | 25.34 | −7.44 |
| miRCURY | MS2 | 32.95 | 33.25 | 26.12 | −6.82 |
| TRIzol LS | MS2 | 33.14 | N.D. | 25.89 | −7.25 |
| Glycogen | 33.93 | N.D. | 26.77 | −7.16 |
Fig. 2.
Consistency of CSF RNA isolation methods. Four replicate isolations were carried out using the same CSF sample for each isolation method. The isolated CSF RNAs were tested using the miR-15/107 TLDA analysis. Cq data of each array were plotted against each other, and the coefficient of correlation (R2) of the two data sets calculated. A representative plot from each method was shown. RNA isolations within each method (a) were highly consistent, with slightly higher variance between the different isolation methods (b) with variance being greater at low-quantity (high-Cq) miRNAs
Table 2.
Consistency of RNA isolation by each method (n = 4)
| Average Cq | STDEV | STDEV range | |
|---|---|---|---|
| miRNeasy | 25.66 | 0.65 | 0.21–1.94 |
| miRCURY | 26.08 | 0.77 | 0.14–3.04 |
| TRIzol LS | 27.09 | 0.48 | 0.16–2.31 |
In summary, the different methods appear to each have separate strengths and weaknesses (Table 3). Overall, TRIzol LS provided slightly more consistent results but it involves a phenol/chloroform step and is technically challenging to obtain a “clean” prep with that method. By contrast, miRCURY stands out for its easy and fast isolation procedure, good quality/quantity RNA, and a lack of organic solvent step in the process. We selected the miRCURY method with MS2 RNA carrier for further studies.
Table 3.
Isolation method comparison
| Isolation method | RNA | Easiness | Organic solvent | Time |
|---|---|---|---|---|
| miRCURY | Good | +++ | No | Fast |
| miRNeasy | Good | ++ | Yes | Fair |
| Trizol LS | Good | − | Yes | Lengthy |
Preamplification Increases the Sensitivity and Uniformity of CSF miRNA Detection
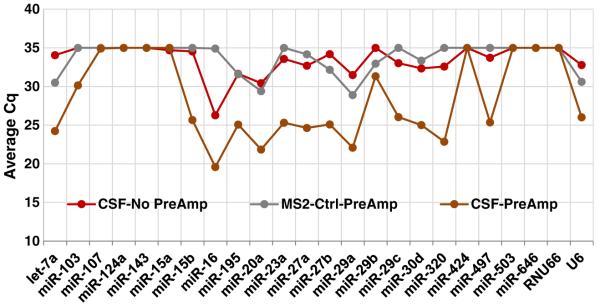
We employed TaqMan® miRNA single-tube assays and TLDA panel to assess CSF miRNAs. Mindful of the challenge of combining both high technical sensitivity and low sample-to-sample variability, TaqMan® assays were tested with or without preamplification. Preamplification is an intermediate step after reverse transcription and before qPCR in which cDNA can be enriched over several orders of magnitude, theoretically without disturbing the relative expression of the different miRNAs (Suppl. Fig. 1 and [28]). For detecting minute quantities of miRNA, preamplification greatly enhances the detection power of qPCR and data uniformity [28]. As showed in Fig. 3, Cq of most miRNAs in the miR-15/107 TLDA panel fell to the background levels without preamplification. In contrast, preamplification improved detection sensitivity by several Cq values. Thus, for most of the miRNA species, preamplification is a necessary step when assessing CSF RNA using RT-qPCR.
Fig. 3.
Preamplification increases the detection sensitivity MiR-15/107 TLDA analysis of cDNA template generated from the same RT product with or without preamplification are shown. The Cq of most miRNAs from the templates without preamplification (red dots and line) in miR-15/107 TDLA cards fell to the background (gray dots and line) level (Cq = 35); in contrast, preamplification array (brown dots and line) improved detection by several Cq values
CSF miRNA Exploratory Profiling Using TaqMan® Array Human MicroRNA A+B Cards Set v3.0
In order to better control technical variables and to assess the repertoire of miRNAs detectable with TaqMan® RT-qPCR assays in CSF, we ran a set of discovery (exploratory) profiling arrays using clinical CSF samples from four controls and four AD patients. Total RNA was extracted from CSF using Exiqon’s miRCURY™ RNA Isolation Kit (Biofluids). MS2 RNA was used as additive and C. elegans Cel-miR-39 was spiked in to monitor the isolation efficiency. TaqMan® RT-qPCR of Cel-miR-39 using the same volume (3 μl) of isolated RNAs gave similar Cq values across eight isolates with a standard deviation of 0.2 (Suppl. Table 2), indicating a consistent RNA isolation of all CSF samples. This was also evident by NanoDrop estimation of RNA concentrations.
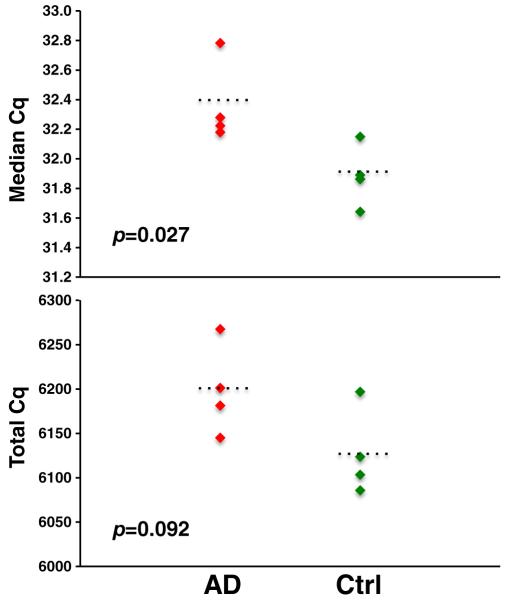
In the TaqMan® RT-qPCR miRNA results, a miRNA with Cq equal to or more than 35 was considered as undetectable. A miRNA was determined to be detectable in CSF if it was present in more than three arrays (out of eight). Using this cutoff, 130 and 63 miRNAs were detected in Card A and Card B, respectively. There was a small but statistically significant difference in median Cq of miRNA expression between the control and AD groups (Fig. 4). Total miRNA content (represented by total Cq values) was also less abundant in AD group (higher Cq value). However, most individual miRNAs were not expressed differently between the samples (using the criterion of fold change ≥ 2, p ≤ 0.05), with the important caveat of the small sample size (Suppl. Excel file 1).
Fig. 4.
CSF miRNA profiling using TaqMan® Array Human MicroRNA A+B Cards (Set v3.0). Eight clinical lumbar puncture-harvested CSF (four ADs and four controls) were subject to miRNA TaqMan® Array. Overall expression level of miRNAs was slightly lower in AD patients (the higher the Cq the lower the expression level)
Most abundant miRNAs in CSF were identified by ranking average Cq values of all samples. The most abundant miRNAs (Table 4) included miR-204, miR-1274B, miR-16, miR-146a, miR-150, miR-223, miR-17, and miR-21 as described previously [13, 19]. The complete list of all detectable miRNAs can be found in supplemental Excel file 2.
Table 4.
Top 50 most abundant miRNAs in CSF identified by TaqMan® miRNA RT-qPCR arrays
| Rank | miRNA | Cq | Rank | miRNA | Cq |
|---|---|---|---|---|---|
| 1 | hsa-miR-204 | 24.4 | 26 | hsa-let-7b | 29.3 |
| 2 | hsa-miR-1274B | 25.3 | 27 | hsa-miR-92a | 29.4 |
| 3 | hsa-miR-17 | 26.6 | 28 | hsa-miR-20a | 29.5 |
| 4 | hsa-miR-106a | 26.8 | 29 | hsa-miR-211 | 29.5 |
| 5 | hsa-miR-19b | 27.0 | 30 | hsa-miR-425-5p | 29.6 |
| 6 | hsa-miR-223 | 27.1 | 31 | hsa-miR-99a | 29.6 |
| 7 | hsa-miR-1298 | 27.3 | 32 | hsa-miR-30b | 29.6 |
| 8 | hsa-miR-720 | 27.5 | 33 | hsa-miR-484 | 29.7 |
| 9 | hsa-miR-21 | 27.6 | 34 | hsa-miR-571 | 29.7 |
| 10 | hsa-miR-661 | 27.8 | 35 | hsa-miR-29a | 29.8 |
| 11 | hsa-miR-146a | 28.1 | 36 | hsa-miR-100 | 29.9 |
| 12 | hsa-miR-150 | 28.2 | 37 | hsa-miR-346 | 29.9 |
| 13 | hsa-miR-30a-5p | 28.5 | 38 | hsa-miR-1274A | 29.9 |
| 14 | hsa-miR-9# | 28.5 | 39 | hsa-miR-338-5P | 30.0 |
| 15 | hsa-miR-24 | 28.6 | 40 | hsa-miR-638 | 30.1 |
| 16 | hsa-miR-320 | 28.6 | 41 | hsa-miR-574-3p | 30.1 |
| 17 | hsa-miR-34b | 28.7 | 42 | hsa-miR-125b | 30.2 |
| 18 | hsa-miR-30c | 28.8 | 43 | hsa-miR-485-3p | 30.2 |
| 19 | hsa-miR-16 | 28.8 | 44 | hsa-miR-146b | 30.3 |
| 20 | hsa-miR-1825 | 28.9 | 45 | hsa-miR-26a | 30.3 |
| 21 | hsa-miR-487a | 29.1 | 46 | hsa-miR-885-5p | 30.3 |
| 22 | hsa-miR-222 | 29.1 | 47 | hsa-miR-143 | 30.4 |
| 23 | hsa-miR-342-3p | 29.1 | 48 | hsa-miR-34a | 30.4 |
| 24 | hsa-miR-30a-3p | 29.2 | 49 | hsa-miR-1275 | 30.5 |
| 25 | hsa-miR-648 | 29.3 | 50 | hsa-miR-145 | 30.5 |
The exploratory profiling also enabled us to select potential CSF-specific “housekeeping” miRNAs for data normalization. Table 5 shows miRNAs displaying relatively constant levels across both the control and AD groups and within the group as judged by their smaller standard deviations.
Table 5.
Top 10 most stable miRNAs in CSF
| miRNA | Rank | Cq-AD | STDEV-AD | Cq-ctrl | STDEV-ctrl | Cq-all | STDEV-all |
|---|---|---|---|---|---|---|---|
| hsa-miR-152 | 88 | 32.0 | 0.14 | 32.0 | 0.19 | 32.0 | 0.16 |
| hsa-miR-618 | 89 | 32.0 | 0.38 | 32.0 | 0.06 | 32.0 | 0.26 |
| hsa-miR-30c | 18 | 28.6 | 0.07 | 29.0 | 0.22 | 28.8 | 0.27 |
| hsa-miR-9# | 14 | 28.6 | 0.31 | 28.4 | 0.26 | 28.5 | 0.28 |
| hsa-miR-208 | 193 | 34.8 | 0.50 | 34.9 | 0.15 | 34.8 | 0.35 |
| hsa-miR-1274B | 2 | 25.3 | 0.20 | 25.4 | 0.50 | 25.3 | 0.35 |
| hsa-miR-30a-5p | 13 | 28.6 | 0.41 | 28.4 | 0.38 | 28.5 | 0.38 |
| hsa-miR-204 | 1 | 24.2 | 0.17 | 24.7 | 0.39 | 24.4 | 0.39 |
| hsa-miR-30b | 32 | 29.5 | 0.42 | 29.7 | 0.39 | 29.6 | 0.40 |
| hsa-miR-338-5P | 39 | 29.9 | 0.37 | 30.1 | 0.48 | 30.0 | 0.42 |
A CSF-miRNA Panel to Study miRNAs in the Context of Alzheimer’s Disease
We chose customized TaqMan® miRNA RT-qPCR low-density array format 48 (Life Technologies) as the platform for studying CSF miRNAs relevant to AD. This PCR-based expression platform can simultaneously analyze eight samples and accommodate a total of 47 miRNAs plus one mandatory control (U6) per sample. The following criteria were used to select miRNAs for the panel (Table 6): (1) miRNA is detectable in CSF based on our profiling and existing literature; (2) miRNA was previously associated with AD (in tissue or CSF or both) or other neurodegenerative pathology; (3) miRNA is predicted to target genes critical to AD pathways (e.g., MAPTau, BACE1); (4) miRNA belongs to miR-15/107 or miR-29 family which are hypothesized to be perturbed and/or involved in neurodegenerative diseases; (5) miRNA is particularly brain-enriched; (6) miRNA is a potential normalizer (CSF housekeeping miRNAs); (7) “contamination” controls; and (8) ubiquitously expressed RNA species. MiRNAs that meet at least one of the above criteria were selected for the panel. These miRNAs plus U6 were custom-made into a low-density array card, and primers for RT and preamplification were prepared by Life Technologies® company.
Table 6.
Specific CSF miRNA panel for studying neurodegenerative diseases
| Assay ID | miRNA | Selection criteria | Note |
|---|---|---|---|
| 000377 | hsa-let-7a | 1, 8 | ND in our array |
| 002619 | hsa-let-7b | 1, 8 | |
| 000379 | hsa-let-7c | 1, 8 | |
| 000382 | hsa-let-7f | 1, 2, 8 | ND in our array |
| 000439 | hsa-miR-103 | 1, 2, 3, 4 | |
| 002169 | hsa-miR-106a | 1, 2, 3, 4 | |
| 000443 | hsa-miR-107 | 1, 2, 3 | ND in our array |
| 000449 | hsa-miR-125b | 1, 2, 5 | |
| 002884 | hsa-miR-1274B | 1, 6 | |
| 002861 | hsa-miR-1298 | 1, 2 | |
| 000457 | hsa-miR-132 | 1, 2, 3 | |
| 000464 | hsa-miR-142-3p | 1, 2 | |
| 002248 | hsa-miR-142-5p | 1 | |
| 000468 | hsa-miR-146a | 1, 2 | |
| 001097 | hsa-miR-146b | 1, 2 | |
| 000473 | hsa-miR-150 | 1 | |
| 002623 | hsa-miR-155 | 1, 2 | |
| 000389 | hsa-miR-15a | 1, 2, 3, 4 | ND in our array |
| 000390 | hsa-miR-15b | 1, 2, 3, 4 | |
| 000391 | hsa-miR-16 | 1, 2, 3, 4 | |
| 002308 | hsa-miR-17 | 1, 3 | |
| 000480 | hsa-miR-181a | 1, 2 | ND in our array |
| 000482 | hsa-miR-181c | 1, 2 | ND in our array |
| 000494 | hsa-miR-195 | 1, 2, 3, 4 | |
| 000396 | hsa-miR-19b | 1, 2, 3 | |
| 000508 | hsa-miR-204 | 1, 3, 6 | |
| 000580 | hsa-miR-20a | 1, 3 | |
| 000397 | hsa-miR-21 | 1, 2, 7 | |
| 002295 | hsa-miR-223 | 1, 2, 7 | |
| 000402 | hsa-miR-24 | 1, 7 | |
| 000408 | hsa-miR-27a | 1, 2 | |
| 002112 | hsa-miR-29a | 1, 2, 4 | |
| 000413 | hsa-miR-29b | 1, 2, 4 | ND in our array |
| 000587 | hsa-miR-29c | 1, 2, 4 | |
| 000416 | hsa-miR-30a-3p | 1, 2, 6 | |
| 000417 | hsa-miR-30a-5p | 1, 2 | |
| 000602 | hsa-miR-30b | 1, 2, 6 | |
| 000419 | hsa-miR-30c | 1, 2, 6 | |
| 000420 | hsa-miR-30d | 1, 2 | |
| 000422 | hsa-miR-30e-3p | 1 | |
| 000426 | hsa-miR-34a | 1, 2 | |
| 002102 | hsa-miR-34b | 1, 2 | |
| 000428 | hsa-miR-34c | 1, 2 | |
| 001043 | hsa-miR-497 | 1, 2, 3, 4 | |
| 000583 | hsa-miR-9 | 1, 2, 5 | ND in our array |
| 001182 | mmu-miR-124a | 1, 2, 5 | ND in our array |
| 001141 | mmu-miR-451 | 1, 2 |
1 miRNA is detectable in CSF based on our profiling and existing literature, 2 miRNAwas previously associated with AD (in tissue or CSF or both) or other neurodegenerative pathology, 3 miRNA is predicted to target genes critical to AD pathways (e.g., MAPTau, BACE1), 4 miRNA belongs to miR-15/107 or miR-29 family which are hypothesized to be perturbed and/or involved in neurodegenerative diseases, 5 miRNA is particularly brain-enriched, 6 miRNA is a potential normalizer (CSF housekeeping miRNAs), 7 contamination controls, 8 ubiquitously expressed RNA species, ND not detected
Performance of CSF-miRNA Panel
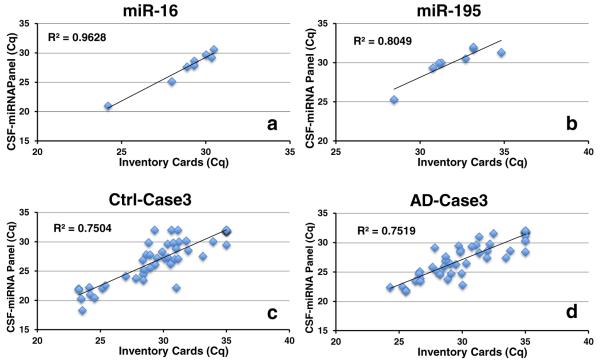
To test the performance of the CSF-miRNA panel, the eight CSF samples that were used for profiling previously (four ADs, four controls) were tested using the novel platform. The CSF-miRNA panel TLDA analysis generated comparable Cq values as that of the inventory TaqMan® array cards. The panel seemed to be more sensitive as the overall average Cq was 2.4 lower than that of the inventory cards (Suppl. Table 3). There was a generally good correlation of miRNAs detected by both methods as indicated by Pearson correlation coefficient R squared (R2) (Fig. 5, Suppl. Table 3). However, several miRNAs showed a discrepancy which could be a result of a difference in primer pool size. Furthermore, primer interaction was observed with the custom-made CSF-miRNA panels (data not shown).
Fig. 5.
Detection of miRNA using CSF-miRNA panel is comparable with inventory TaqMan® Array Cards. The same eight clinical lumbar puncture-harvested CSF RNAs were analyzed using inventory array cards and customized CSF-miRNA TLDA panel. Cq data of the same miRNAs (a, b) and same cases (c, d) generated by CSF-miRNA panel were generally well correlated with that of inventory array cards
Detection of Tissue miRNAs Using CSF-miRNA Panel
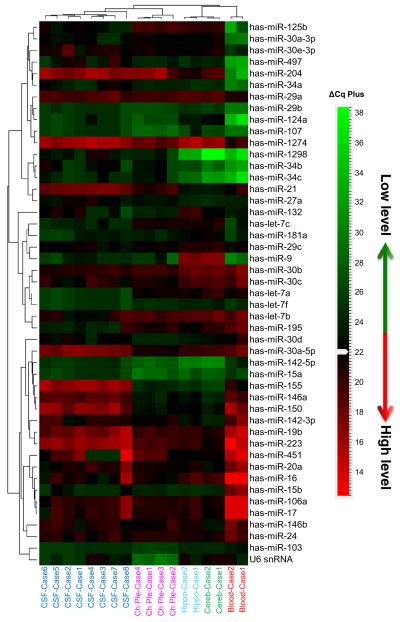
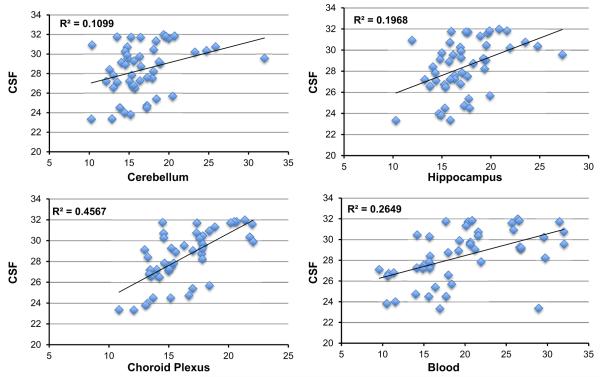
RNA samples derived from the human hippocampus (two cases), cerebellum (two cases), and choroid plexus (four cases) and snap-frozen at autopsy and clinical blood drawn in a clinic and then frozen (two cases) were subjected to CSF-miRNA panel TLDA analysis. Although total RNA is much more abundant in tissue compared with that in CSF and it does not require preamplification to obtain a good readout data from these tissue samples, the RT-qPCR analyses followed the same procedure as that for CSF including the preamplification step to maintain the technical consistency. Most miRNAs in the CSF-miRNA panels were detected in brain tissues and blood samples. As expected, brain-specific or enriched miRNAs in the CSF panel, such as miR-124a, miR-9, and miR-125a, were not detected or with very low levels in the blood samples (Suppl. file 3). Several miRNAs displayed similar expression patterns in CSF and the choroid plexus (Suppl. file 3). For example, miR-1298 was detected in CSF and the choroid plexus, but not detected in the blood and cerebellum and was very low in the hippocampus; miR-34b and miR-34c also had higher expression levels in CSF and the choroid plexus than those in other tested tissues. Neuronal enriched miR-124a has a very low level in both CSF and the choroid plexus, as well as in the blood samples as expected. Several inflammatory/immunity-related miRNAs, such as miR-21, miR-223, miR-150, miR-155, miR-146a, miR-142-5p, and miR-142-3p, were all detected in tested brain tissues, but had relatively high levels in CSF and the choroid plexus. In addition, miR-204 and miR-1274B seemed to have higher levels in all brain tissues including CSF. Hierarchical clustering (Pearson’s distance-Average Linkage) demonstrated that among the tested tissues, CSF miRNAs are clustered closely to the choroid plexus (Fig. 6) indicating a strong relationship to the organ that generates CSF. Correlation coefficient (R2) of miRNA expression pattern between CSF and various tissues (Fig. 7) also indicated a close correlation (R2 = 0.4567) between CSF and choroid plexus miRNA expression. A comparison of all the correlations between CSF and tissues showed that the correlation between miRNAs profiled in CSF and the choroid plexus were statistically significant (p = 0.019) [27]. The close correlation of CSF miRNA profile with the choroid plexus miRNA profile was further confirmed in a separate experiment which was carried out retrospectively after the initial evaluations. Using different CSF and tissue samples (Suppl. Fig. 3, Suppl. file 4), it was demonstrated that the CSF miRNA profile had a much stronger correlation coefficient with that of the choroid plexus (R2 = 0.5694) than with that of the cerebellum (R2 = 0.1583). The difference between these two correlations was statistically significant (p = 0.0005) [27].
Fig. 6.
Hierarchical clustering of miRNA expression in CSF, blood, and tissue RNA was analyzed using CSF-miRNA TLDA. Cq values of each array were normalized by global mean method. Hierarchical clustering of expression patterns was determined by Pearson’s distance-average linkage. The heatmap indicated that miRNA patterns of CSF were relatively similar to that of the choroid plexus, the organ that generates CSF. CSF, blood, hippocampus, and cerebellum samples are marked in the same colors
Fig. 7.
Correlation of miRNAs detected in CSF and tissues using CSF-miRNA panel CSF and tissue RNAs was analyzed using CSF-miRNA TLDA. Average Cq values of each sample group were plotted against each other, and the coefficient correlation (R2) of two data sets were obtained by linear regression
Discussion
Our study introduces a novel miRNA expression profiling platform designed for relatively efficient use in the context of CSF miRNA profiling. We recommend a technical pipeline that includes preanalytical parameters, normalization, and expression profile optimization. The TLDA returns results that are particularly tailored for the study of CSF miRNAs. We found for the first time that CSF miRNA profile is relatively closely correlated with the profile of miRNAs in the choroid plexus, in comparison to the samples of the hippocampus or cerebellum or blood that we evaluated. Since the TLDA is sensitive and characterized by relatively low technical variability and queries miRNAs that previously were shown to be relevant to neurodegenerative diseases, it could provide a robust platform for CSF analyses in conditions such as AD.
There are potential limitations to the present study. We underscore that there is no perfect miRNA profiling technique—because of the low levels of RNA in the CSF, there is a necessary tradeoff of sensitivity and technical variability. We previously have reported some of the technical differences that can be seen when data from different profiling platforms are compared [22, 29, 30]. Further, the use of TLDA cards lacks the ability of some other methods (such as RNA-Seq) to detect novel miRNAs, being limited to the 47 miRNAs on the array card. Yet the panel can be refined to include other newly discovered miRNA biomarkers when necessary. Finally, there are imperfections in the RNA amplification especially when the primers were multiplexed. For all the limitations of any given method, we note that there now have been a number of prior studies of CSF miRNAs and other biofluid miRNA detection methods were reported previously [13–19], providing incentive for the field to achieve a standardized approach.
Among the available miRNA profiling platforms, RT-qPCR is relatively sensitive and economically viable. This method is particularly useful when the RNA input is limited, such as in the case of CSF RNA. There are two distinct miRNA RT-qPCR platforms currently widely used in the field. TaqMan® miRNA RT-qPCR assay (Life Technologies) employs a sequence-specific stem-loop RT primer to generate miRNA cDNA template followed by qPCR using sequence-specific primers and probes for quantification. Another type of platform uses a universal tailing and reverse transcription reaction followed by qPCR with sequence-specific PCR primers. The first method proves to be more specific while the latter is more sensitive, but less specific [31]. We chose TaqMan® miRNA RT-qPCR assay (Life Technologies) as our detection platform for its high specificity. Here, we reported results using TaqMan® Array Human MicroRNA A and B cards to profile eight clinical (four controls and four ADs) CSF miRNAs.
Our qPCR array profiling detected 193 miRNAs, ~25% of miRNA species spotted on A and B cards (which tested a total 766 miRNAs). This is fewer than the detectable miRNAs obtained by Denk and the colleagues who profiled CSF miRNA using similar TaqMan® RT-qPCR-based OpenArray platform [19] which can profile more miRNAs at once (1178 miRNAs of OpenArray vs 766 of miRNA array Card A and B combined). However, most of the detectable miRNA in our profiling were also detected in OpenArray platform. The main purpose of this profiling was to provide necessary information with regard to detectability, abundance, and potential normalizers for the design of an optimized CSF-miRNA panel. Among the detectable miRNAs, several miRNAs, such as miR-1274B, miR-30b, and miR-30c, showed little variation from case to case and were each relatively abundant in CSF (Cq less than 30). These miRNAs were identified as potential normalizers in the panel. Notably, miR-1274B was also suggested as a potential normalizer by Denk et al. [19].
We conclude that TLDA-based PCR assays provide a relatively robust and sensitive platform for CSF miRNA analyses. Most prior studies have focused on “endpoints” rather than optimizing technical parameters. Here, we evaluated separate RNA isolation techniques and RNA carriers. The TLDA panel we propose incorporates the miRNAs highlighted as potentially important by prior studies and provides a context for further study. This constitutes a relatively high throughput assay that enables potential study design with greater numbers of clinical controls—including cognitively intact individuals with and without pathologic changes, as well as dementia controls with a variety of different conditions reflecting the common brain diseases of aging. Finally, we found it interesting that the miRNA profile of CSF more closely resembled (among the samples tested) those of the choroid plexus than those of either brain tissues or blood. MiRNAs are very powerful and promiscuous agents that regulate global gene expression. Our findings raise the provocative hypothesis that the choroid plexus can influence CNS cells via secretion of miRNAs into the CSF, a mechanism that could contribute to both healthy and diseased states in the brain.
Supplementary Material
Acknowledgements
Sincere thanks to the research subjects, clinicians, and staff at the UK-ADC. Funding was provided through NIH grants P30 AG028383, R01 AG 042419, and R21 NS085830.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12035-016-0316-2) contains supplementary material, which is available to authorized users.
References
- 1.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 2.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 7.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert SS, Nelson PT. Studying microRNAs in the brain: technical lessons learned from the first ten years. Exp Neurol. 2012;235:397–401. doi: 10.1016/j.expneurol.2011.12.004. doi: 10.1016/j.expneurol.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson PT, Wang WX, Wilfred BR, Tang G. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta. 2008;1779:758–765. doi: 10.1016/j.bbagrm.2008.03.012. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18:371–390. doi: 10.1111/jcmm.12236. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fourier A, Portelius E, Zetterberg H, Blennow K, Quadrio I, Perret-Liaudet A. Pre-analytical and analytical factors influencing Alzheimer’s disease cerebrospinal fluid biomarker variability. Clin Chim Acta. 2015;449:9–15. doi: 10.1016/j.cca.2015.05.024. doi: 10.1016/j.cca.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Burgos KL, Javaherian A, Bomprezzi R, Ghaffari L, Rhodes S, Courtright A, Tembe W, Kim S, et al. Identification of extra-cellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA. 2013;19:712–722. doi: 10.1261/rna.036863.112. doi: 10.1261/rna.036863.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bekris LM, Lutz F, Montine TJ, Yu CE, Tsuang D, Peskind ER, Leverenz JB. MicroRNA in Alzheimer’s disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013;18:455–466. doi: 10.3109/1354750X.2013.814073. doi: 10.3109/1354750X.2013.814073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zollner H, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 16.Muller M, Kuiperij HB, Claassen JA, Kusters B, Verbeek MM. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging. 2014;35:152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Haghikia A, Haghikia A, Hellwig K, Baraniskin A, Holzmann A, Decard BF, Thum T, Gold R. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79:2166–2170. doi: 10.1212/WNL.0b013e3182759621. doi: 10.1212/WNL.0b013e3182759621. [DOI] [PubMed] [Google Scholar]
- 18.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 19.Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer’s disease. PLoS One. 2015;10:e0126423. doi: 10.1371/journal.pone.0126423. doi: 10.1371/journal.pone.0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS, Abner EL, Markesbery WR. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt FA, Nelson PT, Abner E, Scheff S, Jicha GA, Smith C, Cooper G, Mendiondo M, et al. University of Kentucky Sanders-Brown healthy brain aging volunteers: donor characteristics, procedures and neuropathology. Curr Alzheimer Res. 2012;9:724–733. doi: 10.2174/156720512801322591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, et al. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang WX, Danaher RJ, Miller CS, Berger JR, Nubia VG, Wilfred BS, Neltner JH, Norris CM, et al. Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics. 2014;12:19–30. doi: 10.1016/j.gpb.2013.10.003. doi: 10.1016/j.gpb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 26.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou GY. Toward using confidence intervals to compare correlations. Psychol Methods. 2007;12:399–413. doi: 10.1037/1082-989X.12.4.399. doi: 10.1037/1082-989X.12.4.399. [DOI] [PubMed] [Google Scholar]
- 28.Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143. doi: 10.1093/nar/gkn725. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1:155–161. doi: 10.1038/nmeth717. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 31.Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA. 2012;3:601–616. doi: 10.1002/wrna.1120. doi: 10.1002/wrna.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.