Abstract
The need to overcome human threats from pathogenic microbes, development of nanomaterials have been provoked for a new generation of antimicrobials. In the present study, biosynthesis of silver nanoparticles (AgNPs) was acquired using Comamonas acidovorans extract within 72 h under static condition. Electron microscopy studies revealed that the size of AgNPs was ranging from 6–53 nm and had spherical, oval and irregular shapes with smooth surfaces. Prepared AgNPs interacted with proteins, carbohydrates and other aromatic molecules. Biosynthesized AgNPs were bactericidal, which significantly inhibited pathogenic microbes, i.e., Streptococcus pyogenes, Staphylococcus aureus and Escherichia coli. Higher concentrations of AgNPs (20 μg ml−1) inhibited 92–98% growth of all tested bacteria within 24 h. AgNPs–protein network studies carried out to recognize the protein interactions with AgNPs and to understand probable bactericidal mechanisms. AgNPs may penetrate into cell through membrane proteins and damage them by modifying amino acids. Due to AgNPs–protein interactions, dysfunctions in enzymes obstruct certain metabolic processes, which cause the bacteria to die eventually. In certain pathogenic microbes, cue and cus systems detoxify Ag+ ions, transport through transporter proteins and expel them to the extracellular space, which are mainly responsible for Ag resistance.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0728-3) contains supplementary material, which is available to authorized users.
Keywords: Bactericidal, Comamonas acidovorans, Computational approach, Nanoparticle–protein interaction, Silver nanoparticles
Introduction
In 2015, World Health Organization (WHO) published a report on global antimicrobial resistance surveillance system wherein several microbes, i.e., Escherichia coli, Staphylococcus aureus, Shigella sp., Klebsiella pneumoniae and Salmonella sp. are more highlighted due to their resistance towards one or several antibiotics as they are commonly present in communities and hospitals (World Health Organization 2015). As pathogenic microbes evolve certain machineries for resistance of antibiotics, the rate of antibiotic resistance was increased drastically in last decades (Blair et al. 2015). Due to this reason, the development of new antimicrobial agent becomes a challenging task for researchers. To figure out this problem, nano research is more concerned about the development of effective antibacterial agents (Pelgrift and Friedman 2013).
Since antiquity, silver has been used to prevent gonorrhoeal eye infections, to repair vaginal tears after childbirth and to avoid infection of surgical wounds (Lemire et al. 2013; Bhatia et al. 2016). Due to the large surface-area-to-volume ratio, a nano form of silver is catalytically more active than the bulk material. The nano form of silver is greatly noticed due to the high reactivity and broad antimicrobial spectrum with low doses (Rai et al. 2014). In addition, silver represents less toxicity towards humans at lower concentrations; thus, it has been widely incorporated with drugs under various forms (salts, immobilized ions or nanoparticles) (Le Ouay and Stellacci 2015). Biosynthesis of nanoparticles is an effective alternate that comprises clean, non-toxic and “green” procedures, while physical and chemical synthesis requires higher energy and toxic chemicals (Mohanpuria et al. 2008; Kharissova et al. 2013). Researchers comprehensively studied the physical, chemical and biological properties of biosynthesized AgNPs and confirmed as an efficient antiviral, antibacterial, antifungal, anticancer and anti-proliferative agent (Lara et al. 2011; Kathiravan et al. 2014; Muniyappan and Nagarajan 2014; Yehia and Al-Sheikh 2014; Abdel-Hafez et al. 2016). Several studies have been accomplished to comprehend the bactericidal mechanisms of AgNPs, which are as follows: (1) AgNPs inhibit the cell membrane and membrane proteins; subsequently, they destroy the cell membrane, (2) AgNPs strongly bind with negatively charged proteins and nucleic acids, (3) AgNPs block respiratory chains by binding with enzymes, and (4) AgNPs generate reactive oxygen species, which lead to the structural and functional changes in the cell (Lara et al. 2010; Rai et al. 2014).
Although the higher bactericidal activity of AgNPs was recorded, less information was obtained regarding AgNPs and its interactions with proteins present in Gram negative and Gram positive bacteria. It is necessary to understand the protein that interacted with AgNPs, interaction types and subsequent pathway that leads to the cell death in bacteria. Several pre-computed databases are available that provide the networks of protein–chemicals interactions (Kuhn et al. 2013) and functional annotations of proteins (Ashburner et al. 2000; Kanehisa and Goto 2000; Caspi et al. 2016). In our study, these databases are useful to understand the interactions between AgNPs and involved functional partners of pathogenic bacteria.
The present study is concentrated to evaluate the physico-chemical properties and bactericidal activity of biosynthesized AgNPs. To the best of our knowledge, this is the first attempt, which represents AgNPs–protein interaction network studies in pathogenic bacteria.
Materials and methods
Bacterial strains and chemicals
Comamonas acidovorans (3364) was procured from Microbial Type Culture Collection and Gene Bank, Institute of Microbial Technology (Chandigarh, India). All bacterial strains were maintained and sub-cultured after every week on nutrient agar. Silver nitrate (AgNO3) was procured from Sigma-Aldrich Co. (St. Louis, USA). Agar–Agar and bacteriological media were purchased from Hi-media Labs (Mumbai, India).
Preparation of AgNPs using cell-free extract
The culture of C. acidovorans was grown in nutrient broth at 30 °C for 48 h. Cell-free extract was acquired by centrifuging the grown culture at 10,000 rpm and 4 °C for 15 min in sterile tubes. Cell-free extract was separately added into a sterile flask containing an equal amount of sterile AgNO3 (1 mM). The reduction AgNO3 was incubated for 72 h at room temperature under static condition. Additionally, cell-free extract and AgNO3 samples also incubated individually for 72 h at room temperature to observe color change in the samples. The sample of reaction mixture withdrew after every 12 h, and absorption spectrum of each sample was investigated using UV–Vis spectrophotometer (Shimadzu UV 1800, Japan) in the range of 300–700 nm at a resolution of 2 nm. AgNPs pellet was recovered by centrifuging (15,000 rpm) it for 20 min and repetitively washing with distilled water. After, being air dried centrifuged pellet at 40 °C for 24 h, AgNPs powder proceed for characterization study.
Characterization of AgNPs
The shape and size of AgNPs were detected under transmission electron microscope. To prepare the AgNPs sample for TEM, copper grid (300 mesh size) was dipped in AgNPs solution and dried in air for 10 min. Further, the sample was proceeded for TEM analysis (Tecnai T20, Philips) at 200.0 keV. The surface morphology and size of AgNPs were observed using field emission gun scanning electron microscopy (FEG-SEM) under vacuum (5 × 10−3 Pa) at 20.0 kV with 150,000× magnification. Traces of metals in cell-free extract and AgNPs solution were detected in energy-dispersive X-ray analysis (EDXA) (Nano Nova SEM 450, USA). Crystalline nature of AgNPs powder was detected by X-ray diffractometer (Philips Xpert MPD, Holland). The full widths at half maximum and 2θ values were used in Debye–Scherrer formula to calculate the average crystalline size of AgNPs. The interaction between functional groups of cell-free extract with AgNPs was analysed using FTIR spectroscopy (Spectrum GX, Perkin Elmer, USA). Dried powder (2 mg) of bacterial extract and AgNPs individually mixed with KBr crystals (150 mg) and compressed to form pellet which further analysed in mid-infrared light region of 400–4000 cm−1.
Antibacterial susceptibility assessment
AgNPs were examined for their effect on Gram-negative and Gram-positive pathogenic microbes using well diffusion and growth curve method. In the well diffusion method, AgNPs solution (10 μg ml−1) was compared with AgNO3 (10 μg ml−1) and bacterial supernatant against pathogenic bacteria. The zone of inhibition was measured after 24 h of incubation. In the growth curve method, control and AgNPs solution (5, 10, 15, 20 µg ml−1) containing all bacterial culture was incubated for 24 h to evaluate % inhibition efficacy.
AgNPs–protein interaction study
In silico nanoparticle–protein interaction approach was used to understand the possible interaction mechanism. Twelve Gram-negative and six Gram-positive pathogenic bacterial strains were used for interaction study. STITCH 4.0, a pre-computed database resource, offers a protein–chemical interaction network that incorporates many experimental sources and manually curated evidence with text-mining information and predictions (Kuhn et al. 2013). STITCH score is based on gene ontology, which is grouped by p-value. The annotations of p value and gene ontology were used to understand the biological process, cellular component and molecular function involved in AgNPs interaction using MetaCyc (Caspi et al. 2016). Along with STITCH 4.0, several databases such as KEGG, Transporter Classification Database and Gene Ontology, provide valuable information regarding functional partners and functional annotations (Kanehisa and Goto 2000; Ashburner et al. 2000; Saier et al. 2006).
Results and discussion
Synthesis of AgNPs using cell-free extract
The primary investigation of AgNPs formation was confirmed using visual color change and UV–visible spectroscopy. The color of the reaction mixture gradually altered from pale yellow to dark brown that confirmed the formation of AgNPs after 72 h of incubation. Figure 1 represents the sharp surface plasma resonance peak that evolved at 450 nm after 24 h of incubation. It is a representative characteristic of AgNPs formation. A broad and strong surface plasma resonance peak was observed between 445 to 455 nm after 72 h of incubation. There was no color alteration and absorbance peak observation at 450 nm in the individual solution of cell-free extract and AgNO3 after 72 h of incubation. Similar results were observed by Jeevan et al. (2012) when AgNPs were synthesized using the supernatant of P. aeruginosa culture for 72 h under bright condition. Incubation of AgNO3 with M. dubia leaf extract showed the major absorbance peak at 448 nm that confirmed the formation of AgNPs (Kathiravan et al. 2014).
Fig. 1.
UV–visible spectrum of the AgNPs represented the maximum absorbance peak observed at 450 nm after 72 h and color of bacterial supernatant changed from light yellow to dark brown (inset)
Characterization of AgNPs
The preparation of AgNPs is further confirmed using microscopic, spectroscopic and diffractometric analysis. TEM micrographs display the different size and shape of AgNPs at 50 and 100 nm scale as shown in Fig. 2. The sample of nanoparticles exhibited a difference in their size from 6 to 53 nm. The majority of nanoparticles are spherical, while others are oval-shaped (Fig. 2b, inset). Irregular and larger sized nanoparticles are also visualized in Fig. 2a. Size of spherical and oval-shaped nanoparticles was ranging from 6 to 30 nm, while irregular-shaped particles were comparatively larger (30–52 nm). The selected-area electron diffraction (SAED) pattern of AgNPs detected the plane of nanoparticles (Fig. 2a, inset). The AgNPs were magnified up to 150,000× to observe the surface area and shape of nanoparticles under FEG-SEM. These nanoparticles, ranging from 7 to 50 nm, were visualized with oval, spherical and irregular shapes (Fig. 3a).
Fig. 2.

TEM micrographs of AgNPs at 100-nm (a) and 50-nm scale (b), enlarged view of oval- and spherical-shaped AgNPs (b; inset) and SAED pattern of biosynthesized nanoparticles (a; inset)
Fig. 3.
High-resolution FEG-SEM images of AgNPs at ×150,000 magnification (a), the presence of Ag metal confirmed by EDAX analysis (a, inset) and FTIR images of bacterial supernatant and AgNPs (b)
FEG-SEM and TEM measurements are frequently used to determine the shape, size and morphology of the nanoparticles. In the present study, these techniques confirmed that the prepared nanoparticles had smooth surface edges but they were in different shapes. Less-homogenized spherical AgNPs were synthesized by culture supernatant of S. hygroscopicus (Sadhasivam et al. 2010). The extract of B. amyloliquefaciens was used to synthesize AgNPs, which were spherical and triangular shaped (Wei et al. 2012). Different shapes of nanoparticles such as spherical, oval, triangular and irregular observed when they prepared using biological methods. Sedimentation has been occurring in the late stages of AgNPs formation; so, nanoparticles were agglomerated. This might be the possible reason for the synthesis of large nano-spherical nanoparticles (Ahmad et al. 2013).
The appearance of silver in EDAX spectrum confirmed the presence of Ag metal in AgNPs solution as shown in Fig. 3a (inset). Weight (%) of major element signals of C (63.96), O (5.83), and Na (0.21) was detected in the cell-free extract. The presence of these elements was expected, as they were the major components of bacterial extract. Weights (%) of Ag (21.25), O (49.59), Na (28.24), and P (0.93) were observed in AgNPs powder. The optical absorption peak observed at 3 kV is the representative peak of pure metallic silver nanoparticles due to surface plasma resonance (Jain et al. 2011). Stoichiometric study revealed the higher relative content of Ag+ ions present in AgNPs powder. The XRD pattern revealed the crystalline nature of AgNPs using the peaks at 2θ value of 32.31, 38.54, 45.80, and 46.97. The size of AgNPs at different position is 26.60, 32.59, 51.10 and 52.00 nm. The average crystalline size of AgNPs calculated using Debye–Scherrer’s equation is 40.57 nm. Several researchers studied the average size of AgNPs synthesized using culture supernatant of P. aeruginosa were 50–80 nm. An average size of AgNPs prepared in the present study is 40 nm, which is lower than the previous studies (Jeevan et al. 2012; Jeyaraj et al. 2013).
FTIR measurements were carried out to identify the functional groups involved in the AgNPs reduction (Fig. 3b). Characteristics of FTIR spectra and the shift changes in AgNPs powder is presented in Table 1. The peaks near 3437 cm−1 are attributed to N–H stretching vibrations of a peptide linkage in bacterial extract and AgNPs powder (Kumar and Mamidyala 2011). Bacterial extract and AgNPs powder showed the prominent peak of asymmetrical and symmetrical C–H stretching vibrations (CH3) of carbohydrates and fatty acids at 2930 and 2926 cm−1. The other peaks of carbohydrates and fatty acids were observed at 2856 cm−1 supporting the weak C–H stretching vibrations of aldehyde group. The interaction of the proteins with AgNPs was confirmed by the presence of peaks of carbonyl groups (C=O) and C–N plane vibrations of amines at 1645 and 1639 cm−1 (Jeevan et al. 2012). There are two bands at 1639 and 1544 cm−1 that corresponded to amide I and II bonds of proteins, respectively (Jain et al. 2011). The peaks at 1456 and 1458 cm−1 corresponded to C–H deformation of aliphatic compounds present in carbohydrates and fatty acids. The peaks at 1401 and 1403 cm−1 were assigned to the geminal methyls (Vidhu and Philip 2014). A C–N stretching vibration of primary aliphatic amines evolved at 1119 cm−1 by interacting with Ag+ ions.
Table 1.
Fourier-transform infrared spectral characteristics of bacterial extract and AgNPs powder
| No. | Description | Wavenumber (cm−1) | Differencea | Possible reason for the shift alteration | |
|---|---|---|---|---|---|
| Bacterial extract | AgNPs powder | ||||
| 1 | N–H stretching vibrations of peptide linkage | 3437.83 | 3438.30 | +0.47 | Interaction with proteins |
| 2 | Asym and sym C–H stretching vibration of aliphatics | 2930.32, 2963.11 |
2926.63 | −3.69 | Interaction with fatty acids and carbohydrates |
| 3 | Carbonyl group (C=O), amide I group stretching vibrations | 1645.37 | 1639.15 | −6.22 | Binding with proteins |
| 4 | Amide II group stretching vibrations | 1547.55 | 1544.91 | −2.64 | Interaction with proteins |
| 5 | C–H deformation of CH2 or CH3 in aliphatics | 1456.23 | 1458.49 | +2.26 | Interaction with fatty acids and carbohydrates |
| 6 | Geminal methyls | 1403.54 | 1401.31 | −2.23 | Interaction with methyls |
| 7 | Amide III group stretching vibrations | 1241.89 | 1247.71 | 5.82 | Interaction with proteins |
| 8 | C–N stretching vibrations of primary aliphatic amines | − | 1119.28 | − | Interaction with proteins |
| 9 | P=O stretching in phospholipids and C=O group in polysaccharides | 1080.71 | 1076.26 | −4.45 | Interactions with lipids and carbohydrates |
| 10 | C–O–C and P–O–C bond stretching | 536.77 | 554.70 | +17.93 | Strong interactions of phospholipids, aromatics and sugars |
Asym asymmetrical, sym symmetrical
aShift difference: shift increase +, shift decrease −
The peak of R–CO–NH2 stretching vibrations near 1080 cm−1 revealed the presence of proteins in bacterial extract and AgNPs powder (Jain et al. 2011). A peak of CH=CH2 bond of vinyl compounds evolved at 931 cm−1 by interacting with Ag+ ions. The peak for methylene rocking vibration and out-of-plane C–H bending vibration in mononuclear hydrocarbon was assigned at 712 cm−1 that displayed in the bacterial extract. The peaks for C–O–C and P–O–C groups in phospholipids, aromatics, amino acids (rocking vibrations) and ketones are assigned at 554 and 536 cm−1. An alteration in these peaks (+17.93 cm−1) suggested the strong interaction of aromatics, lipids, amino acids as well as ketones. FTIR study revealed the presence and binding of protein, carbohydrates and aromatics and fatty acids with AgNPs. It is well known that the proteins bind with nanoparticles through cysteine residues or free amino groups (Jeevan et al. 2012). Electrostatic interactions of negatively charged carboxyl groups in proteins also bind with nanoparticles. These biological components may play a significant role in the formation and stabilization of nanoparticles (Wei et al. 2012; Ahmad et al. 2013).
Antibacterial susceptibility assessment
Bactericidal activities of biosynthesized AgNPs revealed that the inhibition was comparatively higher than AgNO3. It inhibited Gram-positive and Gram-negative bacteria very efficiently (Table 2). The inhibition zone of AgNPs was higher in the case of E. coli (22 mm) and S. pyogenes (17.5 mm). AgNPs also inhibited other bacteria, such as S. aureus (17.2 mm), P. aeruginosa (16.7 mm), S. typhi (15.7 mm) and B. subtilis (15.0 mm). Bacterial extract and distilled water were taken as negative controls which did not obtain inhibition zone. The inhibition zone of AgNPs synthesized using S. hygroscopicus observed in case of E. coli and B. subtilis was lower than the present study (Sadhasivam et al. 2010). AgNPs–layered double hydroxide also displayed the similar inhibition efficacy in the case of S. aureus and E. coli (Mishra et al. 2013). Similarly, mean diameters of inhibition zone for AgNPs (average TEM size; 30 nm) against E. coli and S. aureus were 24 and 19 nm, respectively (Ahmad et al. 2013).
Table 2.
Antibacterial activity of AgNPs (10 μg ml-1) and AgNO3 against pathogenic bacteria
| Gram-negative bacteria | Zone of inhibition (mm) | Gram-positive bacteria | Zone of inhibition (mm) | ||
|---|---|---|---|---|---|
| AGNPs | AgNO3 | AGNPs | AgNO3 | ||
| Escherichia coli | 22.0 | 14.0 | Streptococcus pyogenes | 17.5 | 12.4 |
| Psuedomonas aeruginosa | 16.7 | 11.2 | Staphylococcus aureus | 17.2 | 11.9 |
| Salmonella typhi | 15.7 | 10.5 | Bacillus subtilis | 15.0 | 10.8 |
The inhibition profile against pathogenic bacteria was remarkably detected in liquid condition than on agar medium. E. coli, S. aureus and P. aeruginosa were inhibited at 60.39 ± 1.19, 51.49 ± 1.27 and 49.61 ± 1.02%, respectively, after 24 h at even 5 µg ml−1 concentration of AgNPs. All tested Gram-negative and Gram-positive bacteria were highly susceptible to AgNPs at 20 µg ml−1 concentration within 24 h of incubation. The synthesized AgNPs highly decreased the growth of pathogenic bacteria, i.e. E. coli (98.08 ± 1.11%), P. aeruginosa (94.55 ± 0.99%), S. typhi (88.49 ± 1.34%), S. pyogenes (96.66 ± 1.23%), S. aureus (86.48 ± 1.10%), and B. subtilis (90.38 ± 1.54%) within 24 h (Fig. 4). The growth of all bacteria increased marginally after 24–30 h of incubation even in the presence of AgNPs at concentrations of 5–15 μg ml−1. Similar results were observed by Shivaji et al. (2011), when higher concentrations (10 and 20 µg ml−1) of AgNPs completely inhibited Gram positive and negative bacteria within 20 h. The inhibitory action of prepared AgNPs was effective against S. aureus after 24 h than the study carried out by Li et al. (2011).
Fig. 4.
Bactericidal activity of AgNPs evaluated using growth curve method against six bacteria, i.e., Escherichia coli (a), Pseudomonas aeruginosa (b), Salmonella typhi (c), Staphylococcus aureus (d), Streptococcus pyogenes (e), and Bacillus subtilis (f) at 5–20 μg ml−1 concentration
AgNPs–protein interaction study
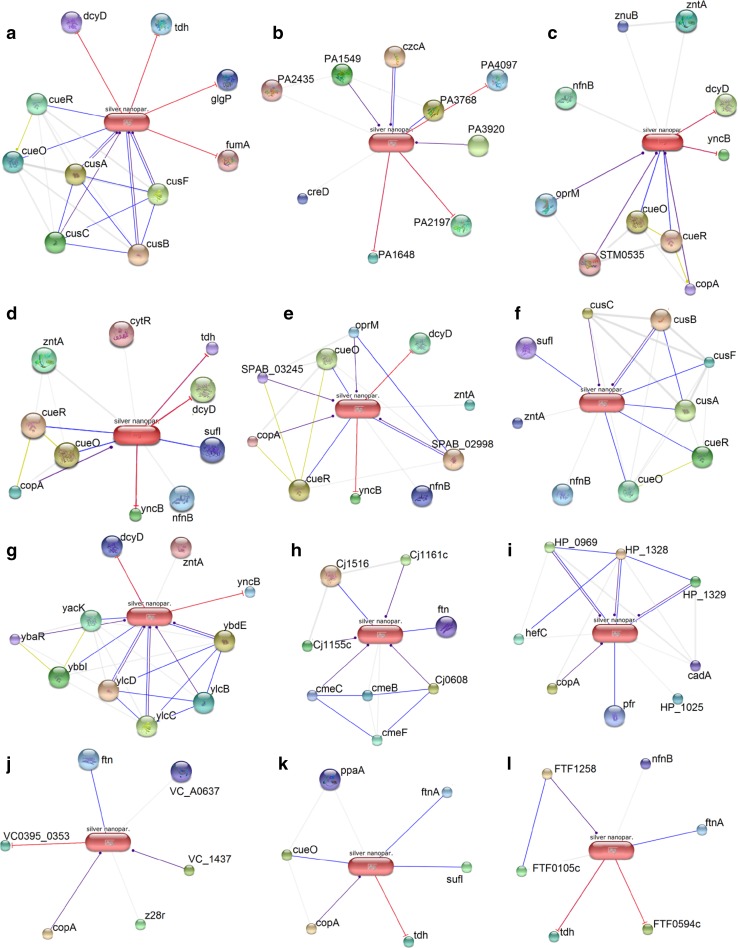
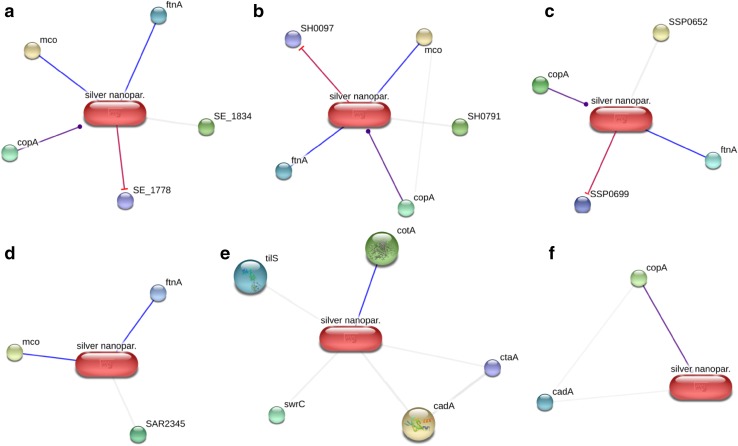
To understand the involvement of functional partners with AgNPs, STITCH 4.0 tool and Gene Ontology provide a better understanding about the confidence score, biological process and molecular functions of proteins (Table 3 and Table 4; Electronic supplementary material). In all pathogenic bacteria, AgNPs bind with the membranous, cytoplasmic and cell periphery proteins. Figures 5 and 6 represent the AgNPs–protein interaction networks of Gram-negative and Gram-positive bacteria, respectively. The confidence score of these functional partners is more than 0.4.
Fig. 5.
AgNPs–protein interaction networks of 12 Gram-negative pathogenic bacteria such as Escherichia coli (a), Pseudomonas aeruginosa (b), Salmonella typhimurium (c), Salmonella typhi (d), Salmonella enterica (e), Klebsiella pneumoniae (f), Shigella sonnei (g), Campylobacter jejuni (h), Helicobacter pylori (i), Vibrio cholerae (j), Proteus mirabilis (k) and Francisella tularensis (l). An individual line between protein and AgNPs represents an interaction node wherein blue line for binding, purple line for catalysis, light green line for expression and red line for inhibition
Fig. 6.
AgNPs–protein interaction networks of six Gram-positive pathogenic bacterial strains like Staphylococcus epidermidis (a), Staphylococcus haemolyticus (b), Staphylococcus saprophyticus (c), Staphylococcus aureus (d), Bacillus subtilis (e) and Streptococcus pyogenes (f). An individual line between protein and AgNPs represents an interaction node wherein blue line for binding, purple line for catalysis and interaction block represented as red line, which shows inhibition
In case of E. coli, all functional partners share the highest confidence score (0.9–1.0). The cus and cue systems respond to copper (Cu) stress by oxidizing and/or expelling Cu+ ions from the cell. Ag+ ion, a mimic of Cu+ ion, acts on the cus as well as cue systems (Singh et al. 2011). CusA, CusB, CusC and CusF are membrane proteins and major components of Ag/Cu efflux system, which may be responsible for Ag resistance in the cell. All functional partners are involved in binding and catalysis of Ag+ ions and also transport Ag+ ions [GO:0010272], which results the expel Ag+ ions from the cell. CueO (multi-copper oxidase) is regulated by CueR (DNA binding transcriptional activator of Cu-responsive regulon genes) and responsible to decrease stress levels of Ag. CueO enzyme binds and transports Ag+ ions through methionine-rich motifs (Singh et al. 2011). The p values for the GO terms of these proteins are statistically significant (p value ≤0.05) as obtained from STRING database. Ag+ ions also inhibit different enzymes such as fumA, glgP, dcyD and tdh, which are involved in citric acid cycle [GO:0006099], starch and sucrose catabolism [GO:0005977], cysteine catabolism [GO:0019447], and threonine catabolism [GO:0006567]. The amino acids, carbohydrates as well as routine metabolic pathways disturb due to the binding of Ag+ ions obstruct the enzyme activities. Experimental data presented by Li et al. (2010) revealed that the dehydrogenase activities of E. coli were rapidly decreased after incubating with AgNPs.
Alcohol dehydrogenase (PA4097) and oxidoreductase (PA1648) were inhibited because of interacting Ag+ ions in P. aeruginosa PAO1. Inhibition of certain cytoplasmic enzymes may lead to discontinue cellular processes, i.e., glycolysis, fatty acid degradation, amino acid metabolism, secondary metabolite synthesis, etc. Metal-ion-transporting ATPases (PA1549, PA3920), cation efflux transporter (czcA) and metallo-oxidative reductase (PA3768) bind with AgNPs, which are responsible for cation transporter activity [GO:0006812] and oxidoreductase activity [GO:0016491]. Kora and Arunachalam (2011) showed the involvement of reactive oxygen species in P. aeruginosa that may generate from the surface of AgNPs and damage cell membrane and membrane proteins.
The functional partners and interaction networks of S. typhimurium, S. typhi, and S. enterica are similar that interact with AgNPs. Ag+ ions bind with dcyD, tdh and yncB (NADP-dependent oxidoreductase) enzymes and obstruct the activities in the cytoplasm. yncB possesses the oxidoreductase activity [GO:0055114] in the cytoplasm. Ag+ ions bind with cueO enzyme, which transports Ag+ ions through methionine-rich motifs (Singh et al. 2011). Ag+ ion-transporting process [GO:0019829] is acted by copA in Salmonella sp. Interaction networks and functional partners of K. pneumoniae 342 and E. coli display higher similarity. Alike E. coli, K. pneumoniae 342 has cus and cue system, which detoxifies and expels Ag+ ions to extracellular space.
Majority of the functional partners present in H. pylori G27 are membranous proteins and enzymes that catalyze Ag+ ions and play important role in Ag resistance. HP_1328, HP_0969, and HP_1329, are membranous proteins and part of the cation efflux system. The efflux protein system is also involved in the resistance of certain metal ions, i.e. Ag, Co, Zn and Cd. Gurunathan et al. (2015) showed the possible bactericidal mechanism of AgNPs in H. pylori wherein increase in reactive oxygen species (ROS), DNA fragmentation, membrane leakage and mitochondria-mediated apoptosis were observed (Gurunathan et al. 2015). Alike H. pylori G27, the functional partners of C. jejuni 11168 are present in membrane. Cj0608 (efflux protein) is present in outer membrane; Cj1516 (periplasmic oxidoreductase) is present in periplasmic space of membrane; and Cj1161c (ATPase), Cj1155c (ATPase) and cmeC (multidrug resistance cmeABC protein) are present in inner side of the membrane. All proteins are involved in Ag resistance in C. jejuni 11168. In case of S. sonnei, yncB and dcyD are inhibited by Ag+ ions, which obstructed oxidoreductase processes and cysteine catabolism. The ylcD, ylcB and ylcC (Cu/Ag efflux system protein) detoxify and transport Ag+ ions to extracellular space. Ag+ ions also inhibit the transcription regulator (ybbI) and stop the transcription activity in S. sonnei.
In case of F. tularensis FSC198, outer membrane efflux protein (FTF1258) could absorb and expel Ag+ ions into extracellular space. Similar to E. coli, Ag+ ions inhibit tdh (threonine 3-dehydrogenase) and obstruct threonine catabolism [GO:0006567]. Binding to ftnA (ferritin family protein) with Ag+ ions may result the interruption in iron homeostasis [GO:0006879]. P. mirabilis shows similar network interactions as tdh and ftnA are inhibited by binding with Ag+ ions. The copA (Cu-transporting P-type ATPase) may act Ag+ ion-transporting activity [GO:0019829], while cueO may be involved in detoxification of Ag+ ions [GO:0010273]. In V. cholerae O395, VC_1437 and copA are cation-transporting E1-E2 family ATPases that catalyze Ag+ ions and show metal-ion-transporting ATPase activity [GO:0019829]. VC0395_0353 (threonine 3-dehydrogenase) and ftnA are inhibited by binding with Ag+ ions.
S. epidermidis 12228, S. haemolyticus, S. saprophyticus, and S. aureus MRSA252 share similar interaction networks with AgNPs. Quinone oxidoreductase (SE_1778), RND superfamily resistance–nodulation–cell division acriflavin: Proton H+ antiporter (SH0097) and NADP-dependent oxidoreductase (SSP0699) inhibit due to the presence of Ag+ ions in the cytoplasm. Based on p value (≤0.05), SE_1778 and SSP0699 exhibit oxidoreductase activity [GO:0016491] as well as SH0097 shares transmembrane transporter activity [GO:0015450]. Multi-copper oxidase (mco) in Staphylococcus sp. acts as oxidation–reduction process [GO:0016491] which is similar to CueO present in Gram-negative bacteria. Binding of mco with Ag+ ions may result into the transportation and detoxification of Ag+ ions. copA catalyzes Ag+ ions into the cytoplasm, which results in cation-transporting ATPase activity [GO:0019829] and expelling of Ag+ ions from cytoplasm to the extracellular space. Ag+ ions interrupt iron homeostasis [GO:0006879] by binding with ftnA. Gordon et al. (2010) revealed that the enzymes present in the respiratory chain, glycolysis, citric acid cycle and iron homeostasis interrupted by incubating silver coordination polymers with Staphylococci. Grigor’eva et al. (2013) showed the accumulation of AgNPs into periplasmic space and the lysis of cytoplasm observed which result in the death of S. aureus.
In B. subtilis, Ag+ ions bind with cotA (Spore Cu-dependent laccase), which is mainly responsible for oxidoreductase activity [GO:0016491] and cell differentiation process [GO:0030435]. Binding of cotA with Ag+ ions may result into transportation and detoxification of Ag+ ions or it may hinder the process of cell differentiation. The copA is only a probable binding interaction observed in S. pyogenes. This enzyme is responsible for Cu/Ag tolerance in both aerobic and anaerobic conditions which mainly transport Ag+ ions [GO:1902601], response to Ag+ ions [GO:0071292] and detoxification of Ag+ ions [GO:0010273]. Overall, the interaction networks presented in Gram negative bacteria are higher than Gram positive bacteria. Less predicted interaction is observed due to the lack of exact inhibition mechanism in Gram positive bacteria.
Previously, researchers studied the bactericidal mechanisms of AgNPs using certain microscopy and spectroscopy techniques. In addition, several studies have attempted to understand the inhibition of cytoplasmic, membranous and extracellular enzymes in specific bacterial strains (Sadhasivam et al. 2010; Li et al. 2011; Singh et al. 2011). The AgNPs–protein interaction approach gives us better understanding regarding the proteins involved in bactericidal mechanism. Due to this approach, we can compare the functional partners present in pathogenic bacteria and also extract the significant information.
As a whole, adaptation and/or resistance mechanisms of AgNPs are observed in several bacterial strains. The expression and regulation of certain proteins, i.e. metal-uptake proteins, efflux proteins, and membrane transporters may cause Ag resistance in bacteria (Lemire et al. 2013). Cus and cue protein systems are actively involved functional partners that are present in most pathogenic bacterial strains (Singh et al. 2011). Though certain Ag resistance systems are observed in bacterial strains, AgNPs work effectively as well as in a systematic manner. Anchoring ability of AgNPs with bacterial cell wall leads to the penetration into cell via transporters and/or co-transport with low–molecular-weight ligands (Rai et al. 2009). After entering into cells, it is supposed that there can be release of Ag+ ions from AgNPs, which are more active and unstable in the cell. Ag+ ions can lead to the production of reactive oxygen species (Kora and Arunachalam 2011). They interact with thiol groups of proteins and enzymes that caused the enzyme dysfunction. Certain metal ion ligand, i.e. Cu+ ion present in copA may be replaced by Ag+ ion, which can lead to the dysfunction of the enzyme (Lemire et al. 2013). Several studies suggested that Ag+ ions also interacted with DNA, which caused the fragmentation of DNA (Gurunathan et al. 2015). Eventually, AgNPs react with multiple enzymes, transporter proteins as well as nucleic acids that lead to the cell death.
Conclusion
Overall, an easy, biological and non-toxic method applied for the preparation of AgNPs using C. acidovorans culture. Even though the prepared AgNPs were small in size and of different shapes, reduction time for preparation was limited to 72 h. Proteins, sugars and other molecules are interacted with AgNPs that might stabilize Ag+ ions. The biosynthesized AgNPs were an efficient bactericidal agent against pathogenic bacteria. Predicted nanoparticles interactions revealed the possible binding of AgNPs with regulators, enzymes and transporter molecules. AgNPs–protein interaction study suggests that AgNPs may damage the cell membrane, pass through certain membrane proteins and transporters and inhibit the activity of cytoplasmic and membranous enzymes, which cause pathogenic bacteria to die in due course.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge SICART, Vallabh Vidyanagar, Anand, for providing the infrastructure, laboratory, instrumental and other necessary facilities for the successful completion of this work.
Compliance with ethical standards
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
Authors do not declare any conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0728-3) contains supplementary material, which is available to authorized users.
References
- Abdel-Hafez SI, Nafady NA, Abdel-Rahim IR, Shaltout AM, Daròs JA, Mohamed MA. Assessment of protein silver nanoparticles toxicity against pathogenic Alternaria solani. 3. Biotech. 2016;6:199. doi: 10.1007/s13205-016-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T, Wani IA, Manzoor N, Ahmed J, Asiri AM. Biosynthesis, structural characterization and antimicrobial activity of gold and silver nanoparticles. Colloid Surf B. 2013;107:227–234. doi: 10.1016/j.colsurfb.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia D, Mittal A, Malik DK. Antimicrobial activity of PVP coated silver nanoparticles synthesized by Lysinibacillus varians. 3. Biotech. 2016;6:196. doi: 10.1007/s13205-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon O, Slenters TV, Brunetto PS, et al. Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrob Agents Ch. 2010;54:4208–4218. doi: 10.1128/AAC.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigor’eva A, Saranina I, Tikunova N, Safonov A, Timoshenko N, Rebrov A, Ryabchikova E. Fine mechanisms of the interaction of silver nanoparticles with the cells of Salmonella typhimurium and Staphylococcus aureus. Biometals. 2013;26:479–488. doi: 10.1007/s10534-013-9633-3. [DOI] [PubMed] [Google Scholar]
- Gurunathan S, Jeong JK, Han JW, Zhang XF, Park JH, Kim JH. Multidimensional effects of biologically synthesized silver nanoparticles in Helicobacter pylori, Helicobacter felis, and human lung (L132) and lung carcinoma A549 cells. Nanoscale Res Lett. 2015;10:1. doi: 10.1186/s11671-015-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Bhargava A, Majumdar S, Tarafdar JC, Panwar J. Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus NJP08: a mechanism perspective. Nanoscale. 2011;3:635–641. doi: 10.1039/C0NR00656D. [DOI] [PubMed] [Google Scholar]
- Jeevan P, Ramya K, Rena AE. Extracellular biosynthesis of silver nanoparticles by culture supernatant of Pseudomonas aeruginosa. Indian J Biotechnol. 2012;11:72–76. [Google Scholar]
- Jeyaraj M, Varadan S, Anthony KJP, Murugan M, Raja A, Gurunathan S. Antimicrobial and anticoagulation activity of silver nanoparticles synthesized from the culture supernatant of Pseudomonas aeruginosa. J Ind Eng Chem. 2013;19:1299–1303. doi: 10.1016/j.jiec.2012.12.031. [DOI] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiravan V, Ravi S, Ashokkumar S. Synthesis of silver nanoparticles from Melia dubia leaf extract and their in vitro anticancer activity. Spectrochim Acta A. 2014;130:116–121. doi: 10.1016/j.saa.2014.03.107. [DOI] [PubMed] [Google Scholar]
- Kharissova OV, Dias HR, Kharisov BI, Pérez BO, Pérez VMJ. The greener synthesis of nanoparticles. Trends Biotechnol. 2013;31:240–248. doi: 10.1016/j.tibtech.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Kora AJ, Arunachalam J. Assessment of antibacterial activity of silver nanoparticles on Pseudomonas aeruginosa and its mechanism of action. World J Microb Biot. 2011;27:1209–1216. doi: 10.1007/s11274-010-0569-2. [DOI] [Google Scholar]
- Kuhn M, Szklarczyk D, Pletscher-Frankild S, et al. STITCH 4: integration of protein–chemical interactions with user data. Nucleic Acids Res. 2013;42:D401–D407. doi: 10.1093/nar/gkt1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CG, Mamidyala SK. Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloid Surface B. 2011;84:462–466. doi: 10.1016/j.colsurfb.2011.01.042. [DOI] [PubMed] [Google Scholar]
- Lara HH, Ayala-Nunez NV, Turrent LCI, Padilla CR. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J Microb Biot. 2010;26:615–621. doi: 10.1007/s11274-009-0211-3. [DOI] [Google Scholar]
- Lara HH, Garza-Trevino EN, Ixtepan-Turrent L, Singh DK. Silver nanoparticles are broad-spectrum bactericidal and virucidal compounds. J Nanobiotechnology. 2011;9:30. doi: 10.1186/1477-3155-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ouay B, Stellacci F. Antibacterial activity of silver nanoparticles: a surface science insight. Nano Today. 2015;10:339–354. doi: 10.1016/j.nantod.2015.04.002. [DOI] [Google Scholar]
- Lemire JA, Harrison JJ, Turner RJ. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11:371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- Li WR, Xie XB, Shi QS, Zeng HY, You-Sheng OY, Chen YB. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biot. 2010;85:1115–1122. doi: 10.1007/s00253-009-2159-5. [DOI] [PubMed] [Google Scholar]
- Li WR, Xie XB, Shi QS, Duan SS, Ouyang YS, Chen YB. Antibacterial effect of silver nanoparticles on Staphylococcus aureus. Biometals. 2011;24:135–141. doi: 10.1007/s10534-010-9381-6. [DOI] [PubMed] [Google Scholar]
- Mishra G, Dash B, Pandey S, Mohanty PP. Antibacterial actions of silver nanoparticles incorporated Zn-Al layered double hydroxide and its spinel. J Environ Chem Eng. 2013;1:1124–1130. doi: 10.1016/j.jece.2013.08.031. [DOI] [Google Scholar]
- Mohanpuria P, Rana NK, Yadav SK. Biosynthesis of nanoparticles: technological concepts and future applications. J Nanopart Res. 2008;10:507–517. doi: 10.1007/s11051-007-9275-x. [DOI] [Google Scholar]
- Muniyappan N, Nagarajan N. Green synthesis of silver nanoparticles with Dalbergia spinosa leaves and their applications in biological and catalytic activities. Process Biochem. 2014;49:1054–1061. doi: 10.1016/j.procbio.2014.03.015. [DOI] [Google Scholar]
- Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliver Rev. 2013;65:1803–1815. doi: 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Rai M, Kon K, Ingle A, Duran N, Galdiero S, Galdiero M. Broad-spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects. Appl Microbiol Biot. 2014;98:1951–1961. doi: 10.1007/s00253-013-5473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhasivam S, Shanmugam P, Yun K. Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms. Colloid Surface B. 2010;81:358–362. doi: 10.1016/j.colsurfb.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Saier MH, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaji S, Madhu S, Singh S. Extracellular synthesis of antibacterial silver nanoparticles using psychrophilic bacteria. Process Biochem. 2011;46:1800–1807. doi: 10.1016/j.procbio.2011.06.008. [DOI] [Google Scholar]
- Singh SK, Roberts SA, McDevitt SF, et al. Crystal structures of multicopper oxidase cueO bound to copper (I) and silver (I) functional role of a methionine-rich sequence. J Biol Chem. 2011;286:37849–37857. doi: 10.1074/jbc.M111.293589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhu V, Philip D. Spectroscopic, microscopic and catalytic properties of silver nanoparticles synthesized using Saraca indica flower. Spectrochim Acta A. 2014;117:102–108. doi: 10.1016/j.saa.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Wei X, Luo M, Li W, et al. Synthesis of silver nanoparticles by solar irradiation of cell-free Bacillus amyloliquefaciens extracts and AgNO3. Bioresource Technol. 2012;103:273–278. doi: 10.1016/j.biortech.2011.09.118. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2015) Global antimicrobial resistance surveillance system: manual for early implementation. World Health Organization
- Yehia RS, Al-Sheikh H. Biosynthesis and characterization of silver nanoparticles produced by Pleurotus ostreatus and their anticandidal and anticancer activities. World J Microb Biot. 2014;30:2797–2803. doi: 10.1007/s11274-014-1703-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.