Abstract
Several plant biotechnology applications are based on the expression of multiple genes located on a single transformation vector. The principles of stable expression of foreign genes in plant cells include integration of full-length gene fragments consisting of promoter and transcription terminator sequences, and avoiding converging orientation of the gene transcriptional direction. Therefore, investigators usually generate constructs in which genes are assembled in the same orientation. However, no specific information is available on the effect of the order in which genes should be assembled in the construct to support optimum expression of each gene upon integration in the genome. While many factors, including genomic position and the integration structure, could affect gene expression, the investigators judiciously design DNA constructs to avoid glitches. However, the gene order in a multigene assembly remains an open question. This study addressed the effect of gene order in the DNA construct on gene expression in rice using a simple design of two genes placed in two possible orders with respect to the genomic context. Transgenic rice lines containing green fluorescent protein (GFP) and β-glucuronidase (GUS) genes in two distinct orders were developed by Cre-lox-mediated site-specific integration. Gene expression analysis of transgenic lines showed that both genes were expressed at similar levels in either orientation, and different transgenic lines expressed each gene within 1–2× range. Thus, no significant effect of the gene order on gene expression was found in the transformed rice lines containing precise site-specific integrations and stable gene expression in plant cells could be obtained with altered gene orders. Therefore, gene orientation and integration structures are more important factors governing gene expression than gene orders in the genomic context.
Keywords: Transgene expression, Multigene transformation, Gene order, Gene orientation, Site-specific integration, Cre-lox
Introduction
Plant cell transformation involves integration of single or multiple gene(s) to express the encoded proteins. Designing DNA constructs consisting of multiple genes is fundamentally tied to the success of plant transformation projects. Multiple genes could be assembled in a single transformation vector for transfer into plant cells. When assembling two or more genes, investigators face the question of the order in which the genes should be assembled to obtain optimum expression upon integration into plant cells. This basic part of the construct design is guided by the principles of stable gene expression in transformed plant cells as the goal of the experiment is mostly to express the gene(s) at high levels. It is well established that a full-length single-copy integration is correlated with high expression levels (Akbudak et al. 2010; Chawla et al. 2006; Srivastava and Ow 2002). Furthermore, tail-to-tail orientation, i.e., two genes having converging transcriptional direction are not desired as antisense transcripts could be generated through the read-through transcription mechanisms leading possibly to silencing of both genes (Luo and Chen 2007; Nicholson and Srivastava 2009; Ulker et al. 2008). A recent paper showed that diverging orientation of genes in which the promoters of the genes are juxtaposed; expression of each gene could be influenced (Qiu et al. 2016), possibly due to the proximity of their enhancers. Therefore, in a multigene construct, all genes should be placed in the same transcriptional orientation (Akbudak et al. 2010; Svitashev et al. 2002). However, whether the order of genes in the assembly also affects gene expression has not been clearly investigated.
When combining genes in a binary vector, the gene order is determined by the T-DNA borders, right border (RB) and left border (LB), since the transfer of T-DNA starts at RB, and most T-DNA fragments are intact at RB but contain variable lengths at the LB end. The significance of gene order would be difficult to determine on non-binary vectors delivered by gene gun, since the whole vector is scrambled into the genome and imprecisely integrated, generating integration structures that alter the gene order designed in the vector (Makarevitch et al. 2003). However, gene gun delivery of minimal cassettes (DNA constructs without the vector backbones) achieves a higher rate of single-copy integration similar to that observed in Agrobacterium-mediated T-DNA delivery (Fu et al. 2000; Jackson et al. 2013).
Furthermore, methods of precise gene integrations are also available that target the delivered DNA to specific sites in the genome, generating predictable integration structures. These methods rely on DNA recombination (site-specific recombination or homologous recombination) to achieve site-specific integration (Albert et al. 1995; Srivastava et al. 2004; Terada et al. 2002). Recombinase-mediated site-specific integration using Cre-lox and FLP-FRT systems is highly efficient (Albert et al. 1995; Day et al. 2000; Kilby et al. 1995; Li et al. 2010; Lloyd and Davis 1994; Srivastava et al. 2004); however, it requires placing a recombination site (lox or FRT) into plant genomes first. Homologous recombination-based gene targeting does not require placement of a recombination sites; however, its observed efficiency in higher plants is much lower even with the newly developed gene editing reagents that induce targeted double-stranded breaks (DSB) that, in turn, trigger homologous recombination (Puchta et al. 1993; Steinert et al. 2016). The newly developed gene targeting and genome editing technologies which make use of site-specific nucleases to induce homologous recombination of delivered DNA do not require such pre-integrated DNAs (Cermak et al. 2011; Feng et al. 2013; Townsend et al. 2009).
Gene stacking or multigenic transformations either by non-targeted approaches (minimal cassettes and T-DNA) or by site-specific integration are important biotechnological tools for engineering useful traits in plants or producing pharmaceutical proteins or metabolites among other applications (Srivastava and Thomson 2016). In all of these approaches, the gene order in the DNA construct is a highly relevant question, which should be experimentally addressed. This study determined the expression of two genes placed in two different orders in the DNA construct relative to the selection marker gene, and determined gene expression upon site-specific integration of these constructs into the rice genome. Site-specific integration conserves gene order determined in the construct upon its integration into plant cell and removes the gene expression variation caused by the genomic positions (position-effect variation) (Albert et al. 1995; Chawla et al. 2006).
In this study, a simple experimental design of rice transformation with two different constructs containing distinct gene orders of green fluorescent protein (GFP) and β–glucuronidase (GUS) genes was used. No significant effect of the gene order in the tested genomic context on gene expression was found. While different genomic contexts could cast different effects on gene expression, it is likely that the effect would be predictably uniform on all genes. Therefore, this study suggests that investigators could develop DNA constructs containing variable gene orders for multigene transformation and expect predicted expression of each gene regardless of the order in which they are assembled as long as the relative orientation of genes in the assembly is the same.
Results and discussion
Molecular design
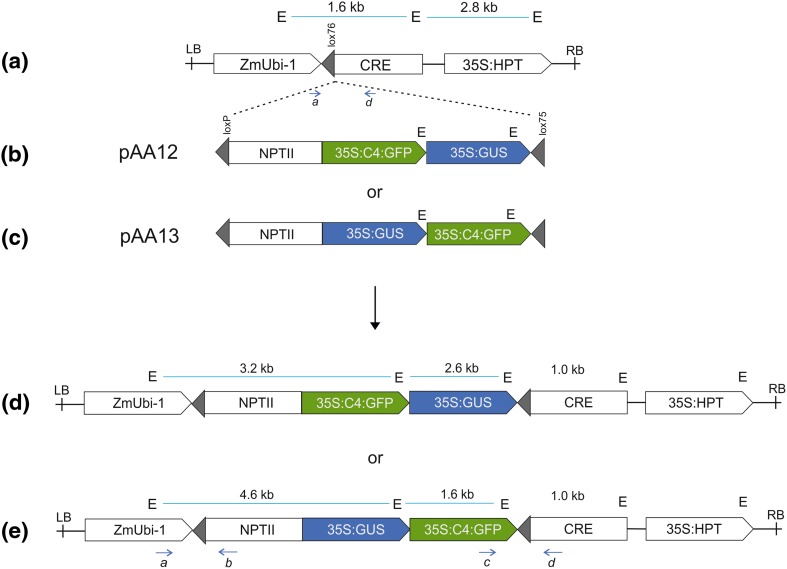
The gene order effect was analyzed through site-specific integration of GFP and GUS genes placed in two possible orders at the T5 locus through Cre-lox-mediated site-specific integration (Fig. 1a). Site-specific integration approach (Srivastava et al. 2004) was used to avoid the gene expression variations caused by genomic position and integration structures. T5 locus contains a single copy of T-DNA, as shown in Fig. 1a, that contains lox76 site for site-specific integration via Cre-lox recombination (Akbudak and Srivastava 2011; Srivastava et al. 2004; Srivastava and Ow 2002). The T-DNA contains a cre gene driven by maize ubiquitin promoter (ZmUbi1) with lox75 site placed between the promoter and the cre coding sequence. Site-specific integration into lox76 will generate an integration structure with a fixed gene orientation and gene order. Two donor constructs, pAA12 and pAA13, were developed that contain promoter-less NPTII gene and the genes-of-interest, GFP and GUS, cloned in two possible orders, GFP-GUS or GUS-GFP (Fig. 1b, c). Site-specific integration (SSI) of these constructs would generate integration structures that differ only in the gene order of two genes without any predictable change in the genomic context or gene orientation. All SSI lines derived from pAA12 contain GFP-GUS cassettes, while those from pAA13 contain the reverse order, i.e., GUS-GFP (Fig. 1d, e). Both pAA12 and pAA13 contain identical regulatory elements for each gene, i.e., 35S:C4 promoter for GFP and 35S promoter for GUS and nos3′ for transcription termination of each gene. The SSI lines developed by gene gun delivery of pAA12 or pAA13 into T5 callus were characterized by PCR and Southern blot and subjected to gene expression analysis by fluorescence or GUS assay.
Fig. 1.
Molecular strategy of site-specific integration (SSI). a Structure of T-DNA used to generate the target site in rice genome (Taipei-309) and termed as T5 site. The T5 site contains a single-copy of the T-DNA that contains lox76 target site (gray triangle) and Cre activity. It also contains 35S:HPT as the selection marker genes. b, c Structures of the two donor vectors, pAA12 and pAA13, designed for site-specific integration into T5 site. Each contains two lox sites (gray triangles), loxP at 5′ end and lox75 at 3′ end, for Cre-lox-mediated site-specific integration into T5 site via lox75 × lox76 recombination. The two vectors contain a promoter-less marker gene, NPT II, for the selection of site-specific integration (SSI) and distinct gene orders of GFP and GUS genes as shown. d, e Integration of the donor constructs via Cre-lox recombination into the target site generates a predictable SSI structure. pAA12 lines contain GFP-GUS order, while pAA13 contain GUS-GFP order within the T5 locus relative to the LB and RB (T-DNA left and right borders). ZmUbi1 maize ubiquitin-1 promoter, HPT hygromycin phosphotransferase gene, 35S cauliflower mosaic virus 35S RNA promoter, C4 maize PPDK promoter, NPT neomycin phosphotransferase gene. Each gene carries a nopaline synthase 3′ transcriptional termination signal (not shown). PCR primer (a–d) sites and their orientation (blue arrows), and EcoRI (E) sites are indicated
Characterization of transgenic callus lines
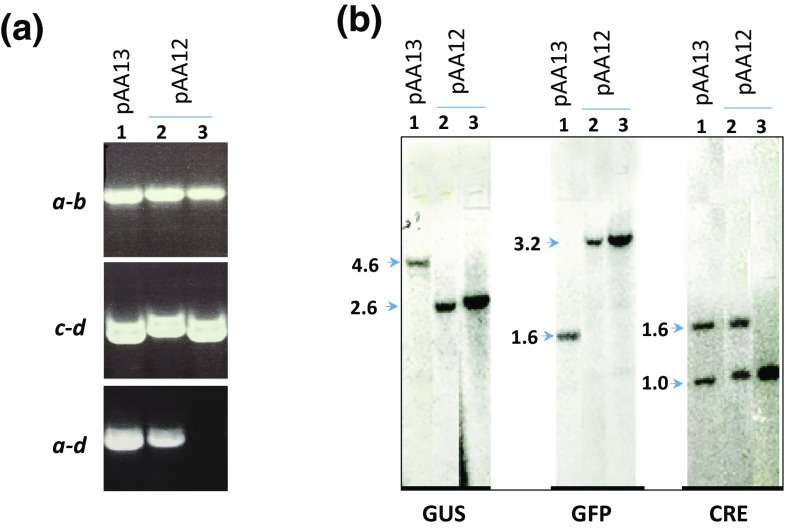
Two different experiments involving bombardment of T5 callus with either pAA12 or pAA13 generated a number of geneticin-resistant lines. Each line was analyzed by PCR using primers a–b and c–d to determine the presence of predicted SSI junctions. In addition, primer pair a–d was used to determine biallelic/monoallelic integrations occurred (Fig. 1a, e). PCR analysis revealed that 23 pAA12 (GFP-GUS) lines and 7 pAA13 (GUS-GFP) lines carried the expected SSI junctions. The rest of the lines were removed from further analysis. The PCR for a–d junction indicated monoallelic integration in all except one pAA12 line, which contained biallelic integration. The presence of SSI junctions indicates precise integration of the construct into T5 locus, and the absence of the parental a–d junction indicates biallelic integration. A representative PCR data is shown in Fig. 2a.
Fig. 2.
Molecular analysis of transgenic lines. a PCR analysis of SSI lines obtained by transformation of T5 callus with pAA12 or pAA13 using primer pairs indicated on each panel, and mapped on Fig. 1. b Southern analysis of SSI lines. Each panel show SSI fragments with GUS, GFP, or CRE probes from EcoRI-digested genomic DNA of (see Fig. 1d, e). These fragment sizes match the predicted sizes EcoRI fragment sizes of SSI locus (Fig. 1d, e). Lane 1 in each panel represents pAA13 line 13.6 (monoallelic), and lanes 2 and 3 represent pAA12 lines, 12.15 (monoallelic) and 12.11 (biallelic), respectively
Subsequently, DNA blot analyses on EcoRI-digested genomic DNA were carried out to confirm the presence of SSI structure, and determine the presence of additional copies of the vectors. Representative Southern blots are given in Fig. 2b. The putative SSI lines could contain random integrations of the vector DNA since ‘illegitimate’ recombination cannot be suppressed. Fourteen out of twenty-three pAA12 lines and six out of seven pAA13 lines showed the predicted hybridization pattern with GUS, GFP, and Cre probes (Fig. 2b). These lines were designated as precise-SSI lines. However, one line each of pAA12 and pAA13 showed an extra band with GUS or GFP probe, indicating the presence of an extra copy (data not shown) (Table 1). The remaining lines showed complex integration pattern indicating the presence of numerous extra copies (data not shown). Of these complex-SSI lines, two complex-SSI of pAA12 and the single complex-SSI of pAA13 were included for expression analysis.
Table 1.
Characterization of transgenic lines and expression analyses
| Vector | Line | Integration pattern | Copy # | PCR | GFPa (µg/mg total protein) | GUSa (nmole/min/mg) | |||
|---|---|---|---|---|---|---|---|---|---|
| GFP | GUS | a–b | c–d | a–d | |||||
| pAA12 | 12.1 | Precise | 1 | 1 | + | + | + | 2.70 | 27.13 |
| 12.5 | Precise | 1 | 1 | + | + | + | 2.78 | 21.44 | |
| 12.8 | Precise | 1 | 1 | + | + | + | 2.76 | 37.81 | |
| 12.11 | Precise | 2 | 2 | + | + | − | 6.61 | 47.86 | |
| 12.12 | Precise | 1 | 1 | + | + | + | 2.51 | 22.80 | |
| 12.13 | Precise | 1 | 1 | + | + | + | 3.29 | 26.60 | |
| 12.15 | Precise | 1 | 1 | + | + | + | 2.86 | 40.29 | |
| 12.16 | Precise | 1 | 1 | + | + | + | 2.29 | 25.08 | |
| 12.17 | Precise | 1 | 1 | + | + | + | 4.04 | 31.67 | |
| 12.26 | Precise | 1 | 2 | + | + | + | 3.05 | 25.13 | |
| 12.30 | Precise | 1 | 1 | + | + | + | 2.38 | 24.43 | |
| 12.32 | Precise | 1 | 1 | + | + | + | 3.31 | 21.99 | |
| 12.33 | Precise | 1 | 1 | + | + | + | 2.32 | 37.42 | |
| 12.24 | Complex | >4 | 2 | + | + | + | 3.62 | 6.21 | |
| 12.35 | Complex | >2 | >2 | + | + | + | 4.03 | 9.16 | |
| pAA13 | 13.2 | Precise | 2 | 1 | + | + | + | 4.85 | 23.27 |
| 13.6 | Precise | 1 | 1 | + | + | + | 2.77 | 25.43 | |
| 13.9 | Precise | 1 | 1 | + | + | + | 2.56 | 25.10 | |
| 13.13 | Precise | 1 | 1 | + | + | + | 2.64 | 23.75 | |
| 13.20 | Precise | 1 | 1 | + | + | + | 2.74 | 23.11 | |
| 13.21 | Precise | 1 | 1 | + | + | + | 2.61 | 22.14 | |
| 13.8 | Complex | >5 | >4 | + | + | + | 2.22 | 6.14 | |
aStandard deviation of mean (sdom; n = 3) in GFP amount and GUS activity were depicted as error bars in Fig. 3
The attempts to regenerate these callus lines failed; however, most lines were maintained for up to 1 year by transferring to fresh selection plates regularly. To assess the potential problem of chimerism, defined as contamination of ‘untransformed’ target cells, we used a molecular approach of evaluating relative intensities of cre gene on Southern blot or PCR. Hybridization with cre probe on a Southern blot displays two distinct bands representing either SSI site (1.0 kb) or target site (1.6 kb). Both 1.0 and 1.6 kb bands are expected from monoallelic SSI, whereas 1.0 kb band would be produced from biallelic SSI (see Fig. 1). Equal intensity of 1.0 and 1.6 kb bands in monoallelic lines suggests the absence of contamination by untransformed target cells (Fig. 2b). In the biallelic SSI line, target site-specific PCR (primers a–d) was conducted to rule out the contamination (Figs. 1a, 2a). These analyses suggested that all SSI lines were homogenous as contamination of untransformed T5 cells was undetectable. Furthermore, PCR and Southern analyses confirmed that all transgenic lines contained site-specific integrations of pAA12 or pAA13 constructs, which rules out the speculation that transgene expression differences could occur due to genomic position or integration structure variations in the precise-SSI lines.
Transgene expression analysis
Fifteen pAA12 and seven pAA13 lines were subjected to GUS and GFP expression analysis (Table 1). Three of these lines, two pAA12 and one pAA13 line, were complex-SSI, while the remaining were precise-SSI containing the predictable structures, as shown in Fig. 1. The GUS activity was assessed using histochemical staining and MUG assay, whereas GFP was quantified by fluorescence spectrometry. All lines stained positive for GUS activity and showed green fluorescence, indicating the expression of GUS and GFP genes.
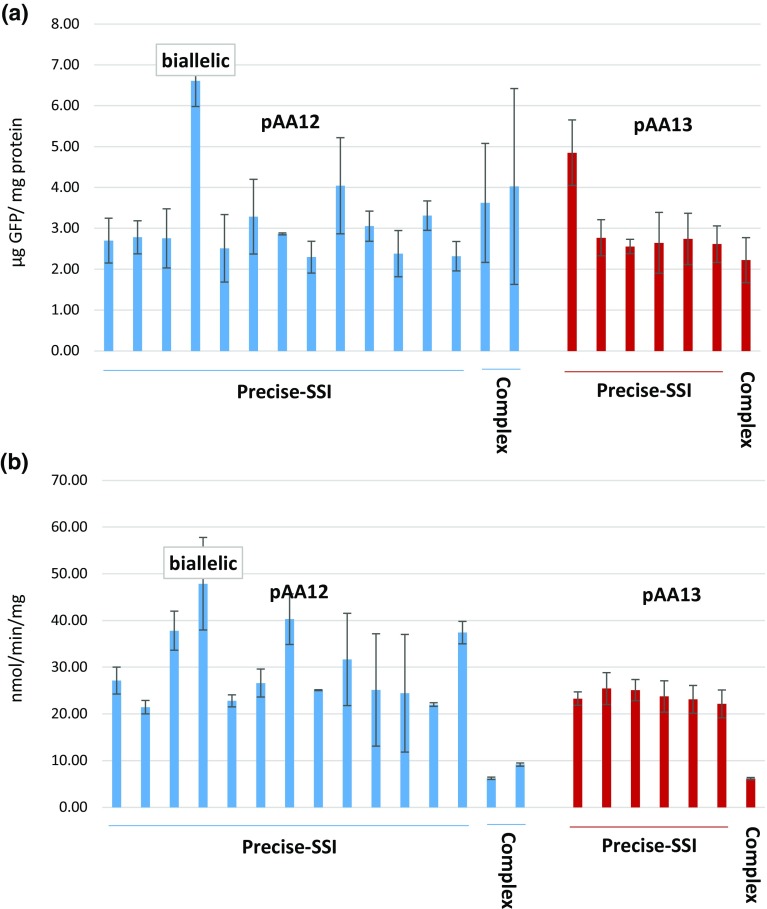
The quantitative measurement of GUS activity by MUG assay indicated high expression in precise-SSI lines, whereas a significant suppression in the complex-SSI lines. In contrast, GFP gene highly expressed in both precise-SSI and complex-SSI lines (Table 1; Fig. 3a, b). This observation could be an artefact based on the functional difference of the two proteins (enzyme vs fluorescent protein) and analytical methods used for their measurements (enzymatic assay vs fluorescence). Higher fluctuations in GUS activity as compared to GFP activity in transgenic rice were also reported by Akbudak et al. (2010). In general, gene suppression could occur in multi-copy lines even if one of the copies represent precise-SSI (Srivastava et al. 2004), and the molecular basis of suppression is most likely RNAi, since segregation of the precise-SSI results in restoration of high gene activity (Chawla et al. 2006).
Fig. 3.
Effect of gene order on gene expression: a GFP and b GUS expression levels in pAA12 (blue bars) and pAA13 (red bars) SSI lines containing the predictable SSI only (precise) or the extra copies of the plasmids in addition to the predictable SSI (complex). Error bars represent standard deviation
Furthermore, one of the SSI lines, 12.11, contained biallelic integration of pAA12 construct. Accordingly, this line expressed highest levels of GFP and GUS activities (Table 1; Fig. 3a, b). Similarly, line 13.2 that contained an extra copy of the GFP gene expressed highest levels of GFP among pAA13 lines. These two lines, therefore, display additive effects of gene copy number. The remaining precise-SSI lines expressed GFP and GUS genes within 1–2× variability (Table 1; Fig. 3a, b). Furthermore, the GFP or GUS expression ranges were similar in SSI lines regardless of the gene order, with the biallelic line, 12.11, showing ≥2× GFP expression level. The GFP expression level in precise-SSI lines was similar to that of complex-SSI, suggesting lack of additive or suppressive effects of the extra copies on GFP gene expression. The GUS expression variation was also minimal ranging between 1 and 2× among SSI lines. These observations match with the previous report that found 2–3× variability in GUS expression among SSI lines carrying a single-copy of the GUS gene (Srivastava et al. 2004).
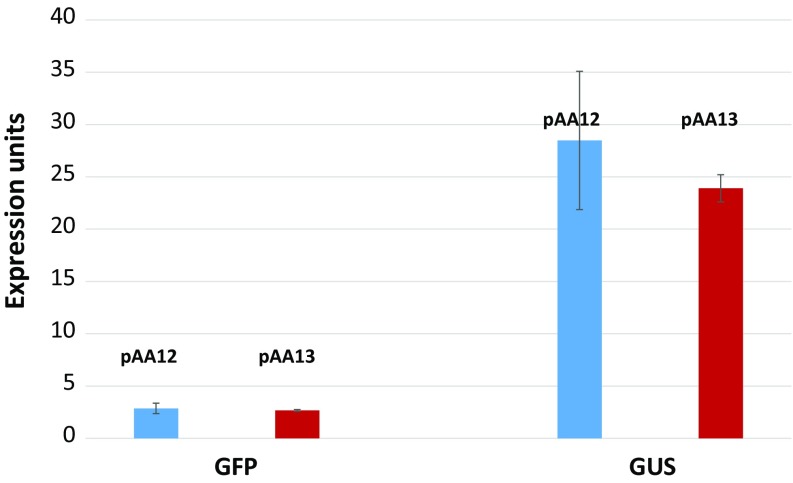
Next, all monoallelic lines were used to calculate average GFP or GUS expression produced by SSI lines carrying distinct gene orders of the two genes. This analysis showed that pAA12 and pAA13 constructs generated similar levels of gene expression of GFP or GUS genes from the T5 locus (Fig. 4). In summary, no significant effect of the gene order in the DNA construct was observed on gene expression and stable gene expression was obtained by both constructs. Therefore, genes could be assembled in any order to obtain optimal expression from a given genomic site provided that they contain full-length single-copy integrations.
Fig. 4.
Effect of gene order: average GFP and GUS expression values obtained from precise-SSI lines of pAA12 and pAA13. Expression units represent GFP and GUS units, as shown in Fig. 3, and error bars represent standard deviation
Materials and methods
Vector construction
The donor vectors were generated in the backbone of pVS55 described by Srivastava et al. (2004). pVS55 (pUC18 backbone) contains lox sites (gray triangles) for Cre-lox-mediated site-specific integration into plant genomes. pVS55 was cut with HindIII to replace the GUS gene with SpeI linker. The XbaI fragments consisting of 35S:C4:GFP and 35S:GUS cassettes were ligated into the SpeI, sequentially, to generate pAA12 and pAA13.
Rice transformation
Rice transformation was done by particle bombardment method using PDS 1000 gene gun (Bio-Rad, Inc.). Rice tissue culture media preparation was essentially following the method described by Hiei et al. (1994). The rice line T5 (Taipei-309) containing a target site, as depicted in Fig. 1a, was used for transformation. The transformation of T5 callus with pAA12 and pAA13 was done according to the protocol described by Srivastava (2013). The bombarded calli were selected on 100 mg/l Geneticin (Gibco BRL, Grand Island, NY, USA) and maintained by regular sub-cultures onto fresh media plates containing Geneticin.
Molecular analysis
Polymerase chain reaction (PCR) was carried out on the genomic DNA with primers ‘a’ (5′-TCTACTTCTGTTCATGTTTGTG-3′), ‘b’ (5′-CTCGATGCGATGTTTCGCTT-3′), ‘c’ (5′-GATTAGAGTCCCGCAATTAT-3′), and ‘d’ (5′-CTAATCGCCATCTTCCAGCA-3′). Taq Polymerase (Promega, Madison, WI) was used for all amplifications following the manufacturer’s recommendation. The reactions consisted of 39 cycles of melting at 96 °C for 1 min, annealing at 58 °C for 1 min, and extension at 72 °C for 1 min, followed by final extension for 7 min at 72 °C. Southern blot analysis was carried out with three 32P-labeled DNA probes (GFP, GUS, and CRE). Approximately 5 µg of genomic DNA digested with EcoRI, fractionated on 0.8% agarose gel, blotted on nylon membrane, was hybridized with the probes using standard Southern hybridization methods.
Expression analysis
GUS activity in calli was detected by the method described by Jefferson (1987). Callus pieces were submerged in the GUS staining solution containing 1 mM X-Gluc (Gold Biotechnologies, St. Louis, MO, USA) and incubated at 37 °C. The quantitative measurement of GUS activity was done using the FluorAce GUS reporter kit (Bio-Rad, Inc.). Total protein of plant extracts was measured using the DC protein assay kit (Bio-Rad Inc.). A standard curve prepared with the dilution series of 4-methylumbelliferone (4-MU) was used to calculate GUS activity. A unit of GUS activity is defined as nmol 4-MU produced per minute from each milligram of the soluble protein (nmol/min/mg). The GFP expression was observed visually under blue light using yellow filter. For the quantitative measurement of GFP, callus was ground in the extraction buffer (10 mM Tris–EDTA, pH 8.0) at 4 °C and centrifuged at 13,000 rpm for 20 min to collect the supernatant. Protein concentrations of the extracts were determined using the DC protein assay kit (Bio-Rad Inc.). GFP quantification was done using VersaFluor fluorometer (Bio-Rad Inc.) fitted with a 490 ± 5 nm excitation filter and a 510 ± 5 nm emission filter. A dilution series (0.1–1 mg/ml) of purified rGFP-S65T protein (Clontech Inc., San Diego, CA, USA) was made in the extraction buffer to generate the standard curve. A unit of GFP is defined as microgram GFP per mg soluble protein (µg/mg total protein)
Acknowledgements
This research was supported by ABI grants and USDA-NIFA award 2014-38821-22460 to V.S. Authors are grateful to Y. Niwa for providing 35S:C4:GFP construct (GenBank Accession # EF 090408.1).
Compliance with ethical standards
Conflict of interest
Authors do not declare any conflict of interest.
References
- Akbudak MA, Srivastava V. Improved FLP recombinase, FLPe, efficiently removes marker gene from transgene locus developed by Cre–lox mediated site-specific gene integration in rice. Mol Biotechnol. 2011;49:82–89. doi: 10.1007/s12033-011-9381-y. [DOI] [PubMed] [Google Scholar]
- Akbudak MA, More AB, Nandy S, Srivastava V. Dosage-dependent gene expression from direct repeat locus in rice developed by site-specific gene integration. Mol Biotechnol. 2010;45(1):15–23. doi: 10.1007/s12033-009-9235-z. [DOI] [PubMed] [Google Scholar]
- Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7(4):649–659. doi: 10.1046/j.1365-313X.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucl Acid Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla R, Ariza-Nieto M, Wilson AJ, Moore SK, Srivastava V. Transgene expression produced by biolistic-mediated, site-specific gene integration is consistently inherited by the subsequent generations. Plant Biotechnol J. 2006;4(2):209–218. doi: 10.1111/j.1467-7652.2005.00173.x. [DOI] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi T, Holappa LD, Albert H, Ow DW. Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Gene Dev. 2000;14(22):2869–2880. doi: 10.1101/gad.849600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Duc LT, Fontana S, Bong BB, Tinjuangjun P, Sudhakar D, Twyman RM, Christou P, Kohli A. Linear transgene constructs lacking vector backbone sequences generate low-copy-number transgenic plants with simple integration patterns. Transgenic Res. 2000;9(1):11–19. doi: 10.1023/A:1008993730505. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Jackson MA, Anderson DJ, Birch RG. Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res. 2013;22(1):143–151. doi: 10.1007/s11248-012-9639-6. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5(4):387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- Kilby NJ, Davies GJ, Snaith MR, Murray JAH. FLP recombinase in transgenic plants—constitutive activity in stably transformed tobacco and generation of marked cell clones in Arabidopsis. Plant J. 1995;8(5):637–652. doi: 10.1046/j.1365-313X.1995.08050637.x. [DOI] [PubMed] [Google Scholar]
- Li B, Li N, Duan XG, Wei AY, Yang AF, Zhang JR. Generation of marker-free transgenic maize with improved salt tolerance using the FLP/FRT recombination system. J Biotechnol. 2010;145(2):206–213. doi: 10.1016/j.jbiotec.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Davis RW. Functional expression of the yeast FLP/FRT Site-specific recombination system in Nicotiana tabacum. Mol Gen Genet. 1994;242(6):653–657. doi: 10.1007/BF00283419. [DOI] [PubMed] [Google Scholar]
- Luo ZH, Chen ZX. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19(3):943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Svitashev SK, Somers DA. Complete sequence analysis of transgene loci from plants transformed via micro-projectile bombardment. Plant Mol Biol. 2003;52(2):421–432. doi: 10.1023/A:1023968920830. [DOI] [PubMed] [Google Scholar]
- Nicholson SJ, Srivastava V. Transgene constructs lacking transcription termination signal induce efficient silencing of endogenous targets in Arabidopsis. Mol Genet Genom. 2009;282:319–328. doi: 10.1007/s00438-009-0467-1. [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double-strand breaks into DNA by a site-specific endonuclease. Nucl Acid Res. 1993;21(22):5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu T, Dong Y, Ren Y, Wang J, Yang M, Zhang J. Effects of the sequence and orientation of an expression cassette in tobacco transformed by dual Bt genes. Plasmid. 2016;89:1–8. doi: 10.1016/j.plasmid.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Srivastava V. Site-specific gene integration in rice. Methods Mol Biol. 2013;956:83–93. doi: 10.1007/978-1-62703-194-3_7. [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW. Biolistic mediated site-specific integration in rice. Mol Breed. 2002;8(4):345–350. doi: 10.1023/A:1015229015022. [DOI] [Google Scholar]
- Srivastava V, Thomson J. Gene stacking by recombinases. Plant Biotechnol J. 2016;14(2):471–482. doi: 10.1111/pbi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Ariza-Nieto M, Wilson AJ. Cre-mediated site-specific gene integration for consistent transgene expression in rice. Plant Biotechnol J. 2004;2(2):169–179. doi: 10.1111/j.1467-7652.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Steinert J, Schiml S, Puchta H. Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep. 2016;35:1429–1438. doi: 10.1007/s00299-016-1981-3. [DOI] [PubMed] [Google Scholar]
- Svitashev SK, Pawlowski WP, Makarevitch I, Plank DW, Somers DA. Complex transgene locus structures implicate multiple mechanisms for plant transgene rearrangement. Plant J. 2002;32(4):433–445. doi: 10.1046/j.1365-313X.2002.01433.x. [DOI] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. Efficient gene targeting by homologous recombination in rice. Nat Biotechnol. 2002;20(10):1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu FL, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouche N, Edwards R, Sanders D, Knight H, Knight MR. Getting the most out of publicly available T-DNA insertion lines. Plant J. 2008;56(4):665–677. doi: 10.1111/j.1365-313X.2008.03608.x. [DOI] [PubMed] [Google Scholar]