Abstract
A method of rapidly differentiating lung tumor from healthy tissue is extraordinarily needed for both the diagnosis and the intraoperative margin assessment. We assessed the ability of fluorescence lifetime imaging microscopy (FLIM) for differentiating human lung cancer and normal tissues with the autofluorescence, and also elucidated the mechanism in tissue studies and cell studies. A 15-patient testing group was used to compare FLIM results with traditional histopathology diagnosis. Based on the endogenous fluorescence lifetimes of the testing group, a criterion line was proposed to distinguish normal and cancerous tissues. Then by blinded examined 41 sections from the validation group of other 16 patients, the sensitivity and specificity of FLIM were determined. The cellular metabolism was studied with specific perturbations of oxidative phosphorylation and glycolysis in cell studies. The fluorescence lifetime of cancerous lung tissues is consistently lower than normal tissues, and this is due to the both decrease of reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) lifetimes. A criterion line of lifetime at 1920 ps can be given for differentiating human lung cancer and normal tissues.The sensitivity and specificity of FLIM for lung cancer diagnosis were determined as 92.9% and 92.3%. These findings suggest that NADH and FAD can be used to rapidly diagnose lung cancer. FLIM is a rapid, accurate and highly sensitive technique in the judgment during lung cancer surgery and it can be potential in earlier cancer detection.
Abbreviations: FLIM, fluorescence lifetime imaging microscopy; HE, haematoxylin and eosin; NADH, reduced nicotinamide adenine dinucleotide; FAD, flavin adenine dinucleotide; TCSPC, time-correlated single photon counting; BEAS-2B, human bronchial epithelial cells
Keywords: Autofluorescence, Lifetime, Lung tissues, Rapid and sensitive diagnosis, Cancer metabolism
1. Introduction
Lung cancer is one of the leading causes of cancer deaths worldwide [1], [2]. Despite undergone pneumonectomy, the overall 5-year survival rate of patients is less than 14% [3], [4]. Complete surgical resection is crucial to increasing the survival rates. The methods of rapidly differentiating tumor from healthy tissue would be extraordinarily beneficial to both the diagnosis and the intraoperative margin assessment. The haematoxylin and eosin (H&E) stained histopathology is the current gold standard to assess and diagnose disease, but can't to be used in the operating room as the long test time (up to one week). Intraoperative frozen section examination could be used to aid the surgeon to distinguish tumor and normal tissues during tumor resection, which still needs the staining process and takes about 20–30 min [5].
The endogenous fluorophores in cells and tissues, such as reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD), play a pivotal role in cellular energy metabolism [6], [7]. Compared to normal cells, many tumor cells have an increased metabolic demand and an elevated rate of aerobic glycolysis for rapid cell division [8]. which usually associates with changes of endogenous fluorophores and can be monitored by fluorescence. Recent studies have revealed the potential of autofluorescence imaging [9], [10], [11] and spectroscopy [12], [13] as diagnostic methods, including oral [8] cervical [14], [15] and breast [16] cancers. Since the irregular tissue surface or endogenous absorbers in tissues, the quantitatively measurement of fluorescence intensity remains a challenge [17]. On the other hand, the fluorescence lifetime of a fluorophore is independent on its concentration or excitation power, and is highly sensitive to its surrounding microenvironment, such as molecular binding, pH, ion concentration and oxygen concentration [12]. Therefore, fluorescence lifetime imaging microscopy (FLIM) can provide clear information of the metabolic difference between tumor and normal tissue.
In this study, we present a novel diagnose method for lung tumor by FLIM. The lifetime images of differentiated lung tumor and normal lung tissue were obtained, and correlated with the diagnoses by H&E stained method. Our results show a rapid, accurate and highly sensitive technique in the judgment during lung cancer surgery and it can be more potential in earlier cancer detection. Moreover, the fundamental of the fluorescent differences between tumor and normal tissue was also studied in vitro.
2. Methods
2.1. Patients eligibility criteria
Between June 2015 and October 2016, thirty-one patients diagnosed as lung cancer and undergoing lung surgery involved in this study. The patients were from Department of Chest, Huashan hospital, Fudan University. This work was under the approval of Institution Review Board in Huashan Hospital. All patients were suspected of lung cancer and staged as I to IIIa, whose performance function scores were between 0 and 2, undergoing chest CT scan, MRI of brain and bone nucleic scan, blood test including blood routine and tumor markers (carcinoma embryo antigen, CEA; neuron-specific enolase, NSE; cytokeratin 19 fragment, CY211). All scan and blood test examinations were prior to any treatment.
2.2. Study design and lung tissue sections preparation
The thirty-one patients were randomly divided into two groups, a testing group including 15 patients and a validation group including 16 patients. Frozen tissue sections or paraffin-embedded tissue sections were obtained during the surgeries with the standard procedures by pathologists [18]. To evaluate the ability of FLIM for lung cancer diagnosis, we first compared FLIM results with H&E stained histopathology for the testing group (n = 15). For each patient of this group, four section samples were obtained. Two adjacent sections were cut from the center of tumor, and the other two sections were obtained at 5 cm away from the tumor margin and treated as normal lung tissues. Among 15 patients, the sizes of 11 patients' tumors were large enough, so that two extra sections could be obtained at the site between normal and cancerous sections (noted as “middle”). The excised samples were placed on glass slides. One section of each part (normal, cancerous and middle) was imaged with FLIM. And the other tissue sections were stained by H&E for pathological examination by a senior pathologist at the Huashan Hospital. Based on the FLIM study of the testing group, a criterion line was proposed to distinguish the normal and cancerous tissues. We then examined frozen sections (n = 41) of the validation group (the remaining 16 patients) with FLIM when we were blinded to the histopathologic diagnosis. Finally, the sensitivity and specificity of FLIM method were quantified through a comparison to the conventional H&E histopathology.
2.3. Fluorescence lifetime imaging for tissue samples
Fluorescence lifetime images of tissue samples were acquired by a single photon counting module on a laser scanning confocal microscope (Olympus, FV300/IX 71, Japan) equipped with a 405 nm picosecond laser (BDL-405-SMC, Becker & Hickl, Germany) and an oil immersion objective (40 ×, NA = 1.2). The autofluorescence signal was obtained with a filter of 430 nm long-pass. Otherwise, band-pass filters of 465–495 nm and 545–580 nm were used to respectively acquire fluorescence signals of NADH and FAD. The lifetime signal was detected by a photomultiplier tube (PMC-100-1, Becker & Hickl) and processed by a time-correlated single photon counting (TCSPC) system (SPC-150, Becker & Hickl). Synchronized fluorescence lifetime data collection was achieved on a pixel-by-pixel basis by using the x and y laser scanning signals generated by the Confocal scan unit. Each FLIM image of 256 × 256 pixels was acquired in 2 min and at least six different areas were imaged for each section. Every area was imaged only once to avoid the photobleaching effect. All the images were obtained under identical experimental conditions. For each FLIM image, time-decay data of each pixel was fitted with multi-exponential decay models using the SPCImage software (Becker & Hickl GmbH). In our study, the data was fit with double or triple exponential function by assessing the minimized χ2 value during the fitting process and the data of all images with χ2 value below 1.3 indicated a good fit. Based on the fitting, the weighted mean lifetime of each pixel is calculated. Then the distribution curve of one FLIM image can be obtained and displayed as a histogram by the SPCImage software. The peak of the distribution curve was analyzed for each FLIM image, and more than six images were studied for each tissue section.
2.4. Cell culture
Human bronchial epithelial cells (BEAS-2B) were obtained from the cell bank of Shanghai Science Academy and cultured under the standard protocols. Cells were plated on petri dishes for microscopic study. To study the cellular metabolism, cells were incubated with 200 μM CoCl2 (Sigma-Aldrich) or 300 μM 3-bromopyruvate (Sigma-Aldrich) for 90 min before observation to inhibit oxidative phosphorylation or glycolysis, respectively. The untreated cells were set as the control group.
2.5. Two-photon fluorescence lifetime imaging for cell samples
The two-photon fluorescence lifetime imaging study was performed using a Leica microscope (TCS SP8 FLIM) equipped with an oil immersion objective (63 ×, NA = 1.4) and a HyD SMD single molecule detector (Leica). The excitation source was a mode-locked Titanium-sapphire laser (Mai-Tai laser DeepSee, 80 MHz, ~ 90 fs) at 750 nm wavelength. Band-pass filters of 435–485 nm and 545–595 nm were used to acquire fluorescence images of NADH and FAD, respectively. Fluorescence decay curves were also calculated using aforementioned multi-exponential decay models for each image by using the available TCSPC software (SymPhoTime 64, PicoQuant GmbH).
3. Results
3.1. Demographics of patients
The patients in this study include 21 adenocarcinoma, 4 squamous carcinoma, 3 adenosquamous carcinoma, 1 small cell carcinoma, 1 atypical carcinoid and 1 large cell carcinoma. The ratio of man/woman was 13/18, whose average age was 61.2 ± 9.7 years old.
3.2. Imaging performance of FLIM
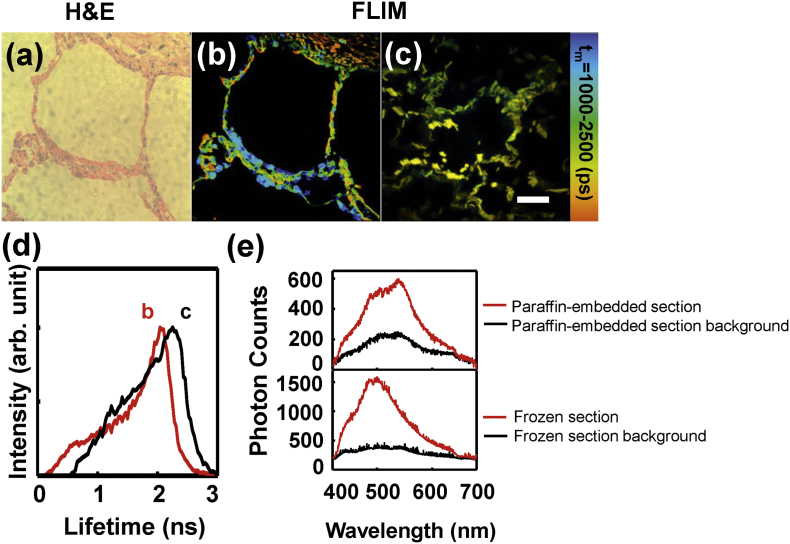
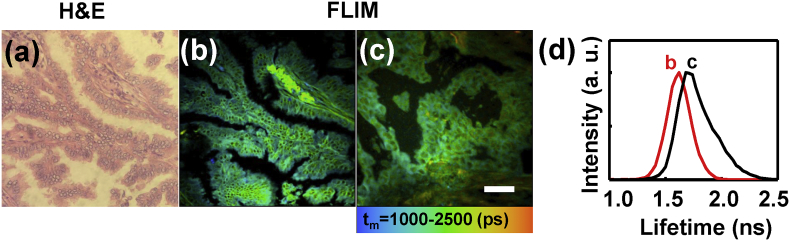
As we all know, the current gold standard for clinical diagnosis of lung cancer is based on the H&E stain for paraffin-embedded tissue sections. Fig. 1(a) demonstrated a representative H&E image of the normal lung tissue paraffin-embedded section, which showed an air-filled alveolar structure of human normal lung tissue and the alveoli was composed of monolayer epithelial cells. For comparison, a FLIM image of an unstained paraffin-embedded section at the same area was shown in Fig. 1(b). It could be seen that the FLIM image exhibited the cellular morphology features as exactly as H&E image. Fig. 1(c) showed the FLIM image of a normal tissue frozen section, in which the tissue structure remained not so intact comparing to paraffin-embedded sections. However, the paraffin itself is fluorescent and has influence on the autofluorescence study. In addition, the paraffin-embedded process usually takes several days, and the fluorescence of endogenous fluorophores would decay with time. Fig. 1(d) showed two lifetime distribution curves of the FLIM images 1(b) and 1(c), which demonstrated that the lifetime of frozen section was distinct from paraffin-embedded section, implying that the fluorescent components in paraffin-embedded section has influence upon the fluorescence lifetime. Otherwise, the components like paraformaldehyd and paraffin wax strongly disrupt the weak endogenous fluorescence of tissues. Similar results were also confirmed for lung cancer tissue sections (Supplementary data Fig. S1). Fig. 1(e) showed the spectra of paraffin-embedded and frozen sections (red line), and the spectra of their background (black line). It can be seen that the background signal of paraffin-embedded section contributed 34.5% intensity to the total spectrum of paraffin-embedded tissue while the background of frozen tissue contributed little. To study the autofluorescence of lung tissues, frozen sections were preferred than paraffin-embedded ones.
Fig. 1.
H&E (a) and FLIM images of normal lung tissue paraffin-embedded section (b) and frozen section (c); (d) fluorescence lifetime distribution curves of FLIM images (b) and (c). (e) autofluorescence spectra of the tissues with paraffin-embedded and frozen sections (red line) and their background spectra (black line). Scale bar is 50 μm.
3.3. FLIM images of frozen lung tissues
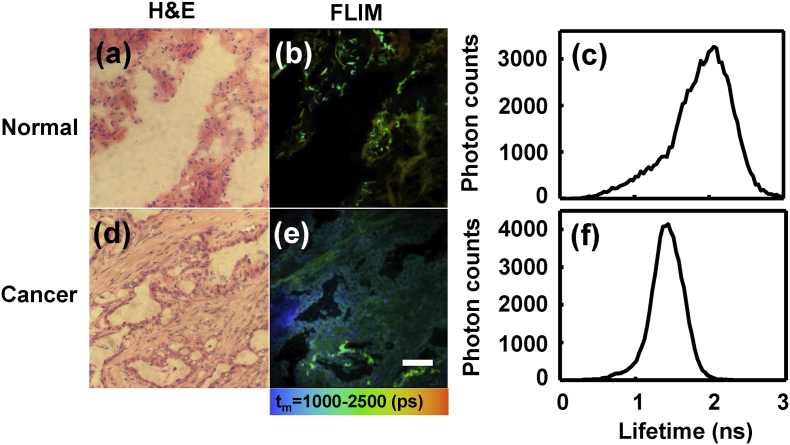
The H&E-stained (Fig. 2(a) and (d)) or FLIM images (Fig. 2(b) and (e)) of the frozen lung tissue sections of one typical patient were shown in Fig. 2. The normal tissue represented longer fluorescence lifetime (orange-yellow) than cancerous tissues (blue-green). Fig. 2(c) and (f) were the corresponding average lifetime distribution curves of Fig. 2(b) and (e). The peaks of the curves were 2.06 ns and 1.40 ns for normal and cancerous tissues, respectively.
Fig. 2.
H&E (a, d) and FLIM (b, e) images of two lung tissue frozen sections: (a, b) normal lung, and (d, e) lung adenocarcinoma. Scale bar is 50 μm. (c) and (f) were the average lifetime distribution curves of (b) and (e), respectively.
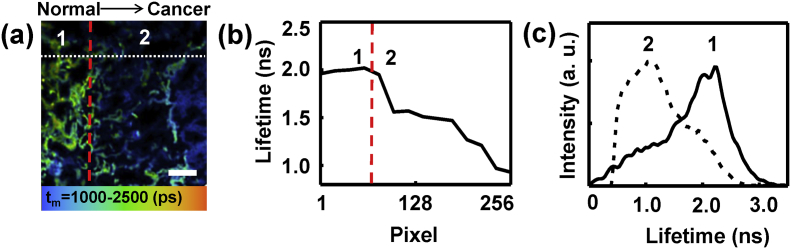
Fig. 3(a) showed the FLIM image of a “middle” lung tissue section, in which the tumor border was marked with a red dash line. Fig. 3(b) showed the lifetimes changed along the white dot line marked in Fig. 3(a). It could be seen that the left part (Area 1 in Fig. 3(a)) has the longer lifetime around 2.0 ns, while the lifetimes in the right part (Area 2 in Fig. 3(a)) decreased sharply at the tumor border then gradually down to lower than 1.0 ns. It suggested that the lung cancer developed from left to right in this case. Fig. 3(c) presented the lifetime distribution of Area 1 (normal side) and Area 2 (cancer side) in Fig. 3(a), in which their peaks were at 2.07 ns and 1.14 ns, respectively. Thus, the average fluorescence lifetime was reduced as normal lung tissues developed to cancerous tissues, which implies the cellular metabolism and microenvironment altered.
Fig. 3.
(a) FLIM image of a “middle” tissue frozen section. Scale bar is 50 μm. (b) The lifetimes changed along the white dot line marked in image (a). (c) The lifetime distribution curves of Area 1 (normal side) and Area 2 (cancer side) in image (a).
3.4. Statistical analysis of lung tissue lifetimes
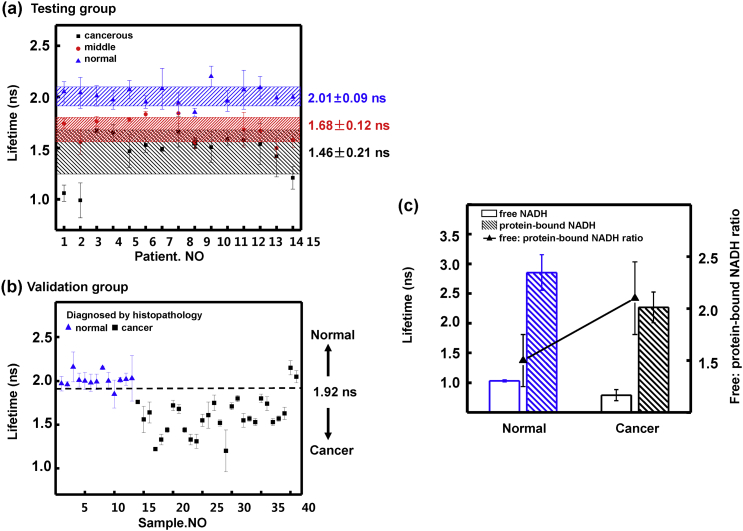
From the testing group, the average lifetimes of normal (n = 15), middle (n = 11) and cancerous lung tissues (n = 15) were presented in Fig. 4(a). Each scatter point was averaged with the weighted mean lifetimes of at least six FLIM images for each section, and the error bars showed the standard deviation due to different scan areas within a section. For each patient, the lifetime of normal section (from 1.85 to 2.20 ns) was obviously longer than middle (from 1.50 to 1.84 ns) and cancerous sections (from 0.99 to 1.67 ns). The heterogeneity between different patients was 0.35, 0.34 and 0.68 ns for normal, middle and cancerous groups, respectively. The larger variability of cancerous groups indicated the variation of cellular metabolism and microenvironment among patients became larger with cancer progression. The average lifetimes for normal, middle and cancerous groups were 2.01 ± 0.09 ns, 1.68 ± 0.12 ns and 1.46 ± 0.21 ns, respectively. The middle group partially overlapped with the cancerous group, which revealed the middle group was abnormal or partially cancerous. To determine the adequate tumor margins, a criterion line could be drawn at 1.92 ns according to the average lifetime of the normal group, which covered 93.3% (14 of 15) normal sections in this study. Based on the criterion line, the tissues with lifetimes higher than 1.92 ns are healthy, and the tissues with lifetimes lower than the line are generally cancerous. The criterion line would be a crucial judgmental tool for lung cancer diagnosis and intraoperative margin assessment.
Fig. 4.
(a) Scatter plot of average lifetimes for 15 patients of the testing group, including normal (blue triangle), middle (red circle) and cancerous (black square) lung tissue sections. (b) Scatter plot of average lifetimes for 41 samples including 13 normal (blue triangle) and 28 cancerous sections (black square) from the validation group, which were diagnosed by H&E histopathological examination. (c) Free and protein-bound NADH lifetimes, and their relative contribution ratio in normal and cancerous lung tissues.
To evaluate the proposed criterion line, we examined 41 frozen sections of the validation group for the 16 patients when we were blinded to the histopathologic diagnosis. In Fig. 4(b), 12 of 13 normal sections and 26 of 28 cancer sections were correct according to the criterion line at 1.92 ns. Compared to the H&E histopathological diagnosis, the sensitivity and specificity of the FLIM method was quantified as 92.9% (26/28) and 92.3% (12/13) respectively. This technique demonstrates high concordance with H&E stain for tumor diagnosis.
3.5. NADH and FAD lifetimes of lung tissues
It is known that NADH emits a fluorescence of 440–530 nm, and FAD emits a fluorescence of 500–600 nm with 405 nm laser excitation by spectral study. To collect the autofluorescence of the tissues as much and fast as possible, the signals of FLIM study in Fig. 1, Fig. 2, Fig. 3 are all the autofluorescence with wavelength longer than 430 nm. To figure out the fundamental of our FLIM data, we examined the lifetimes of NADH and FAD separately of normal and cancerous lung tissues with two different filters. The lifetimes of NADH were 1.48 ± 0.06 ns and 1.22 ± 0.09 ns and those of FAD were 2.82 ± 0.11 ns and 2.61 ± 0.12 ns for normal and cancerous lung tissues, respectively. It seems the lifetimes of NADH and FAD both decreased in cancerous lung tissues. Since the fluorescence of protein-bound FAD is very weak [19] the lifetime of FAD in this study was mainly contributed by free FAD whose lifetime was reported in the range of 2.28–3.13 ns [20], [21]. The decreasing of FAD lifetime was caused by cellular micro-environment change in cancerous lung tissues. For NADH, both free and protein-bound form NADH contributed to the autofluorescence. Free NADH has a lifetime of 0.3–0.8 ns while protein-bound NADH has a longer lifetime of 1.0–6.5 ns [22]. Fig. 4(c) showed free NADH and protein-bound NADH lifetimes of normal and cancerous lung tissues by fitting the FLIM image of NADH with double-exponential decay models. It can be seen that both the free and protein-bound NADH lifetimes decreased in cancerous lung tissues. And the ratio of free to protein-bound NADH increased in cancerous lung tissues. We supposed that the decrease of both NADH and FAD lifetimes may be due to a shift from oxidative phosphorylation to glycolysis, which was further studied by cell models.
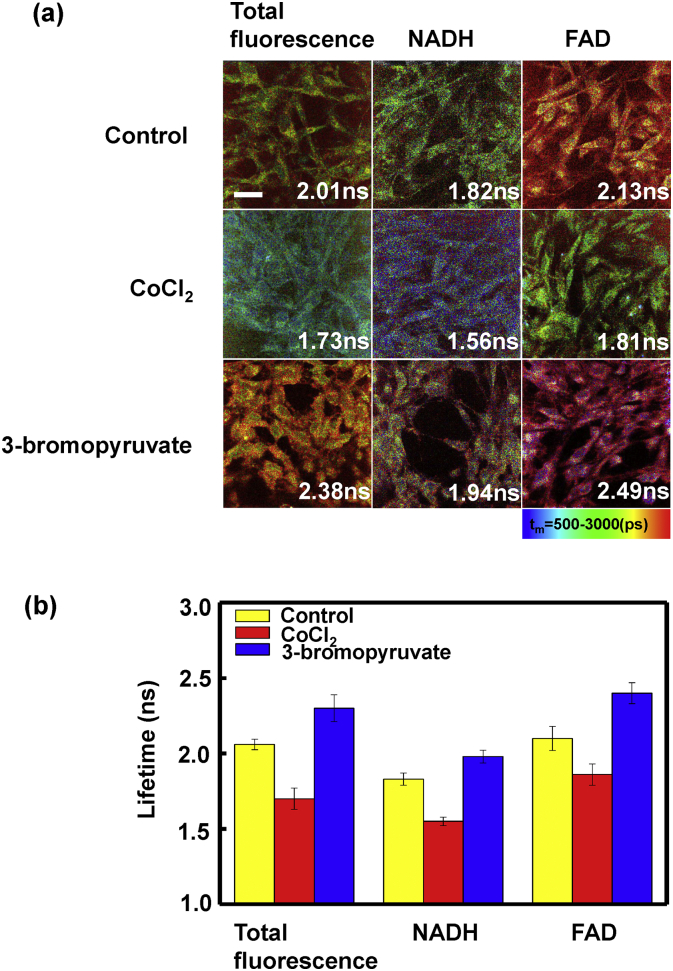
3.6. NADH and FAD lifetimes of BEAS-2B cells
Considering the predictions of neoplastic metabolism and the Warburg effect that cancer favors glycolysis over oxidative phosphorylation [23], we further investigated the NADH and FAD lifetimes of BEAS-2B cells with inhibition of specific metabolic pathways. Fig. 5(a) showed FLIM images of control cells (untreated), cells incubated with CoCl2 or 3-bromopyruvate, from which qualitative differences can be easily seen. CoCl2 inhibits oxidative phosphorylation and 3-bromopyruvate inhibits glycolysis. As shown in Fig. 5, CoCl2 decreased NADH and FAD lifetimes while 3-bromopyruvate elevated them relating to control group. The mutual shifts of metabolism pathways caused a significant lifetime change on two treated groups. From the cell experiment, we can demonstrate that the lifetimes of NADH and FAD decreased with metabolic pathways shifting to glycolysis, which reflected the distribution of NADH and FAD binding sites has changed and gave an explanation of the lung tissue experimental results.
Fig. 5.
(a) FLIM images of BEAS-2B cells: cells were untreated (control), treated with CoCl2 or 3-bromopyruvate. Left column represents the lifetime images of total fluorescence, middle and right columns represent the images of NADH and FAD, respectively. Scale bar is 30 μm. (b) Total fluorescence, NADH and FAD lifetimes of control cells (yellow), cells treated with CoCl2 (red) and treated with 3-bromopyruvate (blue).
4. Discussion
We provide the first ex vivo human lung data to demonstrate the ability of delineating tumor tissue from healthy tissue with FLIM. In our study, the lifetime of cancerous lung tissues is consistently lower than normal tissues, and this is due to the both decrease of NADH and FAD lifetimes. A lifetime decrease in metabolic pathways with neoplastic metabolism is also demonstrated in human bronchial epithelial cells (BEAS-2B) by inhibiting specific metabolic pathways. In general, cancer cells exhibit an elevated level of free NADH. And this is confirmed by metastatic and nonmetastatic murine melanoma cell lines, which show that the average lifetime of NADH is lower in metastatic cells than in nonmetastatic cells. Specifically, nonmalignant cells exhibit average lifetime ranging from 1.4 ns to 1.9 ns while malignant cells lifetime are in the range of 0.5 ns to 0.85 ns [24], [25]. Previous study of NADH lifetime in human breast cells [26] indicated that a decrease in the lifetime of both protein-bound NADH and free NADH is due to hypoxia. Skala et al. used two-photon excited FLIM to study NADH and FAD in the hamster cheek pouch model of oral cancer in vivo. They found a significantly decreased lifetime in both protein-bound NADH and free FAD with increasing cancer grade [27]. In another research of Skala et al, they also demonstrated that the decrease in protein-bound NADH lifetime with dysplasia is due to a shift from oxidative phosphorylation to glycolysis in hamster cheek pouch model [28]. Our results of human lung data are consistent with these recent FLIM studies in animal models or cell lines.
Previous studies reported that high neutrophil: lymphocyte ratio (NLR) value is associated with inflammation, poor survival and a higher risk of distant metastasis [29], [30]. We examined the pathological data of NLR for all the patients before any treatment. There were 4 patients among the total 31 patients having high NLR values (> 5). The “normal” sections of the 4 patients with high NLR were examined by FLIM. Interestingly, it was found that two patients of them (2/4, 50.0%) showed lower lifetimes than other cases. The NLR values of patient No.9 in the testing group and sample No.10 in the validation group were 20.5 and 5.37. And the lifetimes of their “normal” sections were 1.85 ± 0.04 ns and 1.85 ± 0.16 ns respectively (Fig. 4(a) and (b)), which were lower than the proposed criterion line at 1.92 ns and may indicate that the resection margin is still in the cancerous region. Patients with positive margins may lead to second surgery, a higher risk of recurrence and distant metastasis, associated with greater psychological pressure and medical expenses. For the remaining 27 patients with NLR values lower than 5, the average NLR value was 2.07 ± 0.52. The “normal” sections of all the 27 cases (100%) had lifetimes larger than 1.92 ns, which indicated that the sections were in the healthy region based on the FLIM study. Owing to the limited number of patients with high NLR value in this study, we were not able to study the interesting result further. However, we presume that larger tumor margin and surgery area might be more adequate for some patients with higher NLR values.
Current assessment of safe resection margin during lung cancer surgery is still based on the H&E imaging results of frozen sections whose accuracy is highly depended on the pathological diagnosis experience of pathologists. In recent years, several optical imaging techniques such as laser-induced fluorescence imaging (LIF) [31], multiphoton microscopy (MPM) [32], two-photon excited autofluorescence (TPEAF) [10], [33], coherent anti-Stokes Raman scattering imaging (CARS) and second harmonic generation (SHG) [11] have attracted interests for lung cancer and many other diseases. However, these imaging techniques still need experienced pathologists to analyze the cellular structure by the obtained images. FLIM also has the advantages of high-resolution, label-free, noninvasive and real-time imaging. Furthermore, FLIM can provide metabolic information via lifetimes of tissues with high sensitivity, which is objective and not dependent on the image contrast. Therefore, we can distinguish tumor tissue and normal lung tissue based on lifetime values. A criterion line of lifetime at 1920 ps can be given as a supporting approach helping surgeons to assess safe resection margin during the lung cancer surgery.
The FLIM data of the cell experiments are similar between one and two-photons excitations. Although multiphoton FLIM would reach deeper tissue, the excitation laser for one-photon FLIM is a picosecond laser which is of modest cost and portable for the superficial epithelial tumor like non-small cell lung cancer. FLIM without the need of tissue fixation or staining can provide cellular structure images as clearly as H&E images, and give lifetime results sensitively within 2 min, which can dramatically save time comparing to the time-consuming process of H&E-staining. The development of miniaturized endoscopy and scanning system will be necessary for clinical application of early cancer detection. One-photon excited FLIM as a rapid and powerful tool for diagnosis of lung cancer may ultimately be used either to aid surgeries in intraoperative margin assessment or to improve non-invasive screening of lung cancer through endoscope.
The following are the supplementary data related to this article.
Fig. S1.
H&E (a) and FLIM images of lung adenocarcinoma tissue paraffin-embedded section (b) and frozen section (c); (d) fluorescence lifetime distribution curves of FLIM images (b) and (c). Scale bar is 50 µm.
Funding
Supported by National Natural Science Foundation of China (11574056, 61575046, 31500599), Shanghai Municipal Commission of Health and Family Planning (201440407, 201540269), and Shanghai Rising-Star Program (16QA1400400).
Conflicts of interest
The authors disclose no potential conflicts of interest.
Transparency document
Transparency document.
Footnotes
The Transparency document associated with this article can be found, in online version.
Contributor Information
Guifang Wang, Email: wangguifang@fudan.edu.cn.
Lan Mi, Email: lanmi@fudan.edu.cn.
References
- 1.Liu B., Wu X., Liu B., Wang C., Liu Y., Zhou Q., Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. BBA-Mol. Basis Dis. 2012;11:1692–1704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Cathcart M.C., Gately K., Cummins R., Drakeford C., Kay E.W., O'Byrne K.J., Pidgeon G.P. Thromboxane synthase expression and correlation with VEGF and angiogenesis in non-small cell lung cancer. BBA-Mol. Basis Dis. 2014;5:747–755. doi: 10.1016/j.bbadis.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Huang Z., McWilliams A., Lui H., McLean D.I., Lam S., Zeng H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer. 2003;107:1047–1052. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 4.Lam S., MacAulay C., leRiche J.C., Palcic B. Detection and localization of early lung cancer by fluorescence bronchoscopy. Cancer. 2000;89:2468–2473. doi: 10.1002/1097-0142(20001201)89:11+<2468::aid-cncr25>3.3.co;2-m. [DOI] [PubMed] [Google Scholar]
- 5.Maygarden S.J., Detterbeck F.C., Funkhouser W.K. Bronchial margins in lung cancer resection specimens: utility of frozen section and gross evaluation. Mod. Pathol. 2004;17:1080–1086. doi: 10.1038/modpathol.3800154. [DOI] [PubMed] [Google Scholar]
- 6.Ebrecht R., Paul C.D., Wouters F.S. Fluorescence lifetime imaging microscopy in the medical sciences. Protoplasma. 2014;251:293–305. doi: 10.1007/s00709-013-0598-4. [DOI] [PubMed] [Google Scholar]
- 7.Madhuri S., Vengadesan N., Aruna P., Koteeswaran D., Venkatesan P., Ganesan S. Native fluorescence spectroscopy of blood plasma in the characterization of oral malignancy. Photochem. Photobiol. 2003;78:197–204. doi: 10.1562/0031-8655(2003)078<0197:nfsobp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteman S.C., Yang Y., Van Pittius D.G., Stephens M., Parmer J., Spiteri M.A. Optical coherence tomography: real-time imaging of bronchial airways microstructure and detection of inflammatory/neoplastic morphologic changes. Clin. Cancer Res. 2006;12:813–818. doi: 10.1158/1078-0432.CCR-05-0245. [DOI] [PubMed] [Google Scholar]
- 10.Pavlova I., Hume K.R., Yazinski S.A., Flanders J., Southard T.L., Weiss R.S., Webb W.W. Multiphoton microscopy and microspectroscopy for diagnostics of inflammatory and neoplastic lung. J. Biomed. Opt. 2012;17:0360141–0360149. doi: 10.1117/1.JBO.17.3.036014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X., Cheng J., Thrall M.J., Liu Z., Wang X., Wong S.T. Multimodal non-linear optical imaging for label-free differentiation of lung cancerous lesions from normal and desmoplastic tissues. Biomed. Opt. Express. 2013;4:2855–2868. doi: 10.1364/BOE.4.002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suhling K., French P.M., Phillips D. Time-resolved fluorescence microscopy. Photochem. Photobiol. Sci. 2005;4:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- 13.Uehlinger P., Gabrecht T., Glanzmann T., Ballini J.-P., Radu A., Andrejevic S., Monnier P., Wagnières G. In vivo time-resolved spectroscopy of the human bronchial early cancer autofluorescence. J. Biomed. Opt. 2009;14:024011–024019. doi: 10.1117/1.3088100. [DOI] [PubMed] [Google Scholar]
- 14.Drezek R., Brookner C., Pavlova I., Boiko I., Malpica A., Lotan R., Follen M., Richards-Kortum R. Autofluorescence microscopy of fresh cervical-tissue sections reveals alterations in tissue biochemistry with dysplasia. Photochem. Photobiol. 2001;73:636–641. doi: 10.1562/0031-8655(2001)073<0636:AMOFCT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 15.Ramanujam N., Richards-Kortum R., Thomsen S., Mahadevan-Jansen A., Follen M., Chance B. Low temperature fluorescence imaging of freeze-trapped human cervical tissues. Opt. Express. 2001;8:335–343. doi: 10.1364/oe.8.000335. [DOI] [PubMed] [Google Scholar]
- 16.Sharma V., Shivalingaiah S., Peng Y., Euhus D., Gryczynski Z., Liu H. Auto-fluorescence lifetime and light reflectance spectroscopy for breast cancer diagnosis: potential tools for intraoperative margin detection. Biomed. Opt. Express. 2012;3:1825–1840. doi: 10.1364/BOE.3.001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., Phipps J., Elson D.S., Stoy H., Tinling S., Meier J., Poirier B., Chuang F.S., Farwell D.G., Marcu L. Fluorescence lifetime imaging microscopy: in vivo application to diagnosis of oral carcinoma. Opt. Lett. 2009;34:2081–2083. doi: 10.1364/ol.34.002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brun J.-L., Cortez A., Rouzier R., Callard P., Bazot M., Uzan S., Daraï E. Factors influencing the use and accuracy of frozen section diagnosis of epithelial ovarian tumors. Am. J. Obstet. Gynecol. 2008;199:244. doi: 10.1016/j.ajog.2008.04.002. (e241-244. e247) [DOI] [PubMed] [Google Scholar]
- 19.Koziol B., Markowicz M., Kruk J., Plytycz B. Riboflavin as a source of autofluorescence in Eisenia fetida coelomocytes. Photochem. Photobiol. 2006;82:570–573. doi: 10.1562/2005-11-23-RA-738. [DOI] [PubMed] [Google Scholar]
- 20.Thomas P., Pande P., Clubb F., Adame J., Jo J.A. Biochemical imaging of human atherosclerotic plaques with fluorescence lifetime angioscopy. Photochem. Photobiol. 2010;86:727–731. doi: 10.1111/j.1751-1097.2010.00707.x. [DOI] [PubMed] [Google Scholar]
- 21.Heikal A.A. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark. Med. 2010;4:241–263. doi: 10.2217/bmm.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blacker T.S., Mann Z.F., Gale J.E., Ziegler M., Bain A.J., Szabadkai G., Duchen M.R. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014;5 doi: 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J., Tan M., Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356:156–164. doi: 10.1016/j.canlet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan A., Pal P., Durocher G., Villeneuve L., Balassy A., Babai F., Gaboury L., Blanchard L. Steady state and time-resolved fluorescence properties of metastatic and non-metastatic malignant cells from different species. J. Photochem. Photobiol. B. 1995;31:101–112. doi: 10.1016/1011-1344(95)07178-4. [DOI] [PubMed] [Google Scholar]
- 25.Drozdowicz-Tomsia K., Anwer A.G., Cahill M.A., Madlum K.N., Maki A.M., Baker M.S., Goldys E.M. Multiphoton fluorescence lifetime imaging microscopy reveals free-to-bound NADH ratio changes associated with metabolic inhibition. J. Biomed. Opt. 2014;19:086016. doi: 10.1117/1.JBO.19.8.086016. [DOI] [PubMed] [Google Scholar]
- 26.Bird D.K., Yan L., Vrotsos K.M., Eliceiri K.W., Vaughan E.M., Keely P.J., White J.G., Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 27.Skala M.C., Riching K.M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K.W., White J.G., Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skala M.C., Riching K.M., Bird D.K., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K.W., Keely P.J., Ramanujam N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J. Biomed. Opt. 2007;12 doi: 10.1117/1.2717503. (024014-024010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cedres S., Torrejon D., Martinez A., Martinez P., Navarro A., Zamora E., Mulet-Margalef N., Felip E. Neutrophil to lymphocyte ratio (NLR) as an indicator of poor prognosis in stage IV non-small cell lung cancer. Clin. Transl. Oncol. 2012;14:864–869. doi: 10.1007/s12094-012-0872-5. [DOI] [PubMed] [Google Scholar]
- 30.Templeton A.J., McNamara M.G., Seruga B., Vera-Badillo F.E., Aneja P., Ocana A., Leibowitz-Amit R., Sonpavde G., Knox J.J., Tran B. Prognostic Role of Neutrophil-to-Lymphocyte Ratio in Solid Tumors: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2014;106:124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 31.Hariri L.P., Tumlinson A.R., Besselsen D.G., Utzinger U., Gerner E.W., Barton J.K. Endoscopic optical coherence tomography and laser-induced fluorescence spectroscopy in a murine colon cancer model. Lasers Surg. Med. 2006;38:305–313. doi: 10.1002/lsm.20305. [DOI] [PubMed] [Google Scholar]
- 32.Yan J., Zhuo S.M., Chen G., Milsom J.W., Zhang H., Lu J.P., Zhu W.F., Xie S.S., Chen J.X., Ying M.G. Real-time optical diagnosis for surgical margin in low rectal cancer using multiphoton microscopy. Surg. Endosc. 2014;28:36–41. doi: 10.1007/s00464-013-3153-7. [DOI] [PubMed] [Google Scholar]
- 33.Pavlova I., Hume K.R., Yazinski S.A., Peters R.M., Weiss R.S., Webb W.W. Multiphoton microscopy as a diagnostic imaging modality for lung cancer. Int. Soc. Opt. Eng. 2010 doi: 10.1117/12.841017. (pp. 756918-756917) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.