Abstract
Canine babesiosis is a tick borne haemoprotozoan disease caused by large and small intraerythrocytic apicomplexan piroplasms of the genus Babesia spp. The clinical manifestations of the disease vary from inapparent subclinical form to hyperacute shock related haemolytic crisis. Microscopic examination of blood smears from suspected dogs revealed Babesia canis and B. gibsoni in 3.45 and 25.86 % of samples respectively. A seminested PCR based on previously published species specific primers targeting the 18S rRNA gene was utilized to identify the Babesia species infecting dogs of Kerala at the sub species level. The study revealed 57.5 % prevalence of Babesia spp. among dogs. This report also presents the first molecular evidence of Babesia canis vogeli and B. gibsoni among naturally infected dogs in Kerala, South India. Molecular survey revealed a high prevalence of B. gibsoni infections when compared with B. canis vogeli infections among canines of the state. Preliminary survey of the tick population revealed the presence of Rhipicephalus sanguineus, R. haemaphysaloides and Haemaphysalis bispinosa in infected animals. Further studies need be directed towards utilizing the PCR protocol for confirming the vectors of these species in the region.
Keywords: Babesia canis vogeli, B. gibsoni, Seminested PCR, Kerala
Introduction
Canine babesiosis, a tick borne protozoan disease caused by apicomplexan parasites of the genus Babesia, is characterized by fever, anemia and haemoglobinuria. Babesia gibsoni and B. canis comprise the two main species causing natural infections in dogs in a widespread geographic distribution including India. The latter is grouped into three phylogenetic groups and they vary in their geographical distribution, vector specificity and antigenic properties. These subspecies include Babesia canis canis, found in Europe; Babesia canis vogeli, in North and South Africa, North America and Brazil; and B. canis rossi, in South Africa (Uilemberg et al. 1989; Carret et al. 1999; Caccio et al. 2002; Matjila et al. 2004).
The clinical manifestation of the disease varies from subclinical to fatal depending upon pathogenicity of Babesia spp. (Solano-Gallego and Baneth 2011). According to Shaw et al. (2001), Rhipicephalus sanguineus and Haemaphysalis longicornis are the putative vectors of B. canis vogeli and B. gibsoni respectively in India. In India, tick borne diseases have been diagnosed by traditional methods using microscopic observation of the organism in blood smears which, however, does not permit reliable identification of the species. Molecular evidence of B. canis vogeli and B. gibsoni were reported from different cities of India including Delhi, Mumbai, Sikkim and Ladakh (Rani et al. 2011). In South India, sporadic cases of canine babesiosis have been reported in Tamil Nadu (Lakshmanan and John 2007; Sundar et al. 2004; Senthil kumar et al. 2009) and Kerala (Sabu et al. 2002; Karunakaran et al. 2011; Tresamol et al. 2013) using conventional microscopic detection methods. However, there is paucity of information regarding the molecular confirmation of Babesia species in canines from South India including Kerala. Molecular identification of species would help to record the true status of canine babesiosis in the State, besides providing an insight into the possibility of tick vectors infection in dogs in the region. The study was designed to identify the Babesia spp. infecting canines, using confirmatory molecular tool and to compare the results with the conventional microscopic detection methods.
Materials and methods
Sample collection
Blood samples collected from eighty dogs in Thrissur district of Kerala State, showing clinical signs suggestive of babesiosis such as pyrexia, haemoglobinuria, pale mucous membranes, general weakness, anorexia and tick infestation formed the material for the study. Thin peripheral smears were prepared, air-dried and fixed in methanol. The blood smears were subsequently stained following the standard Giemsa’s staining method. Partially engorged ticks were collected manually from the body of dogs suspected for babesiosis at the time of blood collection. They were carried to the laboratory in clean plastic vials covered with a piece of muslin cloth and identified under a stereo-zoom microscope. Ticks were also cleared by boiling in 10 % potassium hydroxide for detailed study. Identification was carried out using standard keys of Sen and Fletcher (1962).
DNA extraction
The blood samples were processed for DNA extraction using QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Germany) following manufacturer’s protocol. The DNA content and purity of the final elutes were estimated using nanospectrophotometer (Nano drop 200 °C, Thermoscientific, USA). The concentration was determined at 260 nm and purity at 260:280 nm ratio. Samples which yielded a ratio between 1.7 and 1.9 were selected for analysis.
PCR protocol
Genus and species specific primers were selected according to Birkenheuer et al. (2003) (Table 1). The PCR was performed in a 25 µl reaction volume containing 2.5 µl of buffer (10×) without MgCl2, 200 µM each of dNTP, 25 pmol each of forward and reverse genus specific primers (OFP and ORP), 1.5 mM of MgCl2, 1U of Taq DNA polymerase and 5.0 µl of template DNA. A thermal cycling program (MJ Mini, Biorad, USA) with initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation (94 °C, 45 s), annealing (59 °C, 45 s) and extension (72 °C, 45 s) and a final extension at 72 °C for 5 min was adopted.
Table 1.
Genus and species specific primers
| Sl no. | Primer sequence | Target organism |
|---|---|---|
| 1 | OFP: GTCTTGTAATTGGAATGATGGTGAC ORP: ATGCCCCCAACCGTTCCTATTA |
Babesia spp. |
| 2 | BCR-F: GCTTGGCGGTTTGTTGC | B. canis rossi |
| 3 | BCC-F: TGCGTTGACGGTTTGACC | B. canis canis |
| 4 | BCV-F: GTTCGAGTTTGCCATTCGTT | B. canis vogeli |
| 5 | BgibAsia-F:ACTCGGCTACTTGCCTTGTC | B. gibsoni |
The amplicons obtained using genus specific primers as per the PCR assay described above were subjected to semi nested PCR for confirmation of the species. For the seminested reaction, the reverse primer ORP was used with BCV-F for B. canis vogeli—specific detection, with BCC-F for B. canis canis—specific detection, with BCR-F for B. canis rossi—specific detection and with BgibAsia-F for B. gibsoni—specific detection under the reaction conditions described above, except for the following: 2.5 µl from the initial reaction was used as DNA template, annealing was at 60 °C for 45 s and the reactions were amplified for 30 cycles.
The amplicons were electrophoresed in 1.5 % agarose gel and sizes were resolved using 100 bp ladder (Fermentas, Lithuania). The amplicons were purified using silica gel purification columns (GeneJET, Thermoscientific), sequenced using Sangers dideoxy chain termination method and the sequences aligned using Sequencher Version 5.0 (SciGenom Labs Pvt Ltd, Cochin) and subjected to sequence analysis using BLASTn.
Results
Microscopic examination revealed Babesia spp. in 26 samples and could be differentiated as B. canis (3.45 %) and B. gibsoni (25.86 %) based on morphological features (Soulsby 1982).
Polymerase chain reaction was carried out with genus specific primers targeting the 18S rRNA gene, on DNA samples (n = 80) collected from babesiosis suspected dogs. Out of the total samples subjected to PCR, 46 (57.5 %) yielded a specific 340 bp amplicon indicating Babesia spp. infection, whereas only 32.5 % were positive by blood smear examination. Twenty-two samples negative by staining were found positive by PCR while two samples positive by staining were negative by PCR.The PCR assay had a sensitivity of 92.13 and a specificity of 59.26 %, when compared to blood smear examination (Table 2).
Table 2.
Comparison of PCR with microscopic examination for detection of canine Babesia spp
| Technique | Microscopy positive | Microscopy negative | Total |
|---|---|---|---|
| PCR positive | 24 | 22 | 46 |
| PCR negative | 2 | 32 | 34 |
| Total | 26 | 54 | 80 |
The amplicons of the genus specific PCR were used as template in a seminested PCR reaction using B. gibsoni species specific primer, BgibAsia-F to amplify partial 18SrRNA gene of the species. We observed a 185 bp amplicon in 40 samples indicating specific amplification of B. gibsoni 18S rRNA gene (Fig. 1). Similarly species specific primer BCV-F was used for the detection of B. canis vogeli and the expected product size was 192 bp (Fig. 2). Of the 46 samples, six were positive for B. canis vogeli infection. None of the samples produced specific amplification patterns with BCC-F and BCR-F primers indicating that B. canis canis and B. canis rossi were not present in the study population.
Fig. 1.
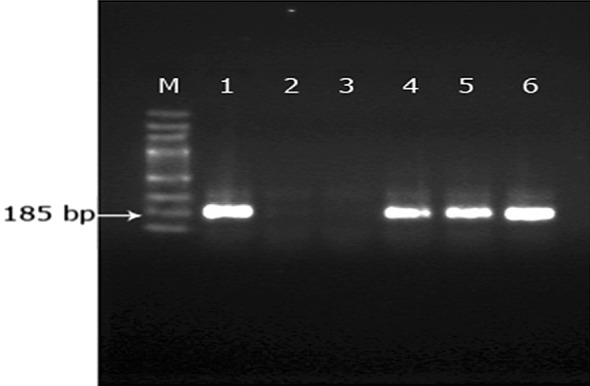
Analysis of seminested PCR products using B.gibsoni specific primers. From left to right 100 bp DNA marker (lane M), B.gibsoni (Lane 1, 4–6), no template control (Lane 2), negative sample (Lane 3)
Fig. 2.
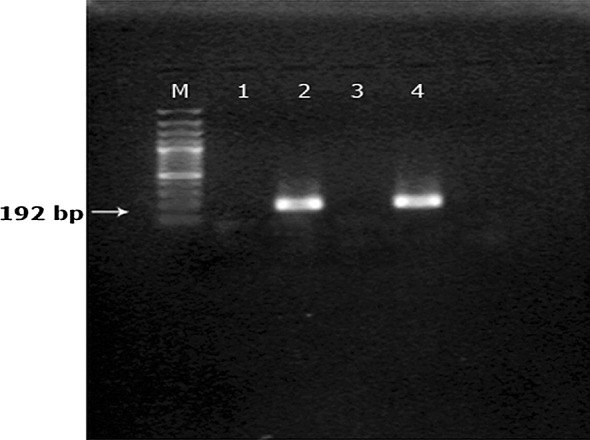
Analysis of seminested PCR products using B.canis vogeli specific primers 100 bp DNA marker (Lane M), no template control (Lane 1), B.canis vogeli (Lane2, 4), negative sample (Lane 3)
Nucleotide blast analysis of sequences data of B. gibsoni and B. canis vogeli amplified with species specific primers revealed 100 and 99 % identity respectively with those of several published sequences of the corresponding species.
From a total of 50 ticks collected from Babesia suspected dogs, 29 (58 %) ticks were identified as Rhipicephalus sanguineus, 20 (40 %) as Haemaphysalis bispinosa and one (2 %) as R. haemaphysaloides (male). Interestingly, all the ticks collected from B. gibsoni positive animals were identified as H. bispinosa.
Discussion
In the present study, seminested PCR assay was used for the detection of Babesia organism by amplifying a fragment of 18S rRNA gene as it was demonstrated to possess a high detection level of piroplasmid infection in symptomatic animals (Criado-Fornelio et al. 2003).Moreover, the first-round PCR products utilizing species specific primers could not be seen as a band after ethidium bromide staining, but the second-round PCR using nested primers produced a visualized band of expected size on each sample as observed by Ano et al. (2001). The authors had also stated that nested PCR approach is useful to detect infection of canine babesiosis with low parasitaemia including carriers because of its high sensitivity.
Out of the 80 samples subjected to PCR, 57. 5 % were found to be positive for Babesia spp. infection, using genus specific primers, suggesting a very high prevalence of babesiosis among canines subjected to the study. Moreover, a significantly higher prevalence of B. gibsoni (50 %) when compared with B. canis vogeli (7.5 %) could be observed utilizing the seminested PCR protocol. The fact that blood smear examination was relied upon for routine diagnosis and that it revealed the presence of Babesia organisms only in 26 samples, led us to conclude that the infection was thoroughly underestimated in the State till date. While comparing the microscopic and PCR observations for B. canis vogeli infection, O’Dwyer et al. (2009) had reported that 8 % of the samples were positive by PCR while only 2 % showed Babesia merozoites on microscopic examination. Extremely low percentage of Babesia infected cells within the peripheral blood might be the reason for the low sensitivity of microscopic detection of organism in blood smears (Fukumoto et al. 2001; M’ghirbi and Bouattour 2008; Buddhachat et al. 2012). At the same time, merit of the microscopic detection of the babesial piroplasms in blood smear depends on the expertise of the individual.
The PCR assay was found to have a sensitivity of 92.31 and a specificity of 59.26 % when compared with examination of stained blood smears. The low specificity appear misleading since PCR has been compared with a less sensitive and specific technique such as staining, the result of which has been considered as true positives. However, the exact sensitivity and specificity of the PCR technique can be determined only by comparing with a more sensitive technique such as cell culture isolation (Lehtinen et al. 2008).
The fact that none of the samples revealed B. canis canis or B. canis rossi infection reaffirms the speculations of Shaw et al. (2001) and confirms for the first time the identity of large babesial piroplasm in dogs of Kerala to be B. canis vogeli. However, contrary to the observations of Shaw et al. (2001), we could not identify H. longicornis from any of Babesia spp. infected dogs. Nevertheless, H. bispinosa was recovered from all B. gibsoni infected dogs necessitating future research into exploring the molecular evidence for vector status of this tick species.
Acknowledgments
The authors acknowledge the technical and financial support provided by Kerala Veterinary and Animal Sciences, University, Wayanad for the study.
References
- Ano H, Makimura S, Rharasawa R. Detection of Babesia species from infected dog blood by polymerase chain reaction. J Vet Med Sci. 2001;63:111–113. doi: 10.1292/jvms.63.111. [DOI] [PubMed] [Google Scholar]
- Birkenheuer AJ, Levy MG, Breitschwerdt EB. Development and evaluation of seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B.canis DNA in canine blood samples. J Clin Microbiol. 2003;41:4172–4177. doi: 10.1128/JCM.41.9.4172-4177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddhachat K, Meesong O, Nganvongpanit K, Osathanunkul M, Chomdej S. Molecular characterization and detection of Babesia canis vogeli in asymptomatic roaming dogs in Chiang Mai, Thailand. Thai J Vet Med. 2012;42:173–178. [Google Scholar]
- Caccio SM, Antunovic B, Moretti A, Mangili V, Marinculic A, Baric RR, Slemenda SB, Pieniazek NJ. Molecular characterization of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Vet Parasitol. 2002;106:285–292. doi: 10.1016/S0304-4017(02)00112-7. [DOI] [PubMed] [Google Scholar]
- Carret C, Alas F, Carey B, Grande N, Precigout E, Moubri K, Schetters TP, Gorenflot A. Babesia canis canis, Babesia canis vogeli, Babesia canis rossi: differentiation of the three subspecies by a restriction fragment length polymorphism analysis on amplified small subunit ribosomal RNA genes. J Eukaryot Microbiol. 1999;46:298–303. doi: 10.1111/j.1550-7408.1999.tb05128.x. [DOI] [PubMed] [Google Scholar]
- Criado-Fornelio A, Martinez-Marcos A, Buling-Sarana A, Barba-Carretero JC. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe part 1. Epizootiological aspects. Vet Parasitol. 2003;113:189–201. doi: 10.1016/S0304-4017(03)00078-5. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Xuvan X, Shigeno S, Kimbita E, Igarashi I, Nagasawa H, Fujisaki K, Mikami T. Development of a polymerase chain reaction method for diagnosing Babesia gibsoni infection in dogs. J Vet Med Sci. 2001;63:977–981. doi: 10.1292/jvms.63.977. [DOI] [PubMed] [Google Scholar]
- Karunakaran S, Pillai UN, Sasidharan HP. Babesia gibsoni infection in a German Shepherd dog. Vet World. 2011;4:269–270. doi: 10.5455/vetworld.4.269. [DOI] [Google Scholar]
- Lakshmanan B, John L. Clinical findings and concurrent infections in canine ehrlichiosis. Indian Vet J. 2007;84:863–864. [Google Scholar]
- Lehtinen LE, Birkenheuer AJ, Droleskey RE, Holman PJ. In vitro cultivation of a newly recognized Babesia sp. in dogs in North Carolina. J Vet Parasitol. 2008;151:150–157. doi: 10.1016/j.vetpar.2007.10.022. [DOI] [PubMed] [Google Scholar]
- M’ghirbi Y, Bouattour A. Detection and molecular characterization of Babesia canis vogeli from naturally infected dogs and Rhipicephalus sanguineus ticks in Tunisia. Vet Parasitol. 2008;152(1-2):1–7. doi: 10.1016/j.vetpar.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Matjila PT, Penzhorn BL, Bekker CP, Nijhof AM, Jongejan F. Confirmation of occurrence of Babesia canis vogeli in domestic dogs in South Africa. Vet Parasitol. 2004;122(2):119–125. doi: 10.1016/j.vetpar.2004.03.019. [DOI] [PubMed] [Google Scholar]
- O’Dwyer LH, Lopes VVA, Rubini AS, Paduan KDS, Ribolla PEM. Babesia spp. infection in dogs from rural areas of Sao Paulo State Brazil. Rev Bras Parasitol Vet. 2009;18:23–26. doi: 10.4322/rbpv.01802005. [DOI] [PubMed] [Google Scholar]
- Rani PAMA, Irwin PJ, Coleman GT, Gatne M, Rraub RJ. A survey of canine tick borne diseases in India. Parasit Vectors. 2011;4:141. doi: 10.1186/1756-3305-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabu L, Baby PG, Ajithkumar S, Devada K. Babesia canis infection in young pups. Intas Polivet. 2002;3:129–130. [Google Scholar]
- Sen SK, Fletcher TB. Veterinary entomology and acarology in India. New Delhi: Indian Council of Agricultural Research; 1962. p. 668. [Google Scholar]
- Senthil Kumar K, Vairamuthu S, Kathiresan D. Prevalence of haemoprotozoans in canines in Chennai city. Tamilnadu J Vet Anim Sci. 2009;5:104–108. [Google Scholar]
- Shaw SE, Day MJ, Birtles RJ, Breitschwerdt EB. Tick borne infectious diseases of dogs. Trends Parasitol. 2001;17:74–80. doi: 10.1016/S1471-4922(00)01856-0. [DOI] [PubMed] [Google Scholar]
- Solano-Gallego L, Baneth G. Babesiosis in dogs and cats-expanding parasitological and clinical spectra. J Vet Parasitol. 2011;181:48–60. doi: 10.1016/j.vetpar.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Soulsby EJL (1982) Helminths, arthropods and protozoa of domesticated animals. Bailliere and Tindall, London, pp 809
- Sundar N, Balachandran C, Senthilvelan A. Incidence of Babesia gibsoni infection in dogs in Tamil Nadu. J Vet Parasitol. 2004;18:79–80. [Google Scholar]
- Tresamol PV, Pillai UN, Anumol J, Devada K, Saseendranath MR. Cerebral babesiosis due to Babesia gibsoni in a dog—a case report. J Vet Anim Sci. 2013;44:85–86. [Google Scholar]
- Uilemberg G, Fransen EEI, Perrie NM, Spanier AAM. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet Quart. 1989;11(1):33–40. doi: 10.1080/01652176.1989.9694194. [DOI] [PubMed] [Google Scholar]


