Abstract
Lysine acetylations are post-translational modifications of cellular proteins, that are crucial in the regulation of many cellular processes. Lysine acetylations on histone proteins are part of the epigenetic code regulating gene transcription and are installed by histone acetyltransferases. Observations that inflammatory lung diseases, such as asthma and chronic obstructive pulmonary disease, are characterized by increased histone acetyltransferase activity indicate that development of small molecule inhibitors for these enzymes might be a valuable approach towards new therapies for these diseases. The 6-alkylsalicylate MG149 is a candidate to explore this hypothesis because it has been demonstrated to inhibit the MYST type histone acetyltransferases. In this study, we determined the Ki value for inhibition of the MYST type histone acetyltransferase KAT8 by MG149 to be 39 ± 7.7 μM. Upon investigating whether the inhibition of histone acetyltransferases by MG149 correlates with inhibition of histone acetylation in murine precision-cut lung slices, inhibition of acetylation was observed using an LC-MS/MS based assay on histone H4 res 4-17, which contains the target lysine of KAT8. Following up on this, upon treatment with MG149, reduced pro-inflammatory gene expression was observed in lipopolysaccharide and interferon gamma stimulated murine precision-cut lung slices. Based on this, we propose that 6-alkylsalicylates such as MG149 have potential for development towards applications in the treatment of inflammatory lung diseases.
Keywords: MG149, 6-alkylsalicylate, acetylation, acetyltransferase inhibitor, lung, inflammation
1. Introduction
Lysine acetylation is an important post-translational modification that is found on numerous cellular proteins, including both histone and non-histone proteins (1,2). Dynamic acetylation and deacetylation is regulated by enzymes called histone acetyltransferases (HATs) as writers and histone deacetylases (HDACs) as erasers. HAT and HDAC activity, and the balance between these activities, plays a crucial role in the regulation of multiple cellular processes (3,4). Moreover, HAT and HDAC activity, and dysregulation of the balance between the activities of these enzymes, has been implicated in multiple pathological conditions such as inflammatory disease and cancer, indicating that these enzymes represent potential drug targets in these diseases (5–8).
The importance of the activity of these enzymes in diseases is exemplified by the inflammatory lung disease asthma in which the balance between HAT and HDAC activity is dysregulated through a shift towards increased HAT activity (9). Chronic obstructive pulmonary disease (COPD), another inflammatory lung disease, is characterized by a loss of HDAC2 expression and activity, which also implies a shift in the balance between HAT and HDAC activity (9). This suggests that restoring the balance between HAT and HDAC activity could be an alternative therapeutic strategy for inflammatory lung diseases such as asthma and COPD. There is an unmet need for this since severe asthmatics display glucocorticoid resistance (10), and since next to this, the effectivity of glucocorticoids in COPD patients is subject to debate (11)(12).
We hypothesize that restoring the balance between HAT and HDAC activity using small molecule inhibitors of HATs may be a starting point for novel treatments for inflammatory lung diseases such as asthma and COPD. Zooming in on the HAT enzymes, these are a disparate group of enzymes from which most isoenzymes can be assigned to 5 main families based on their primary structure homology. Three families that have been studied extensively are the GNAT (GCN5-related N-acetyltransferase) family, represented by KAT2A (also known as GCN5; general control nonderepressible 5) and KAT2B (also known as PCAF; p300/CBP associated factor); the p300/CBP family, including KAT3A (also known as CBP; CREB-binding protein) and KAT3B (also known as p300); and the MYST family including KAT5 (also known as Tip60) and KAT8 (also known as MOF) (4). Over recent years several small molecule HAT inhibitors (HATi) have been described (6), targeting different HAT enzymes. This gives rise to the possibility of studying if these small molecule HATi have potential to achieve inhibition of expression of specific pro-inflammatory genes in model systems for inflammatory lung diseases such as asthma and COPD.
The HATi Anacardic Acid (AA), which is a 6-alkylsalicylate, has been reported to inhibit the HATs KAT8, KAT5, KAT2B and KAT3B (13,14). Interestingly, AA decreased the expression of IL-4, IL-5 and IL-13 in T cells isolated from mice challenged with ovalbumin to model allergic asthma. Upon re-administering these T cells to mice, the balance between HDAC and HAT activities were changed in lung tissue towards more HDAC activity (15). Furthermore, in a mouse model, AA was found to ameliorate lung damage which was induced by exposure of the mice to diesel exhaust particles. This effect was attributed to reduced levels of neutrophils in the lung parenchyma and reduced TNF-α levels in the BALF supernatant (16). These studies demonstrate that 6-alkylsalicylates which inhibit HATs have potential for the treatment of inflammatory lung diseases.
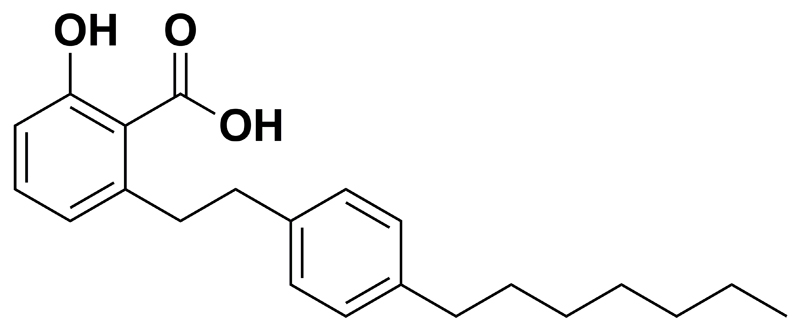
Using AA as a starting point our group developed the 6-alkylsalicylate MG149 (14) (Fig. 1) in which the 15 carbon atom 6-alkyl tail is replaced by a 4-heptylphenethyl substituent, providing a molecule with reduced lipophilicity and flexibility. Next to this, in terms of selectivity, MG149 shows less inhibition for the HATs PCAF and p300 compared to AA measured at concentrations of 200 μM, while retaining inhibitory potency for the MYST type HATs. We envision that this molecule provides a good starting point to investigate the potential of HAT inhibition for inhibition of inflammatory responses in model systems for inflammatory lung diseases. In this study we set out to investigate the potential anti-inflammatory effects of the 6-alkylsalicylate MG149 in model systems for inflammatory lung diseases such as asthma and COPD in connection to its HAT inhibitory activity. As a model system for inflammatory lung diseases murine precision-cut lung slices (PCLS) were selected. The lung tissue structure and cell-cell interactions are maintained in PCLS (17), thus providing a relevant ex-vivo model for lung inflammation. An advantage associated with the use of these type of organ slices is that the amount of required experimental animals can be reduced (18). Since promoting roles for lipopolysaccharide (LPS) and interferon gamma (IFNγ) have been described in asthma and COPD, as reviewed by Boorsma et al. (19), a combined stimulus of LPS and IFNγ was selected as an inflammatory stimulus in PCLS.
Fig. 1.
Chemical structure of MG149
Here, we report the kinetics of inhibition of the MYST HAT family member KAT8 by MG149, and a calculation of the inhibitory constant Ki of MG149 for KAT8. The inhibition of HATs by MG149 could be correlated to inhibition of histone acetylation in murine PCLS upon MG149 treatment, as determined by a mass spectrometry based analysis. This inhibition was observed on histone H4 res 4-17, containing H4 K16 which is the target of KAT8. Finally, we report reduced pro-inflammatory gene expression upon treatment with MG149 in murine PCLS. Taken together, this indicates that 6-alkylsalicylates such as MG149 have potential for development towards applications in the treatment of inflammatory lung diseases.
2. Materials and methods
2.1. General Reagents and Materials
All chemicals and reagents were purchased from Sigma Aldrich (St. Louis, Missouri, USA) unless otherwise stated. MG149 was purchased from Axon Medchem (Groningen, The Netherlands). The purity of MG149 was assessed by HPLC, MS, and NMR by Axon Medchem and was > 99%. Suberoylanilide hydroxamic acid (SAHA) was purchased from Selleckchem (Huissen, The Netherlands). The purity of SAHA was assessed by HPLC, MS, and NMR by Selleckchem and was > 99%.
2.2. Precision-cut lung slices
Precision-cut lung slices (PCLS) were prepared and cultured as described previously (20). All experiments were performed according to national guidelines and upon approval of the experimental procedures by the local Animal Care and Use committee of Groningen University, DEC number 6962A. Viability of MG149 treated PCLS was assessed by the amount of lactate dehydrogenase (LDH) released by the tissue slices into the culture medium. The measurements were performed as described previously (20). LDH release from the PCLS into the incubation medium was plotted relative to maximal LDH release, as determined by lysing 3 slices with 1% Triton X-100 for 30 min at 37°C at the start of the experiments.
2.3. Gene expression analysis in PCLS by RT-q-PCR
For gene expression analysis, PCLS were pre-treated with MG149 at 5 or 10 μM for 16 hrs. Inhibitor stocks were prepared in DMF and were further diluted in culture medium. Vehicle treatment constituted of pre-treatment with 0.2% DMF for PCLS, for 16 hrs. Subsequently, PCLS were stimulated with LPS and IFNγ in continued presence of the inhibitors, with 10 ng/mL LPS (Escherichia coli, serotype 0111:B4; Sigma-Aldrich) and 10 ng/mL IFNγ (cat.#315-05; PeproTech, Hamburg, Germany) for 4 hrs.
Gene expression analysis by RT-q-PCR was performed as described previously (20). The primers for TNFα (Mm00443258_m1), IL1β (Mm00434228_m1), IL6 (Mm00446190_m1), KC (Mm04208136_m1), iNOS (Mm00440502_m1) IL12b (Mm00434174_m1) and GAPDH (Mm99999915_g1) were purchased as Assay-on-Demand (Applied Biosystems).
2.4. Lysine acetyltransferase 8 (KAT8) inhibition assays
Activity of the HAT lysine acetyltransferase 8 (KAT8) was measured using chemical detection of coenzyme A (CoASH) after fluorescent labelling, as described previously (21).
2.5. Histone extraction and Micro BCA™ Protein Assay
For the histone extractions, PCLS were treated with the HAT inhibitor MG149 at 5 or 10 μM, in single treatments or in combination with 2 μM SAHA. PCLS were incubated with the inhibitors for 16 hrs. Subsequently, PCLS were stimulated with LPS and IFNγ in continued presence of the inhibitors as described above for the gene expression analysis. Inhibitor stocks were prepared in DMF and were further diluted in DMEM culture medium. Vehicle treatment constituted 0.2% DMF. PCLS were lysed in ice-cold Dulbecco’s Phosphate-buffered Saline (DPBS; Gibco by Life Technologies) supplemented with protease inhibitors (#88266, Thermo Scientific, Rockford, IL, USA) and sodium butyrate (1 mM final concentration), using glass beads (1.0 mm diameter Cat. No 11079110, BioSpec Products, Breda, The Netherlands) and a Mini-BeadBeater 24 machine (BioSpec Products, Breda, The Netherlands), for 2-3 cycles of 40 seconds, with 30 seconds of incubation on ice in between the cycles. Samples were then sonicated for 15 seconds with 1 second on and 1 second off intervals at amplitude 40% (using a Vibra-Cell VCX 130 from Sonics & Materials, Newtown USA). Histone extractions were then performed as previously described (22). After histone extractions the samples were diluted with PBS (PAA Laboratories GmbH, Austria) to determine the total protein concentration using the micro BCA protein assay according to the manufacturer's instructions (# 23235 Thermo Scientific, Rockford, IL, USA). Absorbance was measured with a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Vermont, USA) at 562 nm. A bovine serum albumin standard (# 23209 2 mg/mL, Thermo Scientific, Rockford, IL, USA,) was used to calibrate the assay.
2.6. Acetylation of histones with acetic anhydride-d6 (in-gel) and nano LC-MS/MS QTOF
The in-gel reactions of histones with acetic anhydride-d6 were performed as described previously (23). Supernatants containing histone peptides were subjected to LC MS/MS analysis as described previously (23) with the following modifications. For the analysis of acetylation status of the histone peptides by LC-MS/MS a quadruple time-of-flight mass spectrometer (QTOF, Bruker MaxisPlus) with a captive spray ionization interface was coupled to a nanoLC system (Dionex Ultimate3000). The auto sampler was held at 5°C, equipped with a 20 µl injection loop and used in the micro liter pick-up mode. A 300 µm x 5 mm trap column and a 75 µm x 150 mm analytical column both filled with C18 Pepmap 100 (Thermo Scienticic), 5 µm and 3 µm respectively were used for peptide forward flush trapping and separation. The following mobile phases were used for LC separations: solvent A, ultrapure water (resistance 18.2 MΩ, Millipore) with 0.1% (v/v) formic acid (FA), and solvent B, acetonitrile (ACN) with 0.1% (v/v) FA. The samples were injected (10 µl) and trapped for 2 minutes (the trapping time was set to a minimal value to prevent the loss of early eluting peptides) at a flow rate of 10 µL/min in a solution of 1% (v/v) ACN and 0.1% (v/v) FA in ultrapure water. The peptide separation was performed at 0.3 µl/min using a linear gradient from 1-35.5% solvent B in 65 minutes after a 4 min isocratic period at 1%. Hereafter the column was eluted with 90% solvent B for 10 minutes and conditioned with 1% solvent B for another 10 minutes after which it was ready for the next injection. The identification of peptides was based on data collected in auto MS/MS mode (2 GHz) with a maximum of 5 precursors per cycle and an active exclusion of 0.3 min using a mass range of 200-1500 amu, and was done using PEAKS Studio 7.5 (Bioinformatics Solutions Inc., Canada).
2.7. RAW264.7 cell culture and histone extractions
For the RAW264.7 murine macrophages, cell culture and histone extractions were carried out as described previously (23).
2.8. Statistical analysis
We used GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego CA, USA) to perform data analysis in all experiments. Data are presented as mean ± SD from at least 3 independent experiments, and were analyzed by 1-way analysis of variance, followed by Bonferroni post hoc tests in all cases. Significance was assigned at a p value ≤ 0.05.
3. Results
3.1. The 6-alkylsalicylate MG149 inhibits the MYST family HAT KAT8 with a Ki value of 39 ± 7.7 μM
6-alkylsalicylates such as MG149 have been shown to inhibit the MYST family HATs with potencies in the micromolar range (13,14). MYST HATs are bisubstrate enzymes binding to both Ac-CoA and histone lysine residues. Recently, we performed an enzyme kinetic study for KAT8 (21). Knowledge of the catalytic mechanism of KAT8 as well as the binding kinetics allowed for recalculating assay-dependent IC50 values for KAT8 inhibitors to assay-independent Ki values, based on an equation described by Cheng and Prusoff (21,24). Using this knowledge here, we analyzed the binding kinetics of 6-alkylsalicylate MG149 to KAT8, allowing for determination of an assay-independent Ki value.
We demonstrated previously that in the catalytic mechanism of KAT8, the free enzyme (E) becomes acetylated through a transfer of an acetyl group from Ac-CoA to the enzyme in a ping-pong mechanism (21). This acetylated form of the enzyme is referred to as (EX). The histone substrate is able to bind only to (EX), and upon binding of the histone substrate, the catalytically active form (E*XS) is induced. Subsequently, the histone substrate is acetylated and leaves the enzyme, upon which the free enzyme conformation (E) is regenerated. In this mechanism the Km of one substrate depends on the concentration of the other substrate (21). This means that IC50 values of KAT8 inhibitors depend on the concentrations of both Ac-CoA as well as the histone substrate. Therefore, it is important to derive assay-independent Ki values for KAT8 inhibitors such as MG149.
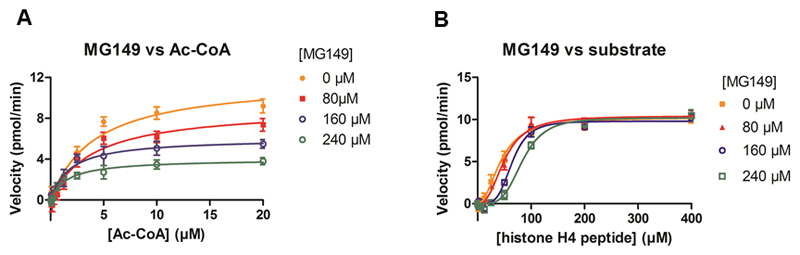
To this end the velocity of KAT8 was determined at different concentrations of Ac-CoA and constant concentration of the histone substrate in the presence of varying concentrations of MG149 (Fig. 2A). There was a clear decrease in Vmax app. and Km app. with increasing concentrations of MG149 (Table 1). The decrease in Vmax app. indicates allosteric binding with respect to the Ac-CoA binding pocket, and the decrease in Km app. suggests that the binding of MG149 stabilizes the binding of Ac-CoA. This suggests a form of uncompetitive inhibition where the substrate must be present for the inhibitor to bind, and supports a mechanism where MG149 binds to (EX), the catalytically inactive form of the enzyme.
Fig. 2.
Inhibitor kinetics of 6-alkylsalicylate MG149 on KAT8. A) The velocity of KAT8 was determined at 0 - 20 µM Ac-CoA in the presence of 0, 80, 160 and 240 µM MG149 respectively. Both Km app. and Vmax app decrease at increasing concentrations of MG149. B) The velocity of KAT8 was determined at 0 - 400 µM of the histone substrate (histone H4 peptide) in the presence of 0, 80, 160 and 240 µM MG149 respectively. The Vmax app. is constant at increasing concentrations of MG149, but the Khalf increases. Data shown are representative curves of 3 independent experiments ± SD.
Table 1.
Vmax app. and Km app. for Ac-CoA at different concentrations of inhibitor MG149. Vmax app., hill slope and khalf for histone H4 peptide at different concentrations of inhibitor MG149. Data shown are representative of 3 independent experiments ± SD.
| [MG149] (µM) | Ac-CoA |
Histone H4 peptide |
|||
|---|---|---|---|---|---|
| Vmax app. (pmol/min) | Km app. (µM) | Vmax app. (pmol/min) | Hill slope | Khalf (µM) | |
| 0 | 12 ± 1.2 | 4.1 ± 1.1 | 10 ± 0.3 | 2.1 ± 0.3 | 43 |
| 80 | 9.1 ± 1.0 | 4.0 ± 1.1 | 10 ± 0.4 | 2.5 ± 0.4 | 50 |
| 160 | 6.0 ± 0.3 | 1.8 ± 0.3 | 9.8 ± 0.3 | 4.2 ± 0.7 | 64 |
| 240 | 4.0 ± 0.3 | 1.6 ± 0.4 | 10 ± 0.3 | 4.0 ± 0.6 | 84 |
In addition, experiments were performed employing different concentrations of the histone substrate, a constant concentration of Ac-CoA and varying the MG149 concentration (Fig. 2B). The curves are sigmoidal in the presence of MG149, indicating cooperativity between the active and inactive forms of the enzyme, and resulting in a Hill coefficient which is not equal to 1. A Khalf can be calculated. The Khalf increases (Table 1), showing that the histone substrate loses affinity for the enzyme at increasing concentrations of MG149. At the same time the Vmax app. remains constant at increasing concentrations of MG149 (Table 1). This indicates that binding of the histone substrate can displace the inhibitor from KAT8.
Based on this, we conclude that the 6-alkylsalicylate MG149 binds according to the same model as previously described for the 6-alkylsalicylate AA (21). Therefore, the Ki value for MG149 was calculated as described previously (21), and found to be 39 ± 7.7 µM (see supporting information Fig. S1).
3.2. The 6-alkylsalicylate MG149 inhibits histone acetylation in murine PCLS
Thus having confirmed that the 6-alkylsalicylate MG149 inhibits the MYST family member KAT8 with a potency in the micromolar range, we moved on to investigate its effect on histone acetylation in murine PCLS stimulated with LPS and IFNγ. Since the HAT KAT8 targets histone H4 K16 (25), we focused in particular on a histone peptide from the N-terminal tail of histone H4 containing this lysine residue (res. 4-17: GKGGKGLGKGGAKR). PCLS were treated with MG149 and LPS and IFNγ, after which histones were extracted, and acetylations were analyzed using an LC-MS/MS based assay. Histone acetylation levels can reproducibly be determined using a mass spectrometry based assay as applied in one of our previous studies (23). This assay follows a previously described approach, in which derivatization of unmodified lysine residues with acetic anhydride-d6 ((CD3CO)2O) is followed by trypsin digestion (26–29). The resulting peptide fragments contain either deuterated or non-deuterated acetyl groups on the lysine residues based on endogenous acetylation levels. This enables quantification of endogenous acetylation levels relative to non-acetylated lysine residues in the corresponding peptides. Compared to antibody based techniques it is more sensitive which allows for small effects on histone acetylation to be observed, circumvents selectivity issues of antibodies, and has the advantage of being able to monitor individual (histone) peptides. Therefore, this method was applied to analyze the effect of MG149 on histone acetylation in PCLS.
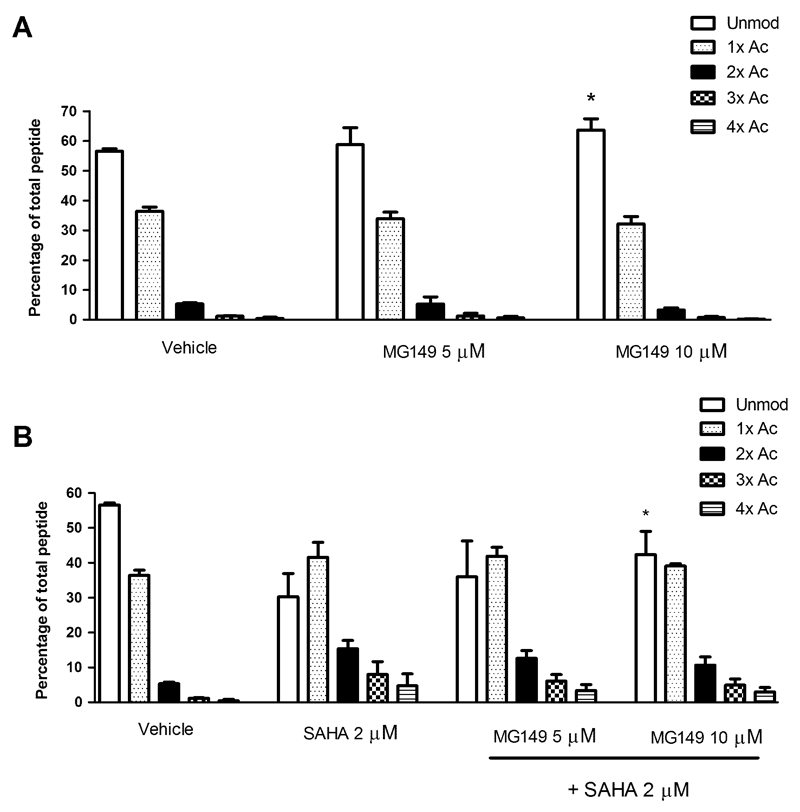
As a control experiment, the effects of the LPS and IFNγ stimulus or the inhibitor solvent DMF (added to the culture medium at 0.2% without MG149 as vehicle treatment) on histone acetylation in PCLS were investigated. Neither was found to affect histone acetylation on histone H4 res. 4-17 under the applied conditions (Fig. S2). Subsequently, PCLS were incubated with MG149 at 5 or 10 μM for 20 hrs (higher concentrations were not studied due to decreased viability as assessed by LDH measurement; see supporting information for the viability at 5 or 10 μM MG149, Fig. S3). Under these conditions, compared to the vehicle treatment, a small but significant inhibition of acetylation was observed on the histone H4 res. 4-17 peptide upon MG149 treatment at 10 μM (Fig. 3A), but not at 5 μM, which indicates a dose dependent effect. An explanation for the small effect on histone acetylation could be that the detection window for inhibition of acetylation is quite small. This could be related to the dynamics and stoichiometry of acetylation as described previously (30)(31). To increase this window the non-selective HDAC inhibitor (HDACi) SAHA was used. In line with our expectations, SAHA increased histone acetylation in PCLS (Fig. 3B). In combined treatment with SAHA, MG149 at 10 μM inhibited histone acetylation compared to treatment with SAHA only (Fig. 3B), and this inhibition of acetylation was larger than the inhibition observed in MG149 single treatment at 10 μM. Next to these findings, under the applied conditions no changes were observed in the acetylation status of other histone peptides that were detectable in our mass spectrometry analysis (Fig. 4). Furthermore, upon attempting to confirm these findings in RAW264.7 murine macrophages as another model system, no changes in acetylation status were found on any of the histone H3 or H4 peptides using this method (Fig S4). Taken together, this indicates that in PCLS treatment with the HATi MG149 can be correlated to inhibition of histone acetylation on one histone peptide of H4 res 4-17.
Fig. 3.
The 6-alkylsalicylate MG149 reduces acetylation status of histone H4 res 4-17 in precision-cut lung slices (PCLS) after 20 hrs of incubation. PCLS were incubated with (A) MG149 at the indicated concentrations for 20 hrs, or (B) 2 μM of the HDAC inhibitor SAHA (to enlarge the detection window for inhibition of acetylation) in combination with MG149 at the indicated concentrations. For the last 4 hrs of the experiment, PCLS were incubated with LPS and IFNγ (10 ng/mL of each) in continued presence of the inhibitor. Histones were then extracted. Histones were resolved by SDS PAGE, and histones H3 and H4 were excised from the gels. Gel pieces were treated with acetic anhydride-d6, followed by trypsin digestion. Resulting peptides were subjected to LC-MS/MS analysis. Data are presented as mean values ± SD of 3 independent experiments. * p < 0.05 compared to vehicle (in A) or SAHA (in B). No significant differences were detected between vehicle treated PCLS and untreated or LPS IFNγ treated PCLS (see Fig S2).
Fig. 4.
Coverage of histone peptides with LC-MS/MS. The peptides indicated in bold are the ones that were detected and analyzed. With this LC-MS/MS method almost all acetylation sites in histone H3 and H4 can be studied. In both histones 2 sites are being missed due to the fact that the peptides containing these lysines are either very small or large, or do not trap well on the LC column. Altogether, most of the important lysines in the N-terminal tails of these histones can be detected.
3.3. The 6-alkylsalicylate MG149 reduces pro-inflammatory gene expression in murine PCLS
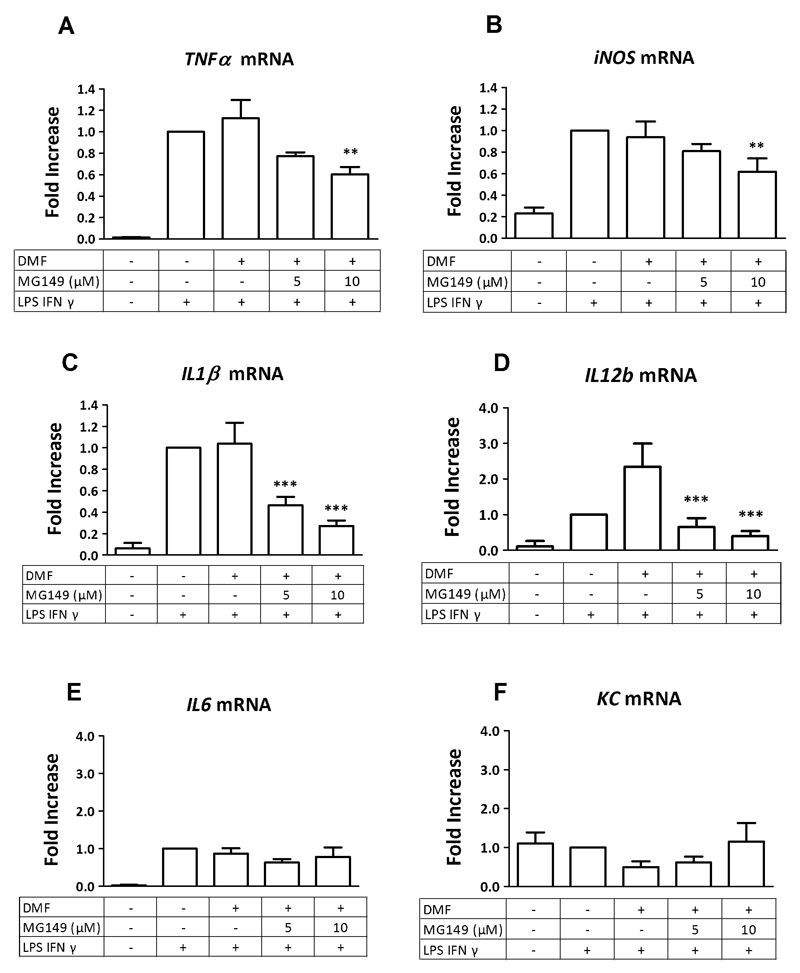
To study whether the inhibition of histone acetylation upon incubation with the 6-alkylsalicylate MG149 correlates with reduced expression levels of pro-inflammatory genes in PCLS, mRNA levels of the pro-inflammatory genes TNFα, iNOS, IL1β, IL6, and KC were monitored using RT-q-PCR. The LPS and IFNγ stimulus in PCLS increased the expression of all these genes compared to untreated cells (Fig. 5A-E), except for KC, where the LPS and IFNγ stimulus did not affect gene expression in PCLS (Fig. 5F). Upon studying the effect of a dilution of the inhibitor solvent DMF (which was diluted into the culture medium at 0.2% as a vehicle treatment without the addition of MG149), no significant changes in gene expression were observed, except for the expression of IL12b, which was increased (Fig. 5D). Importantly, upon MG149 pre-treatment, large reductions in the LPS and IFNγ induced expression levels of IL1β and IL12b were observed (Fig. 5A and D). This reduction was dose-dependent, and was studied at 5 and 10 μM MG149. For TNFα and iNOS, smaller dose-dependent reductions in gene expression were observed at these concentrations (Fig. 5B and C). No changes in the expression levels of IL-6 or KC were observed in PCLS upon MG149 pre-treatment under the same conditions (Fig. 5E and F). Taken together, this indicates that the inhibition of histone acetylation upon MG149 treatment can indeed be correlated with reduced pro-inflammatory gene expression in PCLS.
Fig. 5.
6-alkylsalicylate MG149 dose dependently reduces pro-inflammatory gene expression of A) TNFα B) iNOS C) IL1β and D) IL12b in murine precision-cut lung slices (PCLS). No significant changes were observed on the expression of (E) IL6 or (F) KC. PCLS were pre-treated with MG149 at the indicated concentrations for 16 hours, after which an inflammatory LPS and IFNγ stimulus (10 ng/mL of each) was given for 4 hours in continued presence of the inhibitor (making the total incubation time with the inhibitor 20 hrs). Subsequently, gene expression was analyzed by RT-q-PCR. For vehicle treatment, PCLS were pre-treated with the inhibitor solvent DMF. For IL12b, an increase was seen upon this vehicle treatment (D). Data represent the target gene expression normalized to the reference gene. The values shown are means ± SD of 3-4 independent experiments. *** p < 0.001, ** p < 0.01 and * p < 0.05 compared to vehicle.
4. Discussion
In this study the enzyme kinetics of inhibition of the MYST HAT family member KAT8 by the 6-alkylsalicylate MG149 were investigated. This study demonstrates that the inhibition of HATs by MG149 correlates with inhibition of histone acetylation as well as reduced pro-inflammatory gene expression in murine PCLS, representing an ex vivo model for inflammatory lung diseases. This suggests that 6-alkylsalicylates such as MG149 can be developed towards applications in the treatment of inflammatory lung diseases such as asthma and COPD. Bacterial acute exacerbations of COPD or severe asthma, or acute lung injury, may be relevant areas of application for MG149 based on the relevance of the employed LPS/IFNγ stimulus to these disease conditions.
Here, we demonstrated that the 6-alkylsalicylate MG149 inhibits the MYST family HAT member KAT8 according to a similar mechanism as observed previously for the 6-alkylsalicylate AA (21). These 6-alkylsalicylates bind to the EX form of the enzyme and stabilize this catalytically inactive conformation, thus inhibiting the acetylation of the histone H4 peptide. Knowledge of the catalytic mechanism of KAT8 as well as the binding kinetics allows for recalculating assay-dependent IC50 values for KAT8 inhibitors to assay-independent Ki values, based on an equation described by Cheng and Prusoff (21,24). Using this knowledge, an assay-independent Ki value of 39 ± 7.7 μM was determined for the inhibition of KAT8 by the 6-alkylsalicylate MG149. This can now be directly compared to the 6-alkylsalicylate AA from which MG149 was derived, for which a Ki value of 64 ± 8.9 μM was previously determined for KAT8 inhibition (21). This indicates an improved potency for MG149 with respect to its Ki value for KAT8.
In this study, MG149 demonstrated inhibition of histone acetylation. A small but significant inhibition of histone acetylation was observed upon treatment of the PCLS with 10 μM of MG149. Furthermore, this inhibition of histone acetylation was more pronounced upon combined treatment with SAHA compared to SAHA treatment alone. When studying HATi, a frequently applied method to enable investigation of inhibition of histone acetylation is the simultaneous treatment with a HDACi. This increases the histone acetylation level thus enlarging the window to observe inhibition of histone acetylation. The rationale for combined treatment with HDACi and HATi to increase the detection window to reveal the effect of the HATi under investigation could relate to the dynamics and stoichiometry of histone acetylation, as described previously (30) (31). In this study both methods were used and similar results were found in both cases. Taken together both methods confirm that the HATi MG149 inhibits histone acetylation in PCLS. Our observations are in line with literature in which it has been shown that co-treatments with MG149 and SAHA demonstrated inhibition of histone acetylation compared to SAHA treatment alone in a model using human cancer cell lines MOPL8 and K562 (32). Similar results were obtained for another 6-alkylsalicylate analogue to MG149 in HepG2 cells (33). In both cases western blot was used. This study demonstrates that application of mass spectrometry is more sensitive and enables detection of smaller changes in histone acetylation, which reduces the need to increase the detection window using HDACi. The fact that we did not observe inhibition of histone acetylation in RAW264.7 murine macrophages upon MG149 treatment indicates that this effect could be cell type specific, and could be explained by the presence of various cell types in PCLS, and possibly also due to effects on cell-cell interactions in the intact lung tissue samples.
When studying the potential of HATi for the treatment of inflammatory lung diseases, investigating lung tissue specific effects may be particularly relevant since local administration of small molecule inhibitors in lung tissue is possible and is already applied for inhaled glucocorticoids in the treatment of these diseases. It should be noted that lung tissue specific effects of 6-alkylsalicylates on histone acetylation have not been reported before. Interestingly, it has been demonstrated that Ac-CoA levels vary between different tissue types, and this influences activities of HATs and HDACs (2). This indicates that the effects upon treatments with HATi could differ between different tissues and need to be studied in a tissue specific manner. This study demonstrates the feasibility of studying effects of small molecule inhibitors on histone acetylation specifically in lung tissue using a more accurate mass spectrometry based analysis compared to antibody based techniques, which we believe will have added value for the development of these type of inhibitors towards applications in the treatment of inflammatory lung diseases. Others have not done this using mass spectrometry.
In this study, reduction of pro-inflammatory gene expression was observed in PCLS upon treatment with the 6-alkylsalicylate MG149, supporting the hypothesis that 6-alkylsalicylates have potential for the treatment of inflammatory lung diseases. In lung tissue, effects of MG149 on pro-inflammatory gene expression have not been reported before. However, results obtained with the 6-alkylsalicylate AA in mouse models of lung inflammation are in line with our findings, as exemplified by decreased cytokine expression and a change in the balance of HAT to HDAC activities (15)(16). Furthermore, AA was reported to influence the NF-κB pathway in a study by Sung et al using human cancer cell lines. Important findings of this study include that AA suppressed NF-κB activation upon TNF-α or LPS stimulus, and that AA suppressed acetylation of p65 upon TNF-α stimulus (34). Additionally, for MG149 it is also interesting to note that this inhibitor appears to have selective effects on gene expression, as was shown using a microarray analysis where this inhibitor mainly affected the NF-κB and p53 pathways in MOLP8 and K562 human cancer cell lines (32). Altogether this indicates that 6-alkylsalicylates influence pro-inflammatory events in cellular systems.
Other literature concerning MG149 has also focused on alternative applications. For example, MG149 has been used to study the role of KAT5 in chromosome segregation (35). This indicates that next to applications where 6-alkylsalicylates like MG149 could be used as a drug, these small molecules can also be used as a tool to study the role of MYST type HATs in fundamental cellular processes.
In summary, our findings indicate that the 6 alkylsalicylate MG149 inhibits histone acetylation and expression of several pro-inflammatory genes in murine precision cut lung slices. This sets the stage for further development of this compound class towards applications in the treatment of inflammatory lung diseases such as asthma and COPD. Future directions should focus on improving Ki values and selectivity for the HAT enzymes for these compounds.
Supplementary Material
Acknowledgements
We acknowledge the EU-COST action TD0905 ‘epigenetics from bench to bedside’ for funding a short term scientific mission of TvdB to the group of Axel Imhof, Ludwig Maximilians University, Munich. We acknowledge the EU-COST action CM1406 ‘epigenetic chemical biology’ for building a scientific network. We acknowledge the European Union for funding this project by an ERC starting grant to FJD (309782). Further support was obtained from The Netherlands Organisation of Scientific Research (NWO) by a VIDI grant to FJD (723.012.005). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We acknowledge Prof. Dr. Reinoud Gosens (Department of Molecular Pharmacology, University of Groningen) for support with ex vivo experiments regarding precision-cut lung slices. We acknowledge Andries Heida for support with experiments regarding histone acetylation in RAW264.7 cells.
Abbreviations
- HAT
histone acetyltransferase
- HATi
HAT inhibitor
- HDAC
histone deacetylase
- PCAF
P300/CBP-associated factor
- MOZ
monocytic leukemic zinc finger
- COPD
chronic obstructive pulmonary disease
- PCLS
precision-cut lung slices
- SAHA
Suberoylanilide hydroxamic acid
- LDH
lactate dehydrogenase
- LPS
lipopolysaccharide
- IFNγ
interferon gamma
- TNFα
tumor necrosis factor alpha
- iNOS
inducible nitric oxide synthase
- IL1β
interleukin 1 beta
- IL12b
interleukin 12 subunit beta
- IL6
interleukin 6
- KC
keratinocyte-derived chemokine
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- PMSF
phenylmethylsulfonyl fluoride
- DMF
dimethylformamide
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- KAT8
lysine acetyltransferase 8
- HDACi
HDAC inhibitor
References
- (1).Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006 Aug;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- (2).Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014 Aug;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- (3).Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004 Apr 16;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- (4).Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001 May;58(5–6):693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009 Oct;14(19–20):942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- (6).Dekker FJ, van den Bosch T, Martin NI. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov Today. 2014 May;19(5):654–660. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- (7).Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015 Mar 2;20(3):3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006 Sep;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- (9).Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J. 2005 Mar;25(3):552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- (10).Keenan CR, Salem S, Fietz ER, Gualano RC, Stewart AG. Glucocorticoid-resistant asthma and novel anti-inflammatory drugs. Drug Discov Today. 2012 Sep;17(17–18):1031–1038. doi: 10.1016/j.drudis.2012.05.011. [DOI] [PubMed] [Google Scholar]
- (11).Telenga ED, Kerstjens HA, Postma DS, Ten Hacken NH, van den Berge M. Inhaled corticosteroids in chronic obstructive pulmonary disease: a review. Expert Opin Pharmacother. 2010 Feb;11(3):405–421. doi: 10.1517/14656560903510628. [DOI] [PubMed] [Google Scholar]
- (12).Babu KS, Kastelik JA, Morjaria JB. Inhaled corticosteroids in chronic obstructive pulmonary disease: a pro-con perspective. Br J Clin Pharmacol. 2014 Aug;78(2):282–300. doi: 10.1111/bcp.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003 May 23;278(21):19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- (14).Ghizzoni M, Wu J, Gao T, Haisma HJ, Dekker FJ, George Zheng Y. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur J Med Chem. 2012 Jan;47(1):337–344. doi: 10.1016/j.ejmech.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang HP, Wang L, Fu JJ, Fan T, Wang ZL, Wang G. Association between histone hyperacetylation status in memory T lymphocytes and allergen-induced eosinophilic airway inflammation. Respirology. 2016 Mar 17; doi: 10.1111/resp.12774. [DOI] [PubMed] [Google Scholar]
- (16).Carvalho AL, Annoni R, Torres LH, Durao AC, Shimada AL, Almeida FM, et al. Anacardic acids from cashew nuts ameliorate lung damage induced by exposure to diesel exhaust particles in mice. Evid Based Complement Alternat Med. 2013;2013:549879. doi: 10.1155/2013/549879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Morin JP, Baste JM, Gay A, Crochemore C, Corbiere C, Monteil C. Precision cut lung slices as an efficient tool for in vitro lung physio-pharmacotoxicology studies. Xenobiotica. 2013 Jan;43(1):63–72. doi: 10.3109/00498254.2012.727043. [DOI] [PubMed] [Google Scholar]
- (18).de Kanter R, Monshouwer M, Meijer DK, Groothuis GM. Precision-cut organ slices as a tool to study toxicity and metabolism of xenobiotics with special reference to non-hepatic tissues. Curr Drug Metab. 2002 Feb;3(1):39–59. doi: 10.2174/1389200023338071. [DOI] [PubMed] [Google Scholar]
- (19).Boorsma CE, Draijer C, Melgert BN. Macrophage heterogeneity in respiratory diseases. Mediators Inflamm. 2013;2013:769214. doi: 10.1155/2013/769214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Eleftheriadis N, Neochoritis CG, Leus NG, van der Wouden PE, Domling A, Dekker FJ. Rational Development of a Potent 15-Lipoxygenase-1 Inhibitor with in Vitro and ex Vivo Anti-inflammatory Properties. J Med Chem. 2015 Oct 8;58(19):7850–7862. doi: 10.1021/acs.jmedchem.5b01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wapenaar H, van der Wouden PE, Groves MR, Rotili D, Mai A, Dekker FJ. Enzyme kinetics and inhibition of histone acetyltransferase KAT8. Eur J Med Chem. 2015 Nov 13;105:289–296. doi: 10.1016/j.ejmech.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem. 2008 Nov 28;283(48):33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).van den Bosch T, Boichenko A, Leus NG, Ourailidou ME, Wapenaar H, Rotili D, et al. The histone acetyltransferase p300 inhibitor C646 reduces pro-inflammatory gene expression and inhibits histone deacetylases. Biochem Pharmacol. 2016 Feb 15;102:130–140. doi: 10.1016/j.bcp.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973 Dec 1;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- (25).Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005 Nov;25(21):9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Drogaris P, Villeneuve V, Pomies C, Lee EH, Bourdeau V, Bonneil E, et al. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Sci Rep. 2012;2:220. doi: 10.1038/srep00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hersman E, Nelson DM, Griffith WP, Jelinek C, Cotter RJ. Analysis of Histone Modifications from Tryptic Peptides of Deuteroacetylated Isoforms. Int J Mass Spectrom. 2012 Feb 15;312:5–16. doi: 10.1016/j.ijms.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Villar-Garea A, Israel L, Imhof A. Analysis of Histone modifications by Mass spectrometry. Curr Protoc Protein Sci. 2008;(SUPPL. 51):14.10.1–14.10.14. doi: 10.1002/0471140864.ps1410s51. [DOI] [PubMed] [Google Scholar]
- (30).Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2015 Oct 26;11(10):833. doi: 10.15252/msb.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Legartova S, Stixova L, Strnad H, Kozubek S, Martinet N, Dekker FJ, et al. Basic nuclear processes affected by histone acetyltransferases and histone deacetylase inhibitors. Epigenomics. 2013 Aug;5(4):379–396. doi: 10.2217/epi.13.38. [DOI] [PubMed] [Google Scholar]
- (33).Ghizzoni M, Boltjes A, Graaf C, Haisma HJ, Dekker FJ. Improved inhibition of the histone acetyltransferase PCAF by an anacardic acid derivative. Bioorg Med Chem. 2010 Aug 15;18(16):5826–5834. doi: 10.1016/j.bmc.2010.06.089. [DOI] [PubMed] [Google Scholar]
- (34).Sung B, Pandey MK, Ahn KS, Yi T, Chaturvedi MM, Liu M, et al. Anacardic acid (6-nonadecyl salicylic acid), an inhibitor of histone acetyltransferase, suppresses expression of nuclear factor-kappaB-regulated gene products involved in cell survival, proliferation, invasion, and inflammation through inhibition of the inhibitory subunit of nuclear factor-kappaBalpha kinase, leading to potentiation of apoptosis. Blood. 2008 May 15;111(10):4880–4891. doi: 10.1182/blood-2007-10-117994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mo F, Zhuang X, Liu X, Yao PY, Qin B, Su Z, et al. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol. 2016 Apr;12(4):226–232. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.