Abstract
The MacA-MacB-TolC assembly of Escherichia coli is a transmembrane machine that spans the cell envelope and actively extrudes substrates, including macrolide antibiotics and polypeptide virulence factors. These transport processes are energized by the ATPase MacB, a member of the ATP-binding cassette (ABC) superfamily. We present an electron cryo-microscopy structure of the ABC-type tripartite assembly at near-atomic resolution. A hexamer of the periplasmic protein MacA bridges between a TolC trimer in the outer membrane and a MacB dimer in the inner membrane, generating a quaternary structure with a central channel for substrate translocation. A gating ring found in MacA is proposed to act as a one-way valve in substrate transport. The MacB structure features an atypical transmembrane domain (TMD) with a closely packed dimer interface and a periplasmic opening that is the likely portal for substrate entry from the periplasm, with subsequent displacement through an allosteric transport mechanism.
Keywords: ABC transporter, drug efflux pump, multi-drug resistance, macrolide transporter, toxin transporter
The MacA-MacB-TolC assembly (hereafter, MacAB-TolC) contributes to drug resistance and virulence phenotypes in E. coli and other Gram-negative bacteria that include pathogenic species. These pumps drive not only the efflux of macrolide antibiotics 1, but also the transport of outer membrane glycolipids 2, lipopeptides 3, protoporphyrin 4, and polypeptide virulence factors including the heat-stable enterotoxin II 5. It has been proposed that the pumps are involved in outer membrane maintenance through transport of lipopolysaccharides 6. The transport processes are coupled to input of metabolic energy by the ABC transporter MacB 7,8.
Insight into how ABC transporters energise translocation of compounds comes from structural and functional data for transporters that export substances or import nutrients 9–14. These ABC transporters have TMDs that contain substrate-binding pockets and nucleotide-binding domains (NBDs) that enable binding and hydrolysis of ATP. Some ABC family membranes function as sensors that modulate ion channels 15. The available data for ABC transporters support an ‘alternating access’ mechanism in which the transporter alternates between inward open, occluded, and outward open states to transport substrates across the membrane bilayer. Also, an ‘outward-only’ mechanism has been proposed that can account for transport of substrates by certain ABC exporters 16,17. These exporters, which are capable of transporting large substrates such as lipids and peptides, may intercept their substrates from within the membrane in an outward-facing binding pocket, after which a conformational change decreases affinity for the ligand, causing it to be displaced into the exterior compartment. For both importers and exporters, the conformational changes are governed by ATP binding-associated NBD dimerization, and ATP hydrolysis-associated NBD dissociation 18–20.
MacB is an atypical ABC family transporter, consisting of an N-terminal NBD and four transmembrane (TM) helices. The MacB periplasmic domain is positioned between the first and second TM helices (Supplementary Fig. 1) and is expected to mediate interactions with MacA. Another distinguishing aspect of MacB is that it might accept some substrates such as heat stable enterotoxin II from the periplasm, suggesting that MacB’s transport mechanism is different from conventional models for ABC transporters 5. To gain insight into the details of the interactions between the subunits and how the transporter works in the context of the pump assembly, we undertook structure determination of the complete assembly. Our results show how an ABC transporter is engaged in a tripartite pump complex and reveals that a gating ring in MacA acts as a one-way valve in substrate transport. The MacB structure features an atypical transmembrane domain (TMD). A portal is identified in the periplasm that may serve as the entrance for substrate into the pump.
Results
Engineering stable MacAB-TolC assemblies with functional activity
Preparation of the complete MacAB-TolC assembly for structural studies is challenging, as the complex is liable to dissociate during purification. An engineering approach to stabilise the pump was pursued by fusing components through flexible linkers that were anticipated to maintain the native structure while co-localising the components and so favouring their interactions. We noted that the C-terminus of MacB and N-terminus of MacA are expected to be in proximity on the cytoplasmic side and that MacA has a single N-terminal transmembrane helix. Therefore, the fusion of MacA to the C-terminus of MacB with a flexible poly glycine-serine peptide was anticipated to preserve the proper membrane topology of the components (Supplementary Fig. 1). Another consideration is that the compositional stoichiometry of the MacAB–TolC pump is unclear, with proposed models having a 3:6:2 or 3:6:6 ratio for TolC:MacA:MacB 21–23. The co-expression of the MacB-MacA fusion protein and TolC enable the assembly of a complex with either possible stoichiometries. In the assembly with a 3:6:2 ratio, the four excess, fused subunits of MacB would remain disordered and in close proximity of the complex, but without being recruited into the structured assembly. Such a fusion, constructed with a hexa-histidine tag at the C-terminus of MacA (Supplementary Fig. 1), could be expressed and co-purified with TolC. Complex formation required full length TolC, and was greatly diminished if the C-terminus was truncated, consistent with results from functional analyses 24. Assays show that the fusion complex is likely to be active in vivo to confer macrolide resistance in a drug-hypersensitive E. coli strain lacking expression of the tripartite efflux pump AcrAB-TolC (Supplementary Fig. 2).
We also prepared constructs to stabilise the full pump using disulphide-linkages that were chosen from the proximity of residues MacA Asp271 and MacB Gly465 seen in the cryo-EM structure of the fusion-stabilised pump (described below). Co-expression of these single cysteine mutants without fusion of MacA to MacB showed that MacA D271C could be co-purified with hexa-histidine-tagged MacB G465C, and the interaction was disrupted by reducing agent, indicating disulphide bond formation between the two components. Moreover, the disulphide bond-stabilised MacAB can capture TolC to form a full pump assembly and is fully active in vivo (Supplementary Fig. 2; Supplementary Fig. 3). We used the same procedure to prepare the native MacAB-TolC pump without fusion or disulphide-linkages, but were unable to obtain the full pump assembly. Future work will be required to develop a procedure to capture the full assembly with native, wild type components to corroborate the models presented here.
Quaternary structure of the MacAB-TolC pump
Cryo-EM structure determination by single-particle analysis produced three maps of the constituent parts of the tripartite pump for the fusion-stabilised pump with different overall resolutions (Supplementary Fig. 4). The disulphide-bond stabilised pump yielded a better map for the TolC and MacA portions (Supplementary Fig. 5), whereas the MacB part was better defined in the fusion-stabilised pump. We therefore prepared a hybrid map by combining the MacA-TolC portion (from the disulphide-stabilised pump), and the MP domain of MacA and MacB (from the fusion-stabilised pump). Local resolution analysis of this hybrid map showed a range from 3.0 Å to 8.0 Å (Supplementary Fig. 4; Supplementary Fig. 5). The density readily accommodates the individual crystal structures of TolC, MacA and MacB with some adjustments (see methods section) (Fig. 1; Supplementary Fig. 6). The pump has an elongated shape and contains three protomers of TolC and six of MacA. In the fusion-stabilised pump, a hexamer of MacA can be visualised and each is fused to MacB. It is therefore expected that six MacB protomers are present, but only density for an ordered MacB dimer is observed. The remaining four MacB subunits do not have a structured interaction with the core of the assembly and are disordered and distributed over the periphery of the assembly outside of the central core, which is visible as areas with diffuse density in the images of 2D classification (Supplementary Fig. 4b). In the density map of the disulphide-stabilised pump, only a MacB dimer is observed (Supplementary Fig. 7). Six subunits of MacA are recruited into the assembly, but only two are situated to form a disulphide link to MacB. The other four MacA molecules assemble into the MacA hexamer, but do not form disulphide links with MacB. Thus, the disulphide-stabilised MacAB-TolC pump shows a stoichiometry of 3:6:2 for TolC:MacA:MacB, which is in agreement with the structure obtained for the fusion-stabilised pump.
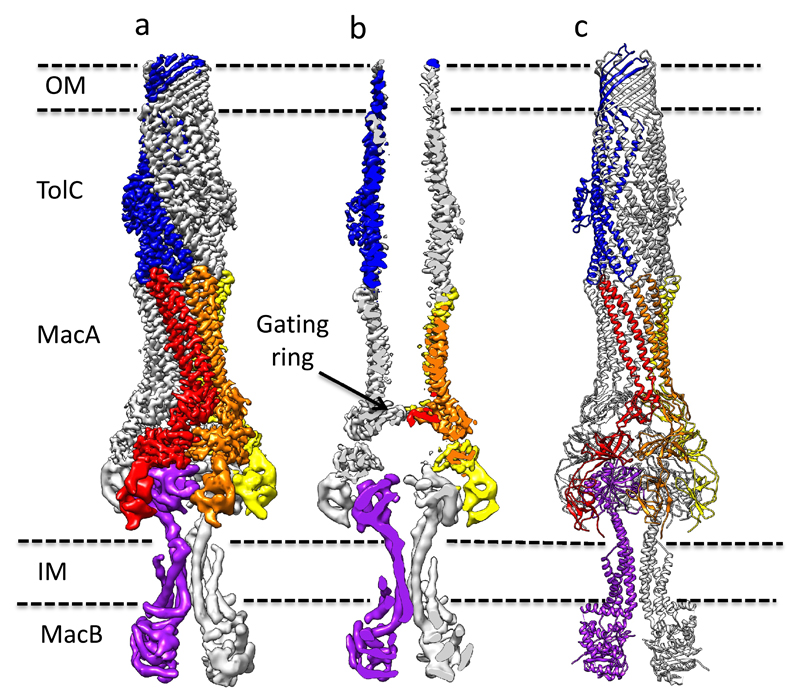
Figure 1. A pseudo-atomic model for the MacAB-TolC pump.
A, The density map for the pump. Five protomers are colour-coded: TolC (blue), MacA (red, orange and yellow), and MacB (purple). b, A sliced view of pump. c, Pseudo-atomic model based on docked crystal structures. A trimer of TolC crosses the outer membrane and its helical end protrudes into the periplasm, where it engages a hexamer of MacA. The β-barrel and membrane proximal (MP) domains of MacA mediate the interaction with the periplasmic domain of MacB, which is a dimer. The map shown in a was prepared by combining maps for the MacB portion (Supplementary Fig. 4d) and the higher resolution map for the MacA/TolC portion (Supplementary Fig. 5b).
The locations of TolC and MacB demark the boundaries of the inner and outer membranes, respectively, and the long axis of the pump assembly through those membranes is roughly 320 Å. This is similar to the dimension seen for another class of envelope-spanning transport machine, namely the AcrAB-TolC multi-drug efflux pump powered by the electrochemical proton gradient 25–27. The NDB domain of MacB extends roughly 44 Å into the cytoplasm.
A MacB dimer in the pump assembly
Consistent with results from biophysical experiments 28 and the crystal structure of a homologue 29, MacB forms a homodimer in the MacAB-TolC assembly. The crystallographic model of the dimeric MacB fits well into the cryoEM map with rigid-body adjustments to the quaternary structure (Fig. 2, Supplementary Fig. 8). As the cryoEM density and crystallographic coordinates were obtained by independent experiments, the good agreement validates the structure models. The region of the map around the MacB protomers clearly resolves three domains and their secondary structural elements: NBD, TMD and periplasmic domain (PLD)(Supplementary Fig. 8). The TMD comprises four TM helices, two of which (TM1 and TM2) have elongated extensions into the periplasm that form the stalk of the globular PLD. TM1 is preceded by an N-terminal helix of roughly 20 residues that skirts along the inner leaflet of the cytoplasmic membrane before making an abrupt turn at nearly a right angle into the interior of the lipid bilayer. This N-terminal helix corresponds to the ‘connecting helix’ found in other ABC transporters 14. The dimer interface in the membrane is packed tightly without space to accommodate a transport substrate. There is no shared TM helix that crosses over between the protomers of the MacB dimer like that seen in the ABC family drug transporter Sav1866 9. Like MacB, the recently solved structure of the heterodimeric ABC transporter ABCG5/8 also reveals a N-terminal NBD and absence of shared TM helices that cross over between the protomers of the dimer; however, its TMD contains 6 TMH and distinct from 4 seen for MacB, and its packing differs as well 14.
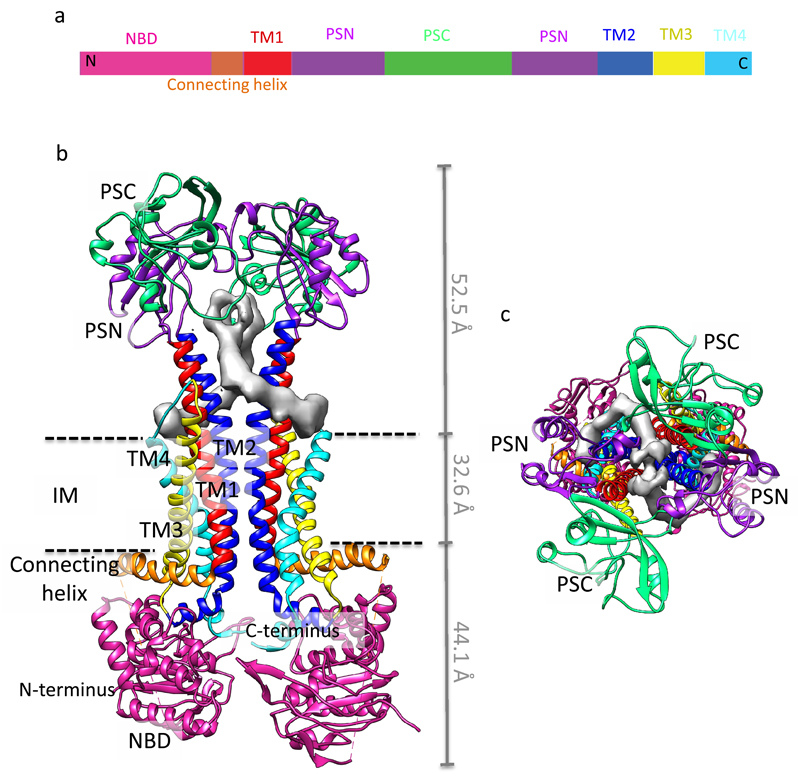
Figure 2. Structure of MacB.
a, Linear representation of MacB. The domains and subdomains in b and c are color-coded as in panel a. b, View into the plane of the membrane. c, View from the periplasm in the perpendicular direction of the membrane. Unidentified, elongated density shown in grey occludes the region between the periplasmic extensions of TM1 and TM2, which is referred to as the MacB portal. The dash lines delimit the boundary of inner membrane. The NBD locates in the cytoplasmic side and the PLD (PSN and PSC) in the periplasmic side. PSN: periplasmic subdomain N; PSC: periplasmic subdomain C; NBD: nucleotide-binding domain; TM: transmembrane helix; IM: inner membrane.
The PLD of MacB bears N-terminal and C-terminal subdomains (PSN and PSC, respectively). It is interesting to note that PSN is a structural homologue of the PN/PC subdomains of the RND transporter AcrB, while PSC has no similarity to AcrB 30. The two NBDs of the MacB dimer are physically separated, and in this state, they cannot be engaged with the nucleotide.
Unexpectedly, the MacB dimer has an opening between the periplasmic extensions of TM1 and TM2. Density was observed inside this opening, which could not be assigned to any of the three protein components making up the pump and does not fit the detergent molecules (DDM or DMNG) used in the sample preparation (Supplementary Fig. 9). The identity of this molecule could not be established by mass spectrometry and is unclear due to the limited local resolution of the map (Fig. 2). This molecule, which occludes the opening, may be a natural transport substrate. Some of the substrates of MacB, such as matured heat-stable enterotoxin II and lipopolysaccharide precursors, may be captured by the transporter from the periplasm 5,6. It seems likely that the opening that we observe is the access point for these and other periplasmic substrates of the MacAB-TolC assembly.
MacA forms a nanotube channel with a gating ring
The cryoEM map clearly reveals the four structural modules of MacA: the α-helical hairpin, lipoyl, β-barrel and membrane proximal (MP) domains (Supplementary Fig. 6a) 31. The density for the N-terminal transmembrane helix is not clear for either the engineered fusion or the cysteine disulphide stabilised assembly, indicating that the helix may not make a defined interaction with the core of the complex. The modular domain organization of MacA is common to the large class of periplasmic proteins that mediate tripartite pump assemblies in Gram-negative bacteria 23. In isolation, MacA forms a hexameric assembly, much like that observed for AcrA in the AcrAB-TolC efflux pump 25,31. The cryoEM map of MacAB-TolC reveals that the same hexameric arrangement occurs in the full pump assembly, with some requirement for structural adjustment for the linker regions between the β-barrel and MP domains (Supplementary Fig. 6a). The helical hairpin regions pack into a cylindrical assembly to form an α-helical barrel quaternary structure 32. The helical hairpin domains engage the open-state TolC through helix-turn-helix motifs, the lipoyl and β-barrel domains form stacked annular rings, and the membrane proximal domain skirts over the surface of the periplasmic domain of MacB, as we will describe further below.
Loops in the MacA lipoyl domains present six glutamine residues (Q209) that form an inter-protomer hydrogen-bonding network (Supplementary Fig. 10a). These residues are not in the same plane, as seen in the crystal structure of E.coli MacA 31. The aperture observed in the cryoEM model is too narrow for substrates to pass readily (Fig. 1b). This glutamine residue is conserved in MacA homologues from divergent species 31. We substituted Q209 to A but found that the mutant was still able to efflux the transport substrate erythromycin in vivo with similar level to the wild type protein, suggesting that substrates can somehow readily pass through the aperture in the native protein (Supplementary Fig. 11). To explore this further, molecular dynamics (MD) simulations were undertaken and show that erythromycin makes favorable interactions with the Q209 hydrogen-bonding network, which partially disrupt the network and enable the substrate to pass through the opening (Supplementary Fig. 12). Steered MD simulations show that inward transfer induces a larger conformational change of the loops than outward transfer. As a consequence, opening the gate in the inward direction requires more non-equilibrium work than in the direction of efflux, similar to a one-way valve (Supplementary Fig. 12). The aperture is specific to the MacA proteins in the MacAB-TolC pump, as there is no such feature in the AcrA of the AcrAB-TolC assembly 25.
Interactions between MacA and MacB
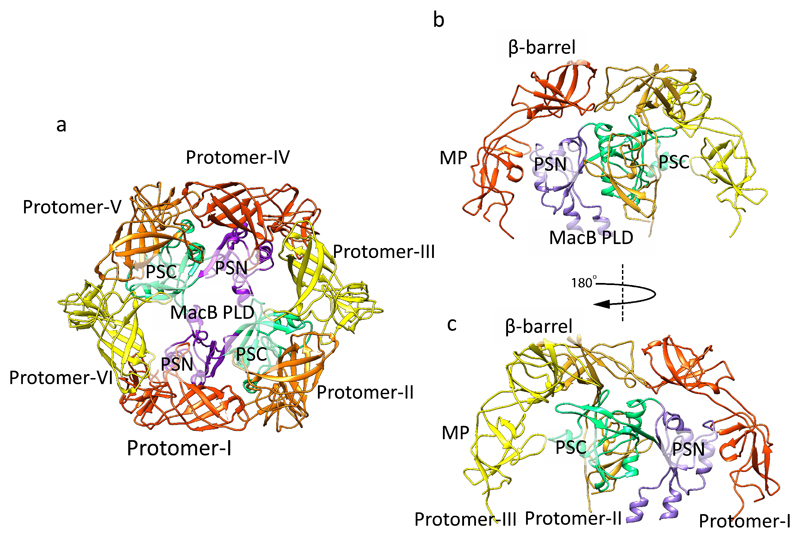
The β-barrel and MP domains of MacA mediate the interaction with the periplasmic domain of MacB, in agreement with the reported functional data 1,33 and the observation that the MP domain is required for MacA and MacB to associate in solution (with nanomolar dissociation constant) 33. Three MacA MP domains contact one MacB protomer, with one MP contacting the PSN and two contacting the PSC. These interactions distort the hexameric ring formed by the MacA MP domains to generate a more oval shape that accommodates the two-fold symmetry of the MacB dimer (Fig. 3a; Supplementary Fig. 7b). The β-barrel domains of MacA dock to the periplasmic domain of MacB (Fig. 3a). Three MacA β-barrel domains contact MacB in non-equivalent ways, with one interacting with the PSN, another contacting the PSC and the third bridging between the PSC and the PSN of the adjacent MacB (Fig. 3a-c). The lipoyl domains form a hexameric ring but do not contact MacB. Comparison of the crystal structure and cryoEM model of MacB indicates movement in the periplasmic domain of MacB to accommodate MacA (Supplementary Fig. 6c).
Figure 3. Interactions between MacA and MacB.
a, Top view of the MacA-MacB interface, viewing from the periplasm in the perpendicular direction of the membrane. Each MacB periplasmic domain interacts with three MacA protomers. These interactions distort the hexameric ring formed by the MacA MP domains to generate a more oval shape that accommodates the two-fold symmetry of the MacB dimer. b and c, Side view of the interactions between three MacA and one MacB. A rotation of 180 degrees relates the view in c relative to b. For clarity, only the β-barrel and MP domains of MacA, and the periplasmic domain of MacB are shown. PLD: periplasmic domain; PSN: periplasmic subdomain N; PSC: periplasmic subdomain C; MP: membrane proximal domain.
Interfacial contacts between TolC and MacA
Using focussed refinement 34 of the disulphide-bond stabilised pump assembly, the MacA-TolC section could be resolved at 3.3 Å resolution, offering unprecedented view of the details of the molecular interfaces (Supplementary Fig. 13). A short helix-turn-helix motif in the α-helical hairpin domain of MacA meshes with the helix-turn-helix motif of TolC in the periplasmic end (Fig. 1; Supplementary Fig. 13), similar to the observations seen for the homologous AcrA interaction with TolC 25,35. The MacA tip regions make slightly different interactions with the intra-protomer and inter-protomer grooves of TolC (Supplementary Fig. 13b,c). Further evidence for this interaction comes from in vitro affinity binding studies of site-directed mutants, in vivo drug resistance measurements 36, and the behaviour of chimeric proteins containing the tip regions of the α-helical barrel of TolC that were found to bind to MacA 37. Lee et al. (2013) explored the functional and physical interaction of two TolC homologues from Vibrio vulnificus with MacA in E. coli using site-directed mutational analyses and chemical cross-linking, and they found that conserved residues at the aperture tip region of the α-hairpin of the TolC proteins play an essential role in the formation of the functional MacAB-TolC pump 38. Taken together, these findings suggest that TolC has conserved tip-to-tip interactions with MacA in Gram-negative bacteria. The interaction between MacA and TolC directly opens the periplasmic end of TolC to form a long, continuous channel that is roughly 25 to 30 Å wide with small variation (Fig. 1b; Supplementary Fig. 6b). Thus, TolC is held in an open state in the pump, a situation which is similar to that observed in the RND-based AcrAB-TolC pump 25.
Discussion
While the MacAB-TolC assembly studied here shares some organizational similarities with the AcrAB-TolC multi-drug efflux pump, there are also some important differences that impact on functionality. Although the periplasmic domains of the inner membrane transporters MacB and AcrB are involved in contact with the periplasmic partner, they interact in completely different ways and have different stoichiometries. In the AcrAB-TolC pump, a trimer of TolC is engaged with a hexamer of AcrA and a trimer of AcrB, yielding a 3:6:3 protomer stoichiometry for TolC:AcrA:AcrB, while the protomer stoichiometry for the MacAB-TolC pump is 3:6:2. Another notable difference between the two pumps is a constriction point in the MacA formed by a conserved glutamine ring in the lipoyl domain. This ring may act as a gate through which substrates may passively move and could help to prevent backflow in situations where the efflux pump operates against a concentration gradient. Thus, this loop may act as a gating ring that favours substrate translocation in the outward direction. A broadly similar gating mechanism may be involved in the capsular polysaccharide transporter Wza, where a hydrogen-bonding ring (formed by residues Y110) may selectively permit phosphorylated polysaccharide to pass through a constrictive barrier (Supplementary Fig. 10b) 39. The absence of a gating ring in the AcrAB-TolC pump might be due to the absence of any continuous opening between the pump and the periplasmic compartment or cytoplasm during the transport process through which substrates might leak.
The MacAB-TolC structure provides some clues as to the transport mechanism. Like the AcrAB-TolC pump, the MacAB-TolC assembly may also accept some transport substrates from the periplasm. One of its substrates, the heat-stable enterotoxin II, is transported as a precursor across the inner membrane to the periplasm by the Sec machinery, where it undergoes maturation 5. This matured form likely enters the MacAB-TolC pump from the periplasmic side 5. The opening we observe in MacB could be the entry point for such substrates, and accordingly we refer to this as a portal. Density that might be attributed to a bound ligand was found at this putative portal of MacB (Fig. 2; Supplementary Fig. 9). For the substrates that gain access from the periplasmic side, MacB possibly uses a transport mechanism that shares certain features of the ‘outward-only’ model advanced for some ABC transporters, whereby the substrate-binding pocket remains in an outward facing conformational state 16,17. The substrates enter the pump through the portal. Conformational changes of this outward facing state are coupled with ATP hydrolysis, resulting in a switch that decreases binding affinity for the substrate and cause it to be jettisoned into the channel of the pump and then transported across the cell envelope to the exterior through the tripartite assembly. In contrast, the ABC transporters adopting an ‘outward-facing’ mechanism capture substrates from the lipid bilayer and move them across the membrane. For MacB, transport through the periplasm would involve allosteric coupling of conformational change propagated from the NBD through the TMD to PLD, which is supported by the finding that interaction of MacA and MacB is stimulated by the presence of ATP and contributes to recruitment of TolC into the pump assembly 7.
Similar to the structure of the human ABCG5/ABCG8 sterol transporter 14, MacB has a tightly packed dimer interface in the TMD, and this interface does not appear to have space required to accommodate a transport substrate. For small substrates like erythromycin and protoporphyrin, MacB may drive the transport through the membrane, but this would require re-organization of the dimer interface during the process. This mode of operation is likely to be similar to the alternating access model for ABC-transporters 14,16. The details of the mechanism of ATP binding and hydrolysis by MacB in the full pump are unclear presently and require further investigation. However, based on the available data, we envisage that the MacAB-TolC ABC-type tripartite assembly is a highly versatile machinery that can accommodate both periplasmic-entry and transmembrane crossing substrates by using different transport mechanisms, both driven by the energy of ATP binding and hydrolysis.
Methods
Construction of vectors for overexpression of MacAB-TolC complex
The macA, macB and tolC genes were amplified from genomic DNA of E. coli strain W3110. The macA gene was first amplified using primer pairs MacAGS_F: 5'- GGTGGGAGCGGCGGTGGTGGCTCTGGTGGCGGCGGTAGTATGAAAAAGCGGAAAACCGTGAAGAAGC -3' and MacAinf_R: 5'-GGTGGTGGTGCTCGAGTTGTGCAGCTCCTGGTTTGGCCTCA -3'; and it was amplified again using primer pairs MacAGSinf_F: 5'- GGCACGAGAGGGATCCGGTGGGAGCGGCGGTGGTGGCTCT -3' and MacAinf_R. The macB gene was amplified using primer pairs MacBinf_F: 5'- AAGGAGATATACATATGACGCCTTTGCTCGAATTAAAGGATATTCGTC -3' and MacBinf_R: 5'- GCTCGAATTCGGATCCCTCTCGTGCCAGAGCATCTACTGGGTCCAGTCG -3'.
The DNA fragment of macB was inserted into the Nde I and BamH I digested pET20b vector using In-Fusion cloning method, generating the construct pET20b-macB; The DNA fragment of polyGlySer-tagged macA was then inserted into the BamH I and Xho I digested pET20b-macB using In-Fusion cloning method, generating the construct pET20b-macB-polyGS-macA-6His.
The tolC gene was amplified using primers TolCinf_F: 5'-AAGGAGATATACATATGAAGAAATTGCTCCCCATTCTTATCGGCC-3' and TolCFLAGXhoI_R: 5'-GAGCTCGAGTCACTTATCGTCGTCATCCTTGTAATCGTTACGGAAAGGGTTATGACCGTTACTGGT -3'; and it was amplified again using TolCinf_F and TolCFLAG_inf_R: 5'-TTGAGATCTGCCATATGTCACTTATCGTCGTCATCCTTGTAATCGTTACG-3'. The resulting DNA fragment of tolC-FLAG was inserted into the Nde I digested pRSFDuet-1 using In-Fusion cloning method, generating the construct pRSFDuet-tolC-FLAG.
The macAB genes were amplified from genomic DNA of E. coli K12 strain W3110 using primers MacAB_F 5'- AAGGAGATATACATATGAAAAAGCGGAAAACCGTGAAGAAGCGT -3' and MacAB_R 5'- GGTGGTGGTGCTCGAGCTCTCGTGCCAGAGCATCTACTGGATCCAG -3', and were inserted into the NdeI and XhoI digested pET20b plasmid using In-fusion ligation kits, generating construct pET20b-macAB6His.
The MacA_D271C and MacB_G465C mutants were produced by site-directed mutagenesis using pET20b-macAB6His as a template and primer pairs MacAD271C_F: 5'- CGACGCCGGAAAAGGTTAACTGCGCTATTTTCTATTACGC -3'/MacAD271C_R: 5'-GCGTAATAGAAAATAGCGCAGTTAACCTTTTCCGGCGTCG -3' and MacBG465C_F: 5'- ATGTCCGGGCGAGTTATGTGCCAGTCGTGGCTTAAC -3'/MacBG465C_R: 5'- GTTAAGCCACGACTGGCACATAACTCGCCCGGACAT -3', generating construct pET20b-MacAD271CMacBG465C_6His
Overexpression and purification of E. coli MacAB fusion with TolC
Genes encoding AcrA and AcrB were deleted from the chromosome of Escherichia coli strain C43 (DE3). The resulting C43 (DE3) ΔacrAB strain was transformed with plasmids pET20b-macB-polyGS-macA-His6 expressing the MacB-MacA fusion with a C-terminal his-tag and pRSFduet-tolC-FLAG expressing full-length TolC with a C-terminal FLAG-tag. A single colony taken from an agar plate with selective antibiotics was used to inoculate 20 ml of LB medium containing carbenicillin at 100 μg ml-1 and kanamycin at 50 μg ml-1 in a 50 ml centrifuge tube. The culture was grown in an orbital shaker at 37 °C, 220 rpm for 4 h. 10 ml of the culture was used to inoculate 1 litre of 2xYT medium with antibiotics in a 2 L baffled flasks. The culture was grown in an orbital shaker at 37 °C, 220 rpm and was induced at absorbance of A600=0.5-0.6 using 0.25 mM IPTG. The temperature was then dropped to 20 °C and the culture incubated overnight. Cells were harvested by centrifugation, and pellets from 10 L culture were re-suspended in 200 ml of lysis buffer composed of 20 mM Tris (pH 8.0), 400 mM NaCl.
To the cell suspension was added 1 tablet of EDTA-free protease inhibitor mixture tablet per 50 ml, lysozyme to a final concentration of 5 mg ml-1, and DNase I to a final concentration of 5 U ml-1. The mixture was incubated at 4 °C for 1 h and then passaged 8 times through a high-pressure homogenizer (Emulsiflex) at 15,000 psi at 4 °C. The lysate was centrifuged at 9,000xg for 30 mins at 4 °C to remove the cell debris, and the supernatant was ultra-centrifuged at 125,775xg for 4 h at 4 °C to pellet the cellular membrane. The cellular membrane pellet from 10 L culture was re-suspended in 200 ml of lysis buffer. EDTA-free protease inhibitor mixture tablet was added to membrane suspension at 1 tablet per 50 ml, and n-dodecyl-β-D-maltoside (DDM) was added to a final concentration of 1.5 % w/v. The mixture was gently stirred at 4 °C for 3 h. The membrane solution was then clarified by ultra-centrifugation at 125,775xg for 30 min at 4°C. Imidazole (5M pH: 7.5) was added to the supernatant to a final concentration of 10 mM, and 100 ml of the mixture was applied onto two collected HiTrap Chelating 1 ml column charged with Ni2+ and equilibrated with lysis buffer containing 20 mM imidazole and 0.05 % DDM. Two columns were washed with 50 ml of lysis buffer supplemented with 0.03 % decyl maltose neopentyl glycol (DMNG) and 50 mM imidazole. The his-tagged MacAB-TolC complex protein was eluted using lysis buffer supplemented with 0.03 % DMNG and 500 mM imidazole. The eluate was buffer exchanged to sample Buffer-I containing 20 mM Tris pH 8.0, 400 mM NaCl, 0.03% DMNG using HiTrap Desalting column.
The MacAB-TolC complex was further purified by ANTI-FLAG M2 affinity resin. 0.5 ml of ANTI-FLAG M2 affinity resin (Sigma, A2220) in a column was washed by three sequential application of 1 ml of 0.1 M glycine HCl, pH 3.5, then the resin was washed with 5 ml buffer containing 20 mM Tris (pH 7.5) and 150 mM NaCl followed by 5 ml of Buffer-I, allowing a small amount of buffer to remain on the top of the column. The ANTI-FLAG M2 affinity resin in the chromatography column was re-suspended in the above buffer-exchanged protein solution. The mixtures were rotated gently at 4 °C for 1 h and loaded on the chromatography column, allowing the residue protein solution to drain. The resin was washed by loading 10 ml of sample buffer-II containing 20 mM NaPi (pH 7.6), 300 mM NaCl and 0.03 % DMNG and was suspended in 0.5 ml of 1 mg ml-1 FLAG-peptide in Buffer-II. The mixture was rotated gently at 4 °C for 30 min, and then was centrifuged at 10,000 rpm in an eppendorf centrifuge for 1 min at 4 °C to spin down the resin. The supernatant was removed with a narrow-end pipette tip and loaded on a mini chromatography column to remove the trace amount of resin, collecting the flow through. The resin was re-suspended in 1 ml of sample Buffer-II and was centrifuged at 10,000 rpm in an eppendorf centrifuge for 1 min at 4 °C to spin down the resin. The supernatant was removed with a narrow-end pipette tip and loaded on a mini chromatography column to remove the trace amount of resin, collecting the flow through. This step was repeated 3 times. Fractions containing purified MacAB–TolC complex were pooled and concentrated to 0.5 mg ml-1 using a Vivaspin column (MWCO: 100 kDa). Amphipol A8-35 (100 mg ml-1) was mixed with the protein solution with a mass ratio of amphipol A8-35 to protein of 4:1. The mixture was incubated at 4 °C for 3 h. Polystyrene beads (Bio-Beads SM-2) were then added to the protein/DMNG/amphipol A8-35 mixture with a mass ratio of Bio-Beads SM2 to detergent of 10:1. The mixture was gently rotated at 4 °C overnight to remove DMNG. The detergent-exchanged MacAB–TolC complex was 5-fold diluted using 20 mM NaPi (pH 7.6), 300 mM NaCl and glutaraldehyde (25 % w/v) was added to a final concentration of 0.1 % w/v. The cross-linking reaction was incubated on ice overnight and then glycine (1 M, pH 7.5) was added to the sample to a final concentration of 80 mM to quench further crosslinking. The mixtures were concentrated to 100 μl using a Vivaspin concentrator (MWCO=100 kDa) and was loaded onto a Superose 6 3.2/300 column equilibrated with Buffer-III containing 20 mM NaPi (pH 7.6), 300 mM NaCl and 0.025 % amphipol A8-35. Fractions containing purified MacAB–TolC complex were pooled and concentrated to 2 mg ml-1 using a Vivaspin column (MWCO: 100 kDa) and embedded in vitreous ice.
Overexpression and purification of disulfide-engineered E. coli MacAB-TolC
The C43 (DE3) ΔacrAB strain was transformed with plasmids pET20b-MacAD271CMacBG465C_6His expressing the MacA_D271C and MacB_G465C mutants with a C-terminal 6xHis-tag on MacB, and pRSFduet-tolC-FLAG expressing full-length TolC with a C-terminal FLAG-tag. The MacAD271CMacBG465C-TolC complex was overexpressed and purified as described above for MacAB fusion-TolC complex without cross-linker treatment. Co-expression of the MacB-MacA fusion and truncated TolC at residue 464 was also tried but did not yield a stable complex.
Cell growth for drug sensitivity assay
Overnight cultures from glycerol-stocks of E. coli ΔacrAB, harbouring pET20b-derived plasmid encoding fused MacA-MacB_E170Q (control) or fused wildtype MacAB in one set of experiments or cysteine mutants MacA_D271C MacB_G465C, MacA_D271C MacB_G465C_E170Q, or wild-type MacAB in another set of experiments, were grown in LB containing 25 mM glucose and 100 µg ml-1 carbenicillin. Cultures were grown for about 16 h at 37°C, and then used to inoculate fresh medium. Once E. coli cells had reached OD600 of 0.8, the cells were diluted to OD600 of 0.06 in fresh medium containing 0.07 mM IPTG in the wells of a 96-well plate to which antibiotics were added as indicated in Supplementary Fig. 2. Growth was followed over time at OD660 at 37 ºC in a Versamax microplate reader (Molecular Devices).
The same experimental procedure was followed for the cell growth-based sensitivity assay with E. coli ΔacrAB, harbouring pET20b-derived plasmid encoding wild-type MacAB, MacA_Q209A MacB, MacA_Q210A MacB, or no MacAB (control) in the presence of 0 – 88.9 µg ml-1 erythromycin (Supplementary Fig. 11).
Electron microscopy data collection
For cryo-EM of the fusion-stabilised pump, aliquots of 3 µl of purified MacAB-TolC in amphipols at a concentration of 2 mg ml-1 were applied to glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3, 300 mesh), blotted with filter paper to remove excess sample, and plunge-frozen in liquid ethane slush using an FEI Vitrobot Mark IV. Zero-energy-loss images of the frozen, hydrated grids were acquired automatically using an FEI Titan Krios electron microscope operated at 300 kV with Leginon on a Gatan K2-Summit detector in super-resolution counting mode 40. A GIF-Quantum energy filter (Gatan) was used with a slit width of 20 eV to remove inelastically scattered electrons. Twenty movie frames were recorded each with an exposure time of 800 ms using a dose rate of ~2.5 electrons Å-2 s-1 (approximately 5 electrons pixel-1 s-1) at a calibrated magnification of 36,764× (yielding a pixel size of 1.36 Å at the sample level). The final dataset is composed of 2,136 micrographs with defocus values ranging from -1.6 to -2.6 µm.
For cryo-EM of the disulphide-stabilised pump, a strategy based on PEGylated gold grids was chosen to improve grid stability and the distribution and number of particles 41,42. The UltrAufoil grids (UltrAuFoil® Holey Gold grid R1.2/1.3, 300 mesh, Quantifoil Micro Tools GmbH) were glow-discharged at 20 mA for 90 s, and immersed in anaerobic ethanol containing 5 mM SPT-0011P6 (SensoPath Technologies). The grids were incubated with the reagent for 2 days at room temperature in an anaerobic glovebox, and were then washed 3 times in absolute ethanol to remove excess reagent and allowed to air-dry. A Vitrobot IV (FEI) was used to prepare grids in a controlled atmosphere (4 °C, 100 % relative humidity). A 2.5 μl aliquot of purified MacAB-TolC at a concentration of 2 mg ml-1 in amphipols was applied, and the grids were blotted at a force of ‘-10’ for 8-16 s before plunging into liquid ethane. The grids were stored in liquid nitrogen before imaging. Zero-energy-loss images of frozen-hydrated MacAB-TolC particles were recorded automatically on an FEI Titan Krios electron microscope at 300 kV with a GIF Quantum energy filter set with a slit width of 20 eV. A Gatan K2-Summit direct electron detector was used in super-resolution counting mode at a nominal magnification of 36,765, corresponding to a calibrated physical pixel size of 1.36 Å, and a dose rate of 2.7 electrons per square angstrom per second (approximately 5 electrons per pixel per second). Exposures of 16 s were dose fractionated into 20 movie frames, leading to an accumulative dose of about 45 electrons/Å2 on the specimen. A total of 869 movie stacks were collected with a defocus range of -1.2 to -2.5 μm.
Image Processing
For the fusion-stabilised pump, all super-resolution frames were corrected for gain reference, binned by a factor of 2 from 0.68 Å pixel-1 to 1.36 Å pixel-1, and motion-corrected using MOTIONCORR43. Aligned frames were then summed into average micrographs, which were then used to estimate the contrast transfer function (CTF) using Gctf 44. All subsequent image-processing steps were performed using RELION 45. Templates for reference-based particle picking were obtained from (reference-free) 2D classification of a subset of manually picked particles. The templates were low-pass filtered to 20 Å to limit reference bias, and used to automatically pick 46 180,602 particles from all micrographs. Selection of suitable 2D classes from a 2D classification with all auto-picked particles yielded 111,656 particles, which were further, classified using 3D classification. An initial model composed of the crystal structures of TolC and MacA (PDB accession numbers 3FPP and 1EK9, respectively) and a homology model of MacB was converted into a density map using e2pdb2mrc.py from EMAN2 47 and low-pass filtered to 60 Å to limit reference bias during classification. The two largest classes, comprising 92,913 particles, were selected for subsequent 3D auto-refinement (without symmetry) to an overall resolution of 6.2 Å. Beam-induced motion correction and radiation-damage weighting (particle polishing) was used to increase the signal-to-noise ratio of the individual particles 48. A further round of 2D classification resulted in a final dataset of 91,162 polished particles.
Initial 3D refinement of the polished particles resulted in a map with an improved overall resolution of 5.2 Å. Masked 3D refinement and classification with partial signal subtraction 34 was used to improve the density for each of the three sub-complexes. In the overall map, the density of the inner membrane protein MacB was relatively weak owing to structural flexibility in this region (as is also visible in the 2D class averages, see Supplementary Fig. 4b). Masked 3D classification with partial signal subtraction and without alignment on this region yielded a more homogeneous subset of 17,154 particles. Masked refinement of this subset lead to a reconstruction of MacB with improved density to a resolution of 5.3 Å (Supplementary Fig. 4c,d). Masked refinements with partial signal subtraction of the MacA and TolC regions were performed imposing C6 and C3 symmetry, respectively, yielding improved maps to 4.0 and 4.6 Å resolution (Supplementary Fig. 4c,d).
For the disulphide-stabilised pump, the software MotionCor2 was used for whole-frame motion correction and dose weighting, CTFFIND4 for estimation of the contrast transfer function parameters, RELION-1.4 and RELION-2.0/beta packages for all other image processing steps. A particle subset was manually picked up to calculate reference-free 2D class averages, which was then used as templates for automated particle picking of the entire data set. The templates were lowpass filtered to 20 Å to limit model bias. 71,462 particles were picked automatically from a total of 792 micrographs with good ice thickness. Then initial runs of 2D and 3D classifications were used to remove the heterogeneous particles, as well as the false positive particles from the auto-picking. 27,614 particles were selected for a first 3D auto-refinement with C3 symmetry. This generated a reconstruction with a resolution of 4.4 Å that clearly showed density for TolC, the α-helical hairpin, lipoyl and β-barrel domains of MacA; however, the density for the other portions of the pump was poor. This particle subset was then used for a focused 3D auto-refinement 49 by applying a soft mask around TolC, the α-helical hairpin, lipoyl and β-barrel domains of MacA, and subtracting the residual signal for the rest of the pump. This yielded a reconstruction with a resolution of 3.3 Å (Supplementary Fig. 5b,c).
This particle subset was also used for a focused 3D classification 34 by applying a soft mask around the MP domains of MacA and the PLD domains of MacB, subtracting the residual signal for the rest part of the pump, and using the orientation information of the particles from the first 3D auto-refinement without perform any alignments. This yielded a homogenous subset of 11,158 particles. This particle subset with partial signal subtraction was submitted to 3D auto-refinement imposing C2 symmetry and using small angular sampling and local search, generating a reconstruction for the MP domains of MacA and the PLD domains of MacB with a resolution of 8.0 Å (Supplementary Fig. 5b,c; Supplementary Fig. 7b).
All resolution estimates were calculated using separately refined half-reconstructions in RELION, using the Fourier shell correlation at 0.143 criterion 50 (Supplementary Fig. 4c; Supplementary Fig. 5c). Local resolution variations were calculated using the RELION wrapper to the ResMap 51 program (Supplementary Fig. 4d; Supplementary Fig. 5b).
Model docking and refinement
The crystal structure of trimeric TolC (PDB code: 1EK9) was docked into the cryoEM map using Chimera. The model was adjusted manually to optimize the local fit to density using Coot. Chain B from the crystal structure of MacA (PDB code: 3FPP) including β-barrel domain, lipoyl domain and α-helical hairpin domain were fitted to the density map by using Chimera. A homology model of the MP domain of MacA was built based on the crystal structure of MexA (PDB code: 2V4D) and was fitted to the density. A homology model of E.coli MacB was built based on the crystal structure of Acinetobacter baumannii MacB 29. The model was docked into the cryoEM map, and the periplasmic and NBD domains of MacB were rigid body fitted to the density by using Chimera. To optimize the local fit to density, the periplasmic domain was further adjusted manually using Coot based on the crystal structure of the periplasmic domain of MacB from Actinobacillus actinomycetemcomitans (PDB code: 3FTJ). The model for the complete pump was refined using REFMAC “jelly-body” refinement using secondary-structure restraints to maintain proper stereochemistry. Ramachandran outliers were corrected semi-automatically in Coot and MolProbity statistics were computed to ensure proper stereochemistry. The model of the whole pump complex was validated by computing a FSC with the density map (Supplementary Fig. 14).
Molecular Dynamics Simulations
All simulations were performed with the software package GROMACS5 52. The amber99sb force field 53 was used for the protein. Erythromycin was parameterized by using amber99sb atom types in conjunction with RESP2 (HF/6-31G(d)) charges 54, as implemented in the Antechamber module of the AMBER12 software package 55. The TIP3P explicit water model 56 was used for solvation of the protein together with Joung and Cheatham ion parameters 57, at a NaCl concentration of 0.15M. Unbiased molecular dynamics simulations of 200 ns length were carried out on the MacA structure obtained by cryo-EM to equilibrate the system. Subsequently, a set of 2×100 individual force-probe (steered) molecular dynamics simulations was performed 58, in which erythromycin was propagated along the pore axis in inward and outward direction by a moving harmonic potential with a force constant of 50 kJ mol-1 Å-2 and at a constant velocity of 2.5 Å ns-1.
Data availability
The cryoEM map and the atomic model have been deposited in the Protein Data Bank and EMDB under accession numbers 5NIK and EMD-3652 for the MacA-TolC section, and 5NIL and EMD-3653 for the MacB section. The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Supplementary information is available for this paper.
Acknowledgements
This work was supported by the Wellcome Trust (B.F.L.), HFSP (B.F.L., H.W.v.V., S. M.), Marie Curie International Outgoing Fellowship (A.W.P.F.), the UK Medical Research Council (MC_UP_A025_1013, to SHWS), Wellcome Trust ISSF award (grant number: WT097818MF), the Scottish Universities’ Physics Alliance (U.Z. and S.L.) and MRC Mitochondrial Biology Unit (Grant number: U105663141). A.N. is the recipient of a Herchel-Smith Scholarship. We thank Jeremy Skepper for help using the EM microscope and imaging facility at Cambridge University, Hong Zhou for providing access to the electron microscope at the University of California at Los Angeles, Jake Grimmett and Toby Darling for support with high-performance computing at the MRC Laboratory of Molecular Biology, Stuart Rankin for help with computing on the High-Performance Computing System at the University of Cambridge and Len Packman for mass spectrometry analyses. We thank the staff of Diamond Light Source for access to the eBIC facility. The EM maps have been deposited in the Electron Microscopy Databank.
Footnotes
Author contributions
D.D., B.F.L. and S.H.W.S. designed the project. D.D. purified the fusion and disulphide-linkage stabilized MacAB–TolC complexes. D.D., A.W.P.F., X.B. and J.N.B. obtained and analysed the single-particle cryo-EM data. U.O. and S.M. built the homology model of MacB. D.D. and B.F.L. devised a model of MacAB–TolC based on the cryo-EM map. A.N. and H.W.v.V. conducted MIC assays on the MacAB–TolC pump. S.L. and U.Z. carried out molecular dynamics simulations of MacA. D.D., B.F.L. and S.H.W.S. wrote the paper. All authors contributed to editing the manuscript.
Additional information
ORCID ID of Dijun Du: 0000-0002-1546-8939
ORCID ID of Ben Luisi: 0000-0003-1144-9877
ORCID ID of Sjors Scheres: 0000-0002-0462-6540
Competing interests
The authors declare no competing financial interests.
References
- 1.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183:5639–44. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallet-Gely I, et al. Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl Environ Microbiol. 2010;76:910–21. doi: 10.1128/AEM.02112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H, Kang H. The PseEF efflux system is a virulence factor of Pseudomonas syringae pv. syringae. J Microbiol. 2012;50:79–90. doi: 10.1007/s12275-012-1353-9. [DOI] [PubMed] [Google Scholar]
- 4.Turlin E, et al. Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. Microbiologyopen. 2014;3:849–59. doi: 10.1002/mbo3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190:7693–8. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu S, Zgurskaya HI. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J Bacteriol. 2013;195:4865–72. doi: 10.1128/JB.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu S, Zgurskaya HI. Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter. Mol Microbiol. 2012;86:1132–43. doi: 10.1111/mmi.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol. 2007;63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. [DOI] [PubMed] [Google Scholar]
- 9.Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–5. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 10.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–10. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutmann DAP, Ward A, Urbatsch IL, Chang G, van Veen HW. Understanding polyspecificity of multidrug ABC transporters: closing in on the gaps in ABCB1. Trends Biochem Sci. 2010;35:36–42. doi: 10.1016/j.tibs.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doshi R, Woebking B, van Veen HW. Dissection of the conformational cycle of the multidrug/lipidA ABC exporter MsbA. Proteins. 2010;78:2867–72. doi: 10.1002/prot.22813. [DOI] [PubMed] [Google Scholar]
- 13.Choudhury HG, et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci U S A. 2014;111:9145–50. doi: 10.1073/pnas.1320506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J-Y, et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature. 2016;533:561–4. doi: 10.1038/nature17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aittoniemi J, et al. SUR1: a unique ATP-binding cassette protein that functions as an ion channel regulator. Philos Trans R Soc B Biol Sci. 2009;364:257–267. doi: 10.1098/rstb.2008.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–93. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 17.Perez C, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–8. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- 18.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33:528–36. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo J-S, Zeltina A, Goetz BA, Locher KP. X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat Struct Mol Biol. 2012;19:1310–5. doi: 10.1038/nsmb.2417. [DOI] [PubMed] [Google Scholar]
- 20.Korkhov VM, Mireku SA, Locher KP. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012;490:367–72. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- 21.Song S, Kim J-S, Lee K, Ha N-C. Molecular architecture of the bacterial tripartite multidrug efflux pump focusing on the adaptor bridging model. J Microbiol. 2015;53:355–64. doi: 10.1007/s12275-015-5248-4. [DOI] [PubMed] [Google Scholar]
- 22.Zgurskaya HI, Weeks JW, Ntreh AT, Nickels LM, Wolloscheck D. Mechanism of coupling drug transport reactions located in two different membranes. Front Microbiol. 2015;6:100. doi: 10.3389/fmicb.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du D, van Veen HW, Luisi BF. Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol. 2015;23:311–9. doi: 10.1016/j.tim.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Yamanaka H, Izawa H, Okamoto K. Carboxy-terminal region involved in activity of Escherichia coli TolC. J Bacteriol. 2001;183:6961–4. doi: 10.1128/JB.183.23.6961-6964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du D, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–5. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daury L, et al. Tripartite assembly of RND multidrug efflux pumps. Nat Commun. 2016;7:10731. doi: 10.1038/ncomms10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J-S, et al. Structure of the tripartite multidrug efflux pump AcrAB-TolC suggests an alternative assembly mode. Mol Cells. 2015;38:180–6. doi: 10.14348/molcells.2015.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HT, et al. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem. 2009;284:1145–54. doi: 10.1074/jbc.M806964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada Ui, Yamashita Eiki, Neuberger Arthur, van Veen Hendrik W, Murakami S. Crystal structure of tripartite-type ABC transporter, MacB from Acinetobacter baumannii. 2016 doi: 10.1038/s41467-017-01399-2. Submitt Artic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, et al. Crystal structure of the periplasmic region of MacB, a noncanonic ABC transporter. Biochemistry. 2009;48:5218–25. doi: 10.1021/bi900415t. [DOI] [PubMed] [Google Scholar]
- 31.Yum S, et al. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol. 2009;387:1286–97. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 32.Calladine CR, Sharff A, Luisi B. How to untwist an alpha-helix: structural principles of an alpha-helical barrel. J Mol Biol. 2001;305:603–18. doi: 10.1006/jmbi.2000.4320. [DOI] [PubMed] [Google Scholar]
- 33.Modali SD, Zgurskaya HI. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 2011;81:937–51. doi: 10.1111/j.1365-2958.2011.07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai X, Rajendra E, Yang G, Shi Y, Scheres SHW. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4 doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong H, et al. Pseudoatomic Structure of the Tripartite Multidrug Efflux Pump AcrAB-TolC Reveals the Intermeshing Cogwheel-like Interaction between AcrA and TolC. Structure. 2016;24:272–6. doi: 10.1016/j.str.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, et al. The tip region of the MacA alpha-hairpin is important for the binding to TolC to the Escherichia coli MacAB-TolC pump. Biochem Biophys Res Commun. 2010;394:962–5. doi: 10.1016/j.bbrc.2010.03.097. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, et al. Functional implications of an intermeshing cogwheel-like interaction between TolC and MacA in the action of macrolide-specific efflux pump MacAB-TolC. J Biol Chem. 2011;286:13541–9. doi: 10.1074/jbc.M110.202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee M, et al. The α-barrel tip region of Escherichia coli TolC homologs of Vibrio vulnificus interacts with the MacA protein to form the functional macrolide-specific efflux pump MacAB-TolC. J Microbiol. 2013;51:154–9. doi: 10.1007/s12275-013-2699-3. [DOI] [PubMed] [Google Scholar]
- 39.Dong C, et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–9. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Meyerson JR, et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci Rep. 2014;4:7084. doi: 10.1038/srep07084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo CJ, Passmore LA. Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–80. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SHW. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–22. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres SHW. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–57. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 50.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–4. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 53.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 55.Case DA, et al. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926. [Google Scholar]
- 57.Joung IS, Cheatham TE. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–41. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grubmüller H, Heymann B, Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–9. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryoEM map and the atomic model have been deposited in the Protein Data Bank and EMDB under accession numbers 5NIK and EMD-3652 for the MacA-TolC section, and 5NIL and EMD-3653 for the MacB section. The data that support the findings of this study are available from the corresponding authors upon request.