Highlight
Barley HvUGT13248 catalyzes conversion of nivalenol (NIV) to its non-toxic form 3-O-glucoside, and in transgenic wheat confers resistance to NIV-producing Fusarium graminearum
Keywords: Fusarium graminearum, Fusarium Head Blight, nivalenol, trichothecene, UDP-glycosyltransferase, wheat.
Abstract
Fusarium Head Blight is a disease of cereal crops that causes severe yield losses and mycotoxin contamination of grain. The main causal pathogen, Fusarium graminearum, produces the trichothecene toxins deoxynivalenol or nivalenol as virulence factors. Nivalenol-producing isolates are most prevalent in Asia but co-exist with deoxynivalenol producers in lower frequency in North America and Europe. Previous studies identified a barley UDP-glucosyltransferase, HvUGT13248, that efficiently detoxifies deoxynivalenol, and when expressed in transgenic wheat results in high levels of type II resistance against deoxynivalenol-producing F. graminearum. Here we show that HvUGT13248 is also capable of converting nivalenol into the non-toxic nivalenol-3-O-β-d-glucoside. We describe the enzymatic preparation of a nivalenol-glucoside standard and its use in development of an analytical method to detect the nivalenol-glucoside conjugate. Recombinant Escherichia coli expressing HvUGT13248 glycosylates nivalenol more efficiently than deoxynivalenol. Overexpression in yeast, Arabidopsis thaliana, and wheat leads to increased nivalenol resistance. Increased ability to convert nivalenol to nivalenol-glucoside was observed in transgenic wheat, which also exhibits type II resistance to a nivalenol-producing F. graminearum strain. Our results demonstrate the HvUGT13248 can act to detoxify deoxynivalenol and nivalenol and provide resistance to deoxynivalenol- and nivalenol-producing Fusarium.
Introduction
Small grain cereals such as wheat and barley are commonly infected by ascomyceteous fungi of the genus Fusarium, causing Fusarium Head Blight (FHB), a plant disease which leads to severe yield losses worldwide and contamination with trichothecene mycotoxins (Goswami and Kistler, 2004; Kazan et al., 2012). Trichothecenes are heat-stable sesquiterpenoid compounds routinely found in grain intended for food and feed use (Gottschalk et al., 2009; Streit et al., 2012), constituting a threat to the health of humans and livestock. Trichothecenes are potent inhibitors of protein biosynthesis in eukaryotic cells and act as virulence factors of FHB disease development (Bai et al., 2002; Jansen et al., 2005; McCormick et al., 2011). Inability to produce trichothecenes in F. graminearum results in strongly reduced disease severity and spread (Proctor et al., 1995; Bai et al., 2002). In turn, successful detoxification of deoxynivalenol is associated with resistance to disease spread within spikes (Lemmens et al., 2005; Li et al., 2015), which is defined as type II resistance (Mesterhazy, 1995).
The Fusarium species causing FHB on wheat and barley produce mainly type A or type B trichothecenes, with the type B trichothecenes deoxynivalenol (DON) and nivalenol (NIV) being the greatest concern in wheat- and barley-growing regions (Alexander et al., 2011). NIV and DON differ only by an additional hydroxyl group at the C4 position in NIV (McCormick et al., 2011). NIV is most prevalent in harvested materials from Asia and South America, whereas DON occurs predominantly in North America and Europe (van der Lee et al., 2015). It is noteworthy that strains producing DON, NIV, and their acetylated derivatives can all be found in the same region (van der Lee et al., 2015), and shifts in chemotype frequency occur and may take place rapidly. For example, a Canadian survey reported a dramatic shift from 15ADON (15-acetyldeoxynivalenol) chemotypes to 3ADON (3-acetyldeoxynivalenol) chemotypes within only 6 years (Ward et al., 2008). Thus, developing germplasm that exhibits resistance to a wide range of trichothecene mycotoxins should provide broad spectrum resistance and reduce the chance of the pathogen undergoing a population shift to a different chemotype composition.
NIV orally ingested by animals is more toxic than DON (Ryu et al., 1988). The European Food Safety Administration established a lower tolerated daily intake (TDI) of 0.7 µg kg–1 body weight for NIV, compared with 1 µg kg–1 for DON (EFSA CONTAM Panel, 2013). The Food Safety Commission in Japan established a TDI for NIV of 0.4 µg kg–1 body weight per day (Food Safety Commission of Japan, 2010). NIV is not a major problem in the USA currently, but NIV-producing F. graminearum strains have been discovered (Gale et al., 2011). In contrast to DON, the United States Food and Drug Administration has not established advisory guidelines for NIV (United States Food and Drug Administration, 2010).
DON resistance can be achieved in plants by the enzymatic conversion of the toxin into the non-toxic DON-3-O-glucoside (D3G) by substrate-specific UDP-glycosyltransferase (UGT) as first demonstrated in Arabidopsis thaliana (Poppenberger et al., 2003). Functional homologs of DON-inactivating UGTs in the agronomically relevant Pooideae subfamily have been identified in Brachypodium distachyon, rice, and barley (Schweiger et al., 2010, 2013; Pasquet et al., 2016). Among these, the highly DON- and F. graminearum-inducible barley HvUGT13248 gene (Gardiner et al., 2010) provided DON resistance when expressed in A. thaliana (Shin et al., 2012), and resistance to DON and DON-producing F. graminearum strains in transgenic wheat (Li et al., 2015). The kinetic properties and crystal structure of the Escherichia coli-expressed and affinity-purified gene product of OsUGT79, a rice gene highly similar to HvUGT13248, have been recently described (Michlmayr et al., 2015; Wetterhorn et al., 2016).
Recently, NIV-3-O-glucoside (NIV3G) has been described to occur in naturally Fusarium-infected barley and other cereals from Finland (Nathanail et al., 2015). The formation of a NIV-glucoside has been demonstrated in NIV-treated wheat (Nakagawa et al., 2011; Yoshinari et al., 2014), and NIV3G was purified from such material. Glycosyltransferases with specificity for NIV therefore exist in different cereal species, and may play a role in defense against NIV producers.
The aim of this study was to determine whether HvUGT13248 can inactivate and provide resistance to NIV. We show that expression of HvUGT13248 in yeast and A. thaliana confers NIV resistance in both model systems and that NIV3G can be efficiently produced by enzymatic synthesis. Stable transgenic events of the susceptible wheat cultivar ‘Bobwhite’ expressing HvUGT13248 confer increased ability to detoxify NIV and increased type II resistance to NIV-producing Fusarium strains.
Materials and methods
Heterologous expression of UGT in yeast
YZGA515 is a toxin-sensitive yeast strain in which three pleiotropic drug resistance genes and a trichothecene-3-O-acetyltransferase have been disrupted (MATa leu2Δ1 trp1Δ63 ura3-52 his3Δ200 lys2-801 ade2-101 pdr5::TRP1 pdr10::hisG pdr15::loxP-KANR-loxP ayt1::URA3), effectively rendering the strain unable to grow on low trichothecene concentrations (Schweiger et al., 2010). The strain was transformed with the previously reported UGT genes DOGT1=AtUGT73C5 (pBP868) (Poppenberger et al., 2003) and HvUGT13248 (pWS1921) (Schweiger et al., 2010), and the empty vector pYAK7 (pBP910; Poppenberger et al., 2003). These UGTs are under control of the strong constitutive pADH1 promotor and are N-terminally fused to a c-Myc epitope tag. Transformants were selected on synthetic complete medium lacking leucine. Stock solutions of 10 g l–1 of DON and NIV were prepared from crystallized toxins, and YPD plates containing 0, 40, 80, and 120 mg l–1 of either toxin were prepared. Exponentially grown yeast strains were rediluted in fresh selective medium to OD 0.05 and 0.005, and 3 µl of suspension cultures harboring either of the three constructs were spotted on plates containing increasing concentrations of NIV and DON.
Recombinant expression and purification of HvUGT13248
HvUGT13248 was expressed in E. coli as a fusion protein with an N-terminal His6-tag and a maltose-binding protein (nHis6-MalE-HvUGT13248). The cDNA was amplified from pWS1921 with the oligonucleotide primers 5'-GATATACATATG GCTGTCCACGACG-3' and 5'-TATATAAAGCTTTCAGCT GGCCTGGATGTC-3' and ligated to pCA02 [a derivative of the pET31b-based pKLD116 (Rocco et al., 2008) containing the pET21d multiple cloning site] using NdeI and HindIII (restrictions sites on primers are underlined). nHis6-MalE-HvUGT13248 was expressed with E. coli SHuffle® T7 Express lysY (New England Biolabs, Frankfurt am Main, Germany).
Protein expression and purification by immobilized metal ion chromatography (IMAC) on Ni2+-charged 5 ml HisTrap Crude FF columns (GE Healthcare, Chalfont St Giles, UK) were performed as recently described for OsUGT79 (Michlmayr et al., 2015). After IMAC, the buffer was changed to 50 mM potassium phosphate pH 7 + 50 mM NaCl + 10% glycerol by gel filtration on Sephadex G25 (GE Healthcare). One-step purified protein was stored in this buffer at −80 °C. Protein concentrations were determined with the Bio-Rad (Hercules, CA, USA) protein assay based on the dye-binding method of Bradford.
Enzyme assays were performed in 100 mM Tris, pH 7 at 37 °C, 2 min reaction time. For kinetic analyses, substrate concentrations ranged from 0.2 mM to 8 mM (NIV) and 0.3 mM to 25 mM (DON). The assays were stopped by transferring 20 μl of sample to 180 μl of methanol. After centrifugation (20 000 g, 5 min) to remove precipitated protein, the samples were further diluted in H2O to an expected concentration range of 1 mg l−1. The concentrations of NIV/NIV3G and DON/D3G were determined by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS, see below). Data regression (Michaelis–Menten equation) was performed with SigmaPlot 11.0 (Systat Software, San Jose, CA, USA). Enzyme activity is reported in μmol min−1 mg−1, which refers to the formation of NIV3G or D3G per mg of protein.
Synthesis and purification of a NIV-3-O-β-d-glucoside standard
NIV3G was produced with OsUGT79 (Michlmayr et al., 2015) in a 44 ml batch containing 1.5 mM NIV, 2.6 mM UDP-glucose, and 0.7 mg ml–1 nHis6-MalE-OsUGT79. The reaction was carried out in 100 mM Tris–HCl pH 7 at 25 °C for 24 h. Purification of NIV3G was carried out using an 1100 series preparative HPLC system equipped with an automatic fraction collector and a multiple wavelength detector (MWD) (all Agilent Technologies, Waldbronn, Germany). A Gemini NX column (150 × 21.2 mm, 5 μm, Phenomenex, Aschaffenburg, Germany) and gradient elution (eluent A, water; eluent B, methanol) was used for the separation of NIV3G from residual glucose and other impurities. The initial condition of 10% B was maintained for 2 min, followed by a linear increase to 60% B within 4 min and to 100% B within 0.1 min. Following a hold time of 1 min at 100%, the initial conditions were achieved with a fast switch to 10% B and the column was equilibrated prior to the next injection. The flow rate was 20 ml min−1 and the injection volume was set to 900 μl. The fractions were collected from 4 min to 6 min with the maximum peak duration of 1.5 min using threshold working mode. The collected fractions were pooled, the organic phase was evaporated on a rotary evaporator at 30 °C, and the remaining water phase was removed by lyophilization. The NIV3G crystals were weighed in a glass vial on a microbalance (16 mg) and stored at −20 °C.
NMR spectroscopy of NIV-3-O-β-d-glucoside
1H and 13C spectra were recorded on a Bruker Avance DRX-400 MHz spectrometer (Bruker, Germany). Data were recorded and evaluated using TOPSPIN 1.3 and TOPSPIN 3.2 (Bruker Topspin, Germany). All chemical shifts are given in ppm relative to tetramethylsilane. The calibration was done using residual solvent signals. Multiplicities are abbreviated as s (singlet), d (doublet), t (triplet), q (quartet), and b (broad signal). Deuterated methanol was purchased from Eurisotop (Gif sur Yvette Cedex, Paris, France).
Translation assays
To determine the in vitro toxicity of DON, NIV, and NIV3G, commercial in vitro transcription/translation systems [TnT® T7 Coupled Wheat Germ Extract System and TnT® T7 Coupled Reticulocyte Lysate System (Promega, Madison, WI, USA)] were used. Transcription/translation reactions were performed as described in Varga et al. (2015) with one minor change being that reactions with rabbit reticulocyte lysate were stopped after 20 min (instead of 24 min) reaction time. At least three independent assays using individual dilutions were performed for each substance.
Quantitative determination using LC-MS/MS
LC-MS/MS analysis was performed on a QTrap 4000 mass spectrometer (Sciex, Foster City, CA, USA). Chromatographic separation was achieved on a Gemini C18 (150 × 4.6 mm, 5 µm; Phenomenex, Aschaffenburg, Germany) at 25 °C with a flow rate of 0.8 ml min–1. The following water–methanol gradient (eluent A, 80:20, v/v; eluent B, 3:97, v/v; both containing 5 mM ammonium acetate) was used: initial conditions at 0% B were held for 1 min, followed by a linear increase to 50% B within 5 min and with a jump to 100% B. After holding 100% B for 2 min, a fast switch to the initial conditions was performed followed by column equilibration until 10 min. Negative electrospray ionization mode with the following source settings: temperature 550 °C, ion spray voltage 4 kV, curtain gas 30 psi (207 kPa of 99.5% nitrogen), source gas one and two both 50 psi (345 kPa of zero grade air), and collision gas (nitrogen) set to high. For quantitation, two selected reaction monitoring transitions per compound were acquired with a dwell time of 25 ms. The acetate adducts of the analytes (m/z 355.1 for DON, m/z 371.1 for NIV, m/z 517.3 for DON3G, and m/z 533.1 for NIV3G) were chosen as precursors, and the declustering potential (DP) was –40 V for DON and NIV, –50 V for DON3G, and –60 V for NIV3G. The following product ions were chosen as quantifier and qualifier, respectively: for DON m/z 59.2 [collision energy (CE) of –40 V] and m/z 265.2 (CE –22 V), for NIV m/z 59.1 and 281.1 (both CE –38 V), for DON3G m/z 427.1 (CE –30 V) and m/z 59.1 (CE -85 V), and for NIV3G m/z 263.0 (CE –30 V) and m/z 443.0 (CE –26 V).
Transgenic Arabidopsis thaliana root growth assay
Arabidopsis thaliana seeds (Shin et al., 2012) were surface sterilized in a 20% bleach, 0.02% Triton X-100, 0.5% sucrose solution for 10 min with continuous shaking, then washed twice using sterilized distilled water, followed by cold treatment in 4 °C for 3 d in the dark. Sterilized seeds were plated on half-strength Murashige and Skoog (MS) medium (2.15 g l–1 MS salt, 0.5 g l–1 MES, 0.5% sucrose, and 10 g l–1 agar) containing varying amounts of NIV.
The square Petri dishes were positioned vertically at room temperature under 16 h light and 8 h dark periods. Pictures of the plates were taken every 24 h starting from 4 d after germination. The longest root of each seedling was measured using ImageJ Software (Schneider et al., 2012).
Greenhouse disease testing of transgenic wheat expressing HvUGT13248 with NIV-producing F. graminearum
Seeds from each wheat genotype (Li et al., 2015) were planted into Sunshine MVP growth medium (Sun Gro Horticulture, Agawam, MA, USA) in 6 inch square plastic pots in a greenhouse. A total of 20–32 seeds were planted for each transgenic event with each pot containing four seeds. Twenty seeds of non-transformed controls (‘Bobwhite’, ‘Sumai 3’, and ‘Wheaton’) were also planted at four seeds per pot. Plants were fertilized with one teaspoon of Osmocote (14-14-14 N-P-K, Scotts Company, Marysville, OH, USA) fertilizer per pot at the three-leaf stage. Plants expressing the transgene were detected based on ELISAs (Agdia Inc., Elkhart, IN, USA) with an NPTII antibody as described in Li et al. (2015). Only transgenic plants expressing NPTII were analyzed further.
At anthesis, one floret of a central spikelet of the main spike was inoculated with 10 µl of macroconidial suspension (105 macroconidia ml–1 and 0.01% Triton X-100) of the NIV-producing F. graminearum strain 02-15 (Gale et al., 2011). Inoculated spikes were covered with transparent plastic bags for 3 d. FHB disease severity was determined as the percentage of spikelets with disease symptoms on the inoculated spikes at 21 d after inoculation. For statistical analysis, Student’s t-tests were used to compare each transgenic line with the non-transformed ‘Bobwhite’ control.
DON, NIV, and ergosterol were measured on NIV-producing F. graminearum-inoculated wheat via GC-MS as described (Dong et al., 2006; Jiang et al., 2006) using whole spikes sampled at 21 d after point inoculation. Nine spikes were sampled from each genotype of wheat tested in the autumn 2013 greenhouse screen.
Conversion of NIV to NIV3G in planta in transgenic wheat expressing HvUGT13248
Transgenic wheat expressing HvUGT13248, HvUGT13248-#19, and the non-transformed ‘Bobwhite’ control were inoculated at anthesis with 10 µl of aqueous NIV solution (2 µg µl–1) between the palea and lemma in the central four florets on the main spike of each plant. In this manner, each spike received 80 µg of NIV. Spikes were sampled at 0, 2, 6, 12, 24, 36, 48, 72, and 96 h, and 14 d after treatment. Nine biological replications were completely randomized during growth and one central spike per replication was used for each time point for each genotype. The four inoculated florets with rachis tissue for each replication were ground in liquid nitrogen, and metabolites were extracted in 4 vols of extraction solvent (50% methanol). NIV and NIV3G levels were ascertained by LC-MS/MS as described above.
Results
HvUGT13248 provides resistance to NIV in yeast
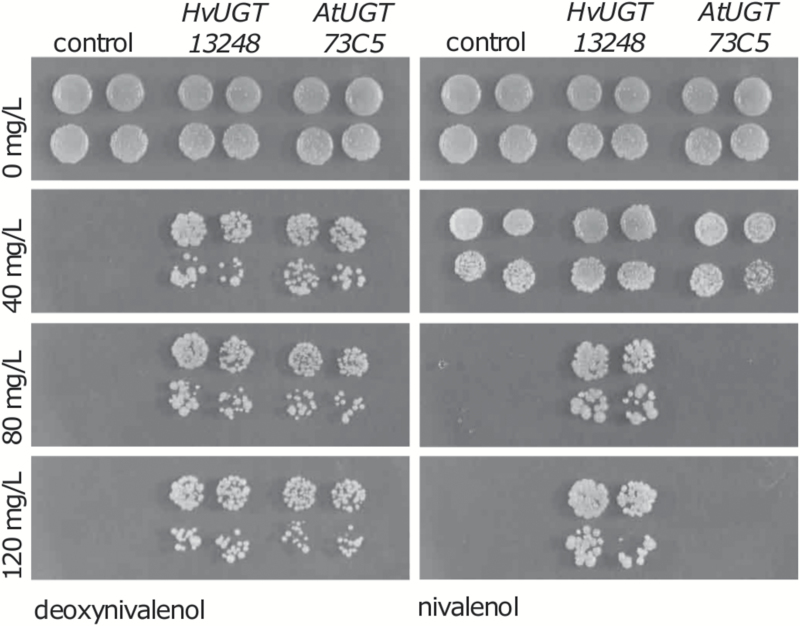
The glycosylation of DON by plant UGTs is well established as a major detoxification process and can be monitored by the increased accumulation of D3G in plant extracts (Shin et al., 2012; Li et al., 2015; Pasquet et al., 2016). Recently, NIV3G has been detected in barley and other cereal species (Nathanail et al., 2015), which may be due to the activity of either a UGT or different enzymes. Toxin-sensitive baker’s yeast transformed with either the barley HvUGT13248 or the A. thaliana AtUGT73C5 gene produce the recombinant proteins, as previously confirmed by western blotting using an antibody detecting the N-terminal c-Myc epitope tag (Schweiger et al., 2010). To test for resistance to DON and NIV, transformants expressing HvUGT13248 or AtUGT73C5 were spotted on agar medium containing increasing amounts of NIV or DON. While both UGTs conferred DON resistance as described before, only the transformants expressing HvUGT13248 grew on plates containing up to 120 mg l–1 NIV (Fig. 1). The results also show that NIV is less toxic for yeast than DON. While 40 mg l–1 DON was completely inhibitory for the strain containing the empty vector, 40 mg l–1 NIV caused only a moderate reduction of growth compared with the control without toxin.
Fig. 1.
Growth of glycosyltransferase-expressing yeast on rich medium containing the indicated concentrations of DON and NIV.
NIV is converted to NIV-3-O-β-d-glucoside by purified HvUGT13248
To investigate the fate of NIV in vitro, we examined the biochemical properties of the HvUGT13248 enzyme towards NIV as a substrate. We expressed HvUGT13248 and a previously characterized highly similar rice UGT, OsUGT79, which also converts DON to D3G in E. coli (Michlmayr et al., 2015). Both enzymes were produced as fusion proteins with an N-terminal His6-tag and a malE domain to improve solubility. While OsUGT79 was highly expressed and easily purified, HvUGT13248 was only very weakly expressed and highly unstable, preventing further purification after the first IMAC purification step. Nevertheless, the analysis of the enzymatic reaction by HPLC-MS showed that both OsUGT79 and HvUGT13248 can metabolize NIV into a substance with a mass expected for a NIV-glucoside. HvUGT13248 yielded a product with an identical MS/MS fragmentation pattern to the product of OsUGT79, demonstrating that both generate the same substance.
For structural elucidation of this glucoside, we therefore employed OsUGT79 to synthesize the compound in preparative amounts. From 20 mg of NIV in a first experiment, 16 mg of NIV3G were purified. The NIV-glucoside from this first reaction was purified by preparative HPLC and subjected to NMR analysis. Theoretically, the glucose could be linked to any of the four hydroxy groups in NIV (C3-OH, C4-OH, C7-OH, or C15-OH). The NMR results confirmed exclusive glucosylation of NIV at the C3 position with glucopyranose linked in β-configuration (see Supplementary Fig. S1 at JXB online).
Both OsUGT79 and HvUGT13248 recombinant fusion proteins were characterized with respect to kinetic properties towards NIV and DON (Table 1). The kcat values for HvUGT13248 might be underestimated, due to lower purity after only one purification step. The results nevertheless indicate that HvUGT13248 has higher affinity and ~5-fold higher catalytic efficiency towards NIV compared with DON.
Table 1.
Biochemical characterization of recombinant UDP-glucosyltransferases from rice (nHis6-MalE-OsUGT79) and barley (nHis6-MalE-HvUGT13248).
| Catalyst | Substrate | K M (mM) | V max (µmol min–1 mg–1) | k cat | k cat/ K M |
|---|---|---|---|---|---|
| HvUGT13248 | Deoxynivalenol | 3.0 ± 0.6 | 0.49 ± 0.04 | >0.78 | >0.26 |
| Nivalenol | 1.2 ± 0.3 | 0.96 ± 0.06 | >1.5 | >1.3 | |
| OsUGT79 | Deoxynivalenola | 0.23 ± 0.06 | 0.36 ± 0.02 | 0.57 | 2.5 |
| Nivalenol | 0.35 ± 0.04 | 0.38 ± 0.01 | 0.60 | 1.7 |
The displayed values are the results of three independent measurements ±SDs.
a Reported in Michlmayr et al. (2015).
NIV3G has minimal inhibitory activity at the ribosomal target
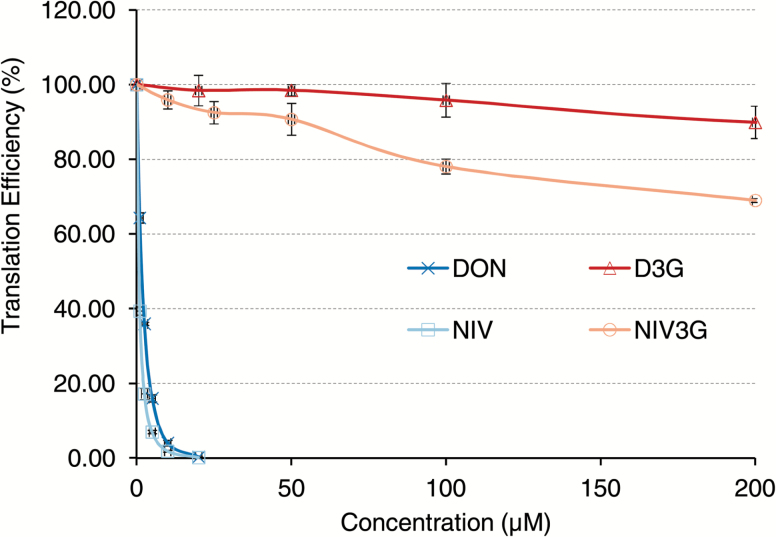
The primary mode of action of trichothecenes is inhibition of eukaryotic protein synthesis. To test whether NIV3G can inhibit animal and plant ribosomes, we utilized coupled in vitro transcription and translation systems, where ribosomes either from rabbit reticulocytes or from a wheat germ extract translate a luciferase reporter gene. As shown in Fig. 2 and Supplementary Fig. S2, both NIV and DON efficiently block translation in both systems. For wheat ribosomes, the IC50 for DON was 1.6 µM, and 0.7 µM for NIV. Likewise, in rabbit reticulocytes, the IC50 values for DON and NIV were 1.4 µM and 0.8 µM, respectively. Thus, NIV is slightly more inhibitory for ribosomes than DON for both animal and plant ribosomes. NIV-glucoside showed strongly reduced inhibitory activity (Fig. 2; Supplementary Fig. S2) in both systems, as previously described for D3G (Poppenberger et al., 2003). At the highest NIV3G concentration (200 µM) used in the translation assays, ~30% inhibition was observed in the wheat germ assay (Fig. 2). The IC20 values for NIV and NIV3G were reached at 0.3 µM and 90 µM, respectively, in the wheat germ assay, corresponding to an ~300-fold reduction in translation inhibition. Similarly, in the rabbit reticulocyte system, 600-fold more NIV3G than NIV is required to result in 20% translation inhibition (Supplementary Fig. S2). Results of molecular modeling (Pierron et al., 2016) suggest that the addition of the bulky glucose group should completely prevent interaction with the ribosomal binding site. The slight translation inhibition at extremely high NIV3G levels (200 µM or ~95 mg l–1) could theoretically be caused by partial hydrolysis of the glucoside during the assay, as previously observed for acetylated trichothecene (NX-2; Varga et al., 2015). We therefore analyzed samples from the end-points of the assay (stopped by precipitation with acetonitrile). However, no NIV was detectable in any of the samples (data not shown). We can therefore exclude that the minor residual inhibitory effect was due to hydrolytic cleavage of NIV3G.
Fig. 2.
NIV-3-glucoside is less toxic than NIV and DON on wheat ribosomes.
Increased NIV resistance in transgenic A. thaliana expressing HvUGT13248
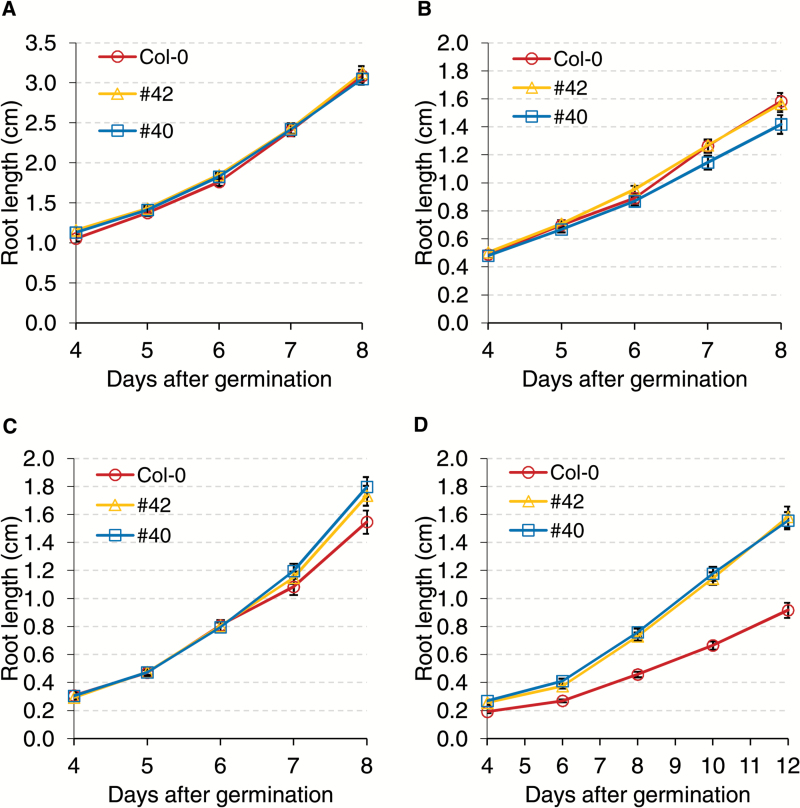
To investigate whether HvUGT13248 provides resistance to NIV in plants, we tested transgenic A. thaliana expressing HvUGT13248 on NIV-supplemented growth medium (Fig. 3; Supplementary Fig. S3). Two transgenic lines (#40 and #42) previously reported by Shin et al. (2012), along with a non-transformed Col-0 control, were grown on half-strength MS medium supplemented with 0 mg l–1 to 100 mg l–1 NIV. The results show that in the control treatment (0 mg l–1 NIV), root lengths of the transgenic lines were not significantly different from the non-transformed Col-0 (Fig. 3A: Supplementary Fig. S3). When grown on medium containing 100 mg l–1 NIV, the roots of both transgenic lines were significantly longer than those of Col-0 (Fig. 3D; Supplementary Fig. S3). Therefore, transgenic A. thaliana expressing HvUGT13248 show increased resistance to high concentrations of NIV.
Fig. 3.
Root growth of transgenic Arabidopsis thaliana expressing HvUGT13248 in the Col-0 background on half-strength MS medium containing (A) 0 mg l–1 NIV, (B) 20 mg l–1 NIV, (C) 40 mg l–1 NIV, and (D) 100 mg l–1 NIV.
Transgenic wheat expressing HvUGT13248 exhibits type II resistance to NIV-producing F. graminearum
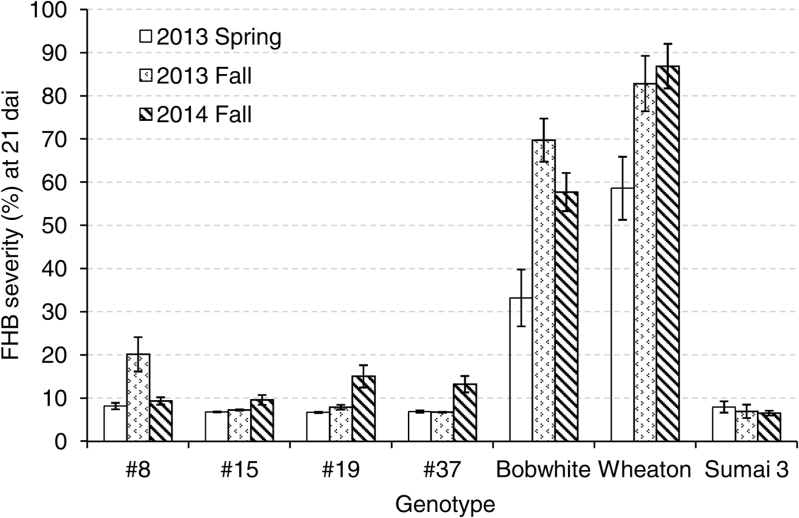
To test NIV resistance in transgenic wheat expressing HvUGT13248, we performed point inoculation assays with a NIV-producing F. graminearum strain (Gale et al., 2011). Transgenic wheat carrying HvUGT13248 (independent transgenic lines #8, #15, #19, and #37 previously described in Li et al., 2015), ‘Bobwhite’, ‘Sumai 3’, and ‘Wheaton’ were evaluated for type II resistance. We repeated this disease screen in the greenhouse three times and, for each screen, 20–32 plants of each transgenic event were grown. The susceptible check ‘Wheaton’ exhibited FHB severity levels that ranged between 58.6 ± 7.3% and 86.9 ± 5.1%, while the resistant check ‘Sumai 3’ exhibited FHB severity levels that ranged between 6.5 ± 0.6% and 8.0 ± 1.3%, indicating that the environments for FHB severity screening were successful and discriminative (Fig. 4; Supplementary Table S1). The checks also demonstrate differences in FHB severity between the trials, which is a frequent observation when assaying FHB severity (e.g. Okubara et al., 2002; Li et al., 2015). The transgenic lines significantly reduced FHB severity compared with the non-transformed ‘Bobwhite’ control (Fig. 4; Supplementary Table S1). For the three greenhouse trials, transgenic line #8 showed FHB severity of 8.2–20.2%, #15 showed 6.8–9.6% severity, #19 showed 6.7–15.1% severity, and #37 showed 6.8–13.2% severity. All four transgenic lines exhibited a significant reduction of FHB severity relative to ‘Bobwhite’, ranging from 71.1% to 90.3%. Three of the transgenic lines (#15, #19, and #37) showed FHB severity in more than one trial at levels similar to the resistant line ‘Sumai 3’. NIV and ergosterol contents were also measured on whole spikes by GC-MS during the 2013 greenhouse trial (Table 2). All four transgenic lines accumulated significantly less NIV and ergosterol compared with ‘Bobwhite’. DON did not accumulate to detectable levels in the transgenic lines or ‘Sumai 3’; however, in ‘Bobwhite’ and ‘Wheaton’, we observed a small amount of DON accumulation. These results indicate that the NIV-producing strain also produces a trace amount of DON, and when inoculated on genotypes (e.g. ‘Wheaton’ and ‘Bobwhite’) that have less capacity to detoxify trichothecenes, a small amount of DON is detected. Taken together, transgenic wheat expressing HvUGT1348 exhibited high levels of type II resistance to a NIV-producing F. graminearum strain and lowered NIV content.
Fig. 4.
FHB severity of transgenic wheat expressing HvUGT13248 at 21 d after point inoculation with NIV-producing F. graminearum strain #02-15 in three greenhouse trials. Lines #8, #15, #19, and #37 were transgenic wheat expressing HvUGT13248, and ‘Bobwhite’ was the non-transformed control. ‘Wheaton’ was the susceptible check and ‘Sumai 3’ was the resistant check.
Table 2.
Trichothecene accumulation and ergosterol content in NIV-producing F. graminearum-inoculated wheat spikes
| Genotype a | Ergosterol (mg l –1) | NIV (mg l –1) | DON (mg l –1) |
|---|---|---|---|
| #8 | 5.38 ± 1.58*** | 4.70 ± 0.50*** | NDb |
| #15 | 1.37 ± 0.50*** | 2.00 ± 0.31*** | ND |
| #19 | 1.85 ± 0.05*** | 3.03 ± 0.58*** | ND |
| #37 | 1.08 ± 0.20*** | 1.80 ± 0.06*** | ND |
| Bobwhite | 49.53 ± 6.95 | 29.57 ± 5.82 | 0.28 ± 0.06 |
| Wheaton | 54.88 ± 4.23 | 47.43 ± 2.16 | 0.29 ± 0.02 |
| Sumai 3 | 0.90 ± 0.02 | 0.58 ± 0.02 | ND |
Values provided are the means ±SE.
a #8, #15, #19, and #37 were transgenic lines, and ‘Bobwhite’ was the non-transformed control. ‘Sumai 3’ was the resistant check and ‘Wheaton’ was the susceptible check.
b ND, not detected (<0.05 mg l–1).
*** indicate significance at the 0.001 level compared with the non-transformed ‘Bobwhite’ control (Student’s t-test).
Increased NIV to NIV3G conversion in transgenic wheat
To determine if the enhanced type II resistance conferred by HvUGT13248 was indeed due to increased glycosylation, we monitored the NIV and NIV3G concentrations in transgenic line #19 and the ‘Bobwhite’ non-transgenic control from 0 d to 14 d after NIV application. Two central spikelets on the main spike of transgenic line #19 and non-transformed ‘Bobwhite’ were inoculated with NIV at 40 µg per spikelet (80 µg or 256.41 nmol NIV per spike). Treated spikelets together with the connecting rachis tissue were collected at 10 time points: 0, 2, 6, 12, 24, 36, 48, 72, and 96 h, and 14 d after NIV application. NIV and NIV3G concentrations were measured by LC-MS/MS.
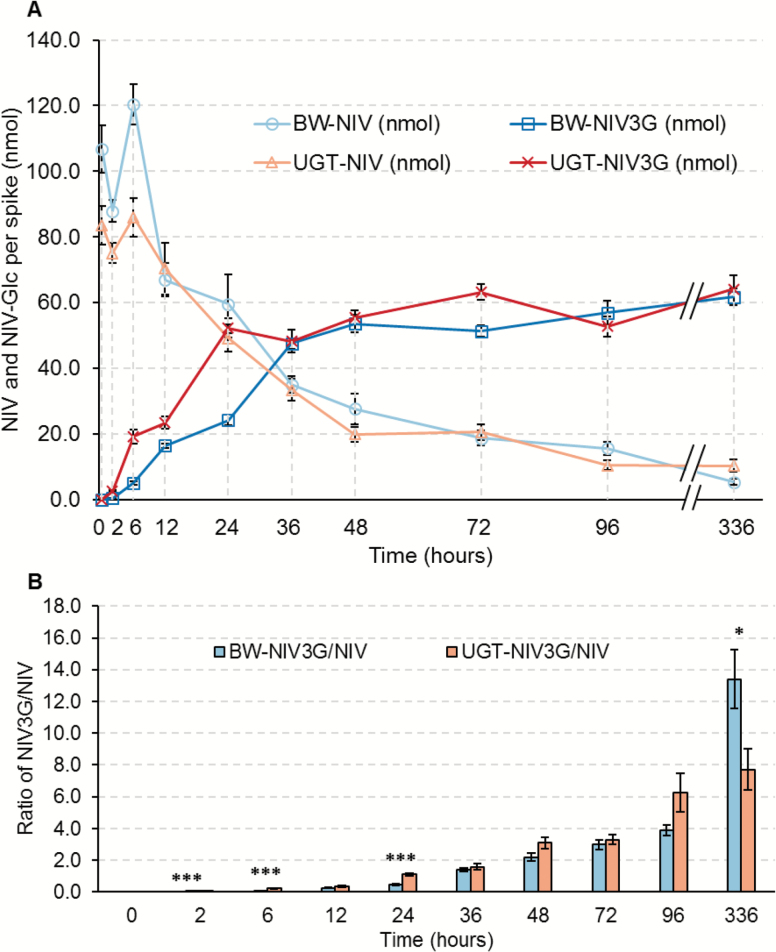
NIV concentration decreased and NIV3G concentration increased in both transgenic and non-transformed wheat (Fig. 5; Supplementary Table S2), indicating that both genotypes possess NIV-glycosyltransferase activity. However, the conversion was faster in transgenic line #19 than in ‘Bobwhite’ at early time points. Before 6 h after NIV injection, NIV concentrations in line #19 were significantly lower than in ‘Bobwhite’, while NIV3G contents were significantly higher than in ‘Bobwhite’. NIV3G concentrations continued to be significantly higher in line #19 at 12 h and 24 h after treatment. Average NIV3G/NIV ratios in the transgenic lines were significantly higher than in ‘Bobwhite’ during the first 24 h after NIV application, except for the 12 h time point. After 36 h, the differences between transgenic and non-transformed lines were not significant, indicating that the greatest impact of HvUGT13248 overexpression on NIV to NIV3G conversion is at early time points.
Fig. 5.
HvUGT13248 promotes NIV to NIV3G conjugation in transgenic wheat. (A) NIV and NIV3G concentrations in ‘Bobwhite’ (BW) and transgenic line #19 at 0, 2, 6, 12, 24, 36, 48, 72, 96, and 336 h after NIV treatment. ‘BW-NIV’ and ‘UGT-NIV’ are the NIV content in BW or transgenic event #19 at each time point, respectively. ‘BW-NIV3G’ and ‘UGT-NIV3G’ are the NIV3G content in BW or transgenic line #19 at each time point, respectively. (B) Fold change of the molar ratio of NIV3G to NIV concentrations in BW and transgenic line #19 at each time point.
Discussion
The HvUGT13248 gene product can metabolize different trichothecene toxins
Family 1 UGTs comprise a large family of genes (Caputi et al., 2012). UGTs play important roles in diverse biological processes (Ross et al., 2001) and UGT genes seem to evolve rapidly by amplification and gene death; thus, it is non-trivial to identify true orthologs in related plant genomes (Schweiger et al., 2013). Moreover, individual plant UGTs may accept more than one substrate. For example, A. thaliana AtUGT73C5, the first UGT described to inactivate DON (Poppenberger et al., 2003), can also glycosylate brassinosteroids (Poppenberger et al., 2005), the structurally unrelated Fusarium toxin zearalenone (Poppenberger 2003), and regioselectively also quercetin at certain hydroxyl groups (Lim et al., 2004). Surprisingly, despite this broad activity towards multiple structurally unrelated substrates, AtUGT73C5 does not confer NIV resistance (Fig. 1), even though only the presence of the C4-OH distinguishes NIV from DON. In contrast, the rice OsUGT79 can metabolize both DON and NIV, but not T-2 toxin (Wetterhorn et al., 2016). Likewise, the barley HvUGT13248 also efficiently metabolizes NIV and DON. Our NMR results show that the mode of NIV detoxification is analogous to that of DON, by exclusive formation of NIV3G. The HvUGT13248 enzyme has higher affinity (Km value in Table 1) for NIV and about five times more efficiency (kcat/Km in Table 1) than with DON as the substrate.
Inactivation of the Brachypodium gene Bradi5g03300, which is most similar to HvUGT13248, causes reduced resistance to Fusarium infection (Pasquet et al., 2016). Assuming that HvUGT13248 is also a highly relevant detoxification enzyme in barley, the observation of a much lower affinity of the enzyme towards DON than NIV suggests that the switch from the ancestral NIV production to DON production in Fusarium by mutation of TRI13 (Brown et al., 2002; Lee et al., 2002) might have allowed DON partially to escape detoxification, providing a selective advantage to DON producers on barley. DON and NIV chemotype strains co-exist in the field and seem to be maintained by balancing selection (Ward et al., 2002). We have also identified a glucosyltransferase from Brachypodium which confers resistance to NIV but is nearly inactive with DON (G. Wiesenberger et al., unpublished results). Therefore, it is conceivable that differences in the detoxification capacity in various host plants exist, making production of either DON or NIV advantageous. Interestingly, NIV production has been described to be a virulence factor on maize (Maier et al., 2006), while disruption of DON production had little effect, which could be explained by high DON detoxification capability. Yet, DON producers are in general also predominant in maize grown in Europe, North America, and China (Qiu and Shi, 2014; Kuhnem et al., 2015; Pasquali et al., 2016), while in South American-grown maize high frequencies of NIV-producing F. meridionale and F. boothii were found (Sampietro et al., 2012). Interestingly, strains have been reported that were genotyped as NIV producers, but produced up to 20% DON besides NIV, indicating that not only is complete loss of function by disrupting the TRI13 coding region possible, but potentially also down-regulation of the TRI1 expression level. Overall, it seems likely that genotypic variation within different crop species with respect to detoxification capacity is high, and drawing conclusions based on one or a few cultivars is premature. Currently, information on the detoxification capability of different cultivars is lacking in most crop plants infected by Fusarium ssp.
Higher intrinsic plant resistance to NIV than to DON
Overexpression of HvUGT13248 provided resistance to NIV in A. thaliana, alleviating the detrimental effect of the toxin on root length at 100 mg l–1 NIV (Fig. 3). However, compared with similar assays with DON (Shin et al., 2012), we found that the minimal concentration required to observe root inhibition in the wild type Col-0 is dramatically higher for NIV (20 mg l–1 no inhibition, Fig. 3B) than for DON (0.5 mg l–1, Shin et al., 2012). This is consistent with a previous report of relatively lower phytotoxicity of NIV compared with DON in an A. thaliana leaf assay (Desjardins et al., 2007). Similar results have been previously reported in wheat (Shimada and Otani, 1990), which showed severe wheat root growth inhibition by DON, while observing no such differences for NIV at the same concentrations.
In contrast, NIV has higher cytotoxicity than DON when applied orally to experimental animals, and is ~1.5- to 1.7-fold more toxic to Caco-2 human cells (Alassane-Kpembi et al., 2013). NIV was also shown to have greater impact than DON on the pig intestinal mucosa, both in vitro and in vivo (Cheat et al., 2015). Our data obtained with the in vitro translation systems (Fig. 2; Supplementary Fig. S2) show that this discrepancy between plants and animals is not due to differences at the ribosomal target. NIV is slightly more inhibitory than DON for both rabbit and plant ribosomes. The difference in toxicity in plants may be in part at the level of uptake or drug efflux (e.g. substrate specificity for ABC transporters). Yet, it seems likely that the ability to glycosylate these toxins plays an important role. Interestingly, there is also a difference in animals. While DON is converted into glucuronides in pigs (and other experimental animals and humans) and in vitro by different UDP-glucuronosyltransferases (Maul et al., 2015), no evidence for formation of NIV-glucuronide was detected in a pig feeding study (Hedman et al., 1997) and in mice treated with NIV (Poapolathep et al., 2003), which might be a reason for the higher toxicity of NIV in animals.
Our data suggest that the higher basal resistance of wheat to NIV compared with DON directly correlates with the higher levels of NIV3G found in the susceptible wheat cultivar ‘Bobwhite’ compared with D3G following treatment with the respective toxin. Apparently, wheat has endogenous UGTs to inactivate NIV but a much lower capacity to detoxify DON. Constitutive expression of HvUGT13248 clearly increased resistance to a NIV-producing F. graminearum, although the capacity to detoxify externally applied NIV was high in both parental and transgenic wheat. The transgenic lines converted 62.6% of the administered NIV into the glucoside 24 h after treatment, while ‘Bobwhite’ metabolized only 22.7%. By 36 h after NIV treatment, transgenic wheat converted 57.7% of the toxin to NIV3G, while non-transformed ‘Bobwhite’ converted 44.5% (Fig. 5; Supplementary Table S2). In comparison, in a similar DON treatment experiment, ‘Bobwhite’ metabolized only 2.2% DON to D3G 24 h after treatment (Li et al., 2015).
NIV-producing F. graminearum inoculated on transgenic wheat resulted in a lower NIV content (and reduced fungal biomass) and a higher NIV3G to NIV ratio compared with non-transformed ‘Bobwhite’. One can expect that this is also the case in natural infection with NIV producers in the field. NIV3G is a neglected ‘masked mycotoxin’ of unknown toxicological relevance. Yet, based on results of the reticulocyte lysates and experience from D3G in mice and pigs, resorbed NIV3G itself will not be toxic for mammalian ribosomes, and the back-conversion of NIV3G to NIV by bacterial glucosidases should also take place rather late in the intestinal tract as demonstrated for D3G (Nagl et al., 2014), so that most of the released NIV should not be resorbed but excreted via feces (Gratz et al., 2016). The lack of an analytical standard for NIV3G and the large amounts needed for toxicological studies have to date prevented research on this issue. So far only small amounts of NIV3G have been purified from NIV-contaminated wheat (9 mg from 12 kg of starting material; Yoshinari et al., 2014). We showed that NIV3G can be efficiently synthesized enzymatically in vitro with the rice enzyme OsUGT79. As the reaction can be driven to completion by cofactor recycling (as shown for D3G; Michlmayr et al., 2015) and the reaction mix is much less complex, purification from this source is much easier. The described method can easily provide gram amounts of NIV3G for animal feeding studies, which will allow experimental testing of our predictions.
Rapid trichothecene detoxification is key to FHB resistance
The interaction between the fungal pathogen and the plant host is very dynamic. Trichothecenes play an important role in the disease spread during FHB development (Bai et al., 2002), and reducing trichothecenes is important to type II resistance (Li et al., 2015). Gene expression profiles have shown that F. graminearum induces trichothecene biosynthesis genes as early as 48 h after germination in wheat (Lysøe et al., 2011), and Boenisch and Schäfer (2013) reported similar timing based on microscopic examination of reporter genes. In barley and Brachypodium, the DON-inactivating UGTs are highly inducible by the toxin (Gardiner et al., 2010; Schweiger et al., 2013). Upon perception of the toxin, it depends on how rapidly the already partially inhibited ribosome can translate the induced UGT transcript into an active detoxification enzyme. If the toxin can be efficiently neutralized, the plant can contain the pathogen, limiting its spread. If the toxin diffusing ahead of the infection zone blocks or at least strongly reduces or severely delays translation, the plant is susceptible and the pathogen can spread throughout the spike. Consistent with this model, transgenic wheat constitutively overexpressing HvUGT13248 efficiently converted NIV to NIV3G, leading to high levels of resistance to disease spread after inoculation with a NIV-producing F. graminearum (Fig. 4; Table 2; Supplementary Table S1). The ability of the susceptible ‘Bobwhite’ and the transgenic resistant line to convert comparable amounts of NIV to NIV3G at 36 h after administering the toxin and later suggests that resistance against NIV-producing chemotypes is constituted at earlier time points.
With constitutive overexpression, transgenic wheat expressing HvUGT13248 provides resistance to both DON and NIV and therefore does not seem to be at risk of being easily overcome by a shift in chemotype composition, and thus is an excellent candidate gene for FHB control also in regions of the world where NIV-producing species are highly prevalent.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. NMR data of NIV-3-O-β-d-glucoside.
Fig. S2. NIV-3-glucoside is less inhibitory than NIV for rabbit reticulocyte ribosomes.
Fig. S3. Root growth of Col-0 wild-type and transgenic A. thaliana expressing HvUGT13248 on half-strength MS medium containing 0 mg l–1 NIV at 7 d and 100 mg l–1 NIV at 14 d after germination. Scale bars=2 cm.
Table S1. Summary of transgenic wheat expressing HvUGT13248 in greenhouse point-inoculation tests with the NIV-producing F. graminearum strain.
Table S2. HvUGT13248 converts NIV to NIV3G faster in transgenic wheat than in non-transformed ‘Bobwhite’.
Supplementary Material
Acknowledgements
This material is based upon work supported by the US Department of Agriculture, under Agreement no. 59-0206-4-021. This is a co-operative project with the US Wheat and Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the US Department of Agriculture. The work performed in Austria was funded by the FWF special research project Fusarium (F3708, F3706, F3711, and F3715) and the Vienna Science and Technology Fund (LS12-021). Furthermore, we thank the Austrian Federal Ministry of Science, Research and Economy, the Austrian National Foundation of Research, Technology and Development, as well as the BIOMIN Holding GmbH for funding the Christian Doppler Laboratory for Mycotoxin Metabolism. We also thank H. Corby Kistler for the NIV-producing Fusarium graminearum strain.
Glossary
Abbreviations:
- D3G
DON-3-O-β-d-glucoside
- DON
deoxynivalenol
- FHB
Fusarium Head Blight
- NIV
nivalenol
- NIV3G
NIV-3-O-β-d-glucoside
- UGT
UDP-glycosyltransferase.
References
- Alassane-Kpembi I, Kolf-Clauw M, Gauthier T, Abrami R, Abiola FA, Oswald IP, Puel O. 2013. New insights into mycotoxin mixtures: the toxicity of low doses of Type B trichothecenes on intestinal epithelial cells is synergistic. Toxicology and Applied Pharmacology 272, 191–198. [DOI] [PubMed] [Google Scholar]
- Alexander NJ, McCormick SP, Waalwijk C, van der Lee T, Proctor RH. 2011. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genetics and Biology 48, 485–495. [DOI] [PubMed] [Google Scholar]
- Bai GH, Desjardins AE, Plattner RD. 2002. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153, 91–98. [DOI] [PubMed] [Google Scholar]
- Boenisch MJ, Schäfer W. 2011. Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biology 11, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genetics and Biology 36, 224–233. [DOI] [PubMed] [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. 2012. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. The Plant Journal 69, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Cheat S, Gerez JR, Cognié J, Alassane-Kpembi I, Bracarense AP, Raymond-Letron I, Oswald IP, Kolf-Clauw M. 2015. Nivalenol has a greater impact than deoxynivalenol on pig jejunum mucosa in vitro on explants and in vivo on intestinal loops. Toxins 7, 1945–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, McCormick SP, Appell M. 2007. Structure–activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. Journal of Agricultural and Food Chemistry 55, 6487–6492. [DOI] [PubMed] [Google Scholar]
- Dong Y, Steffenson BJ, Mirocha CJ. 2006. Analysis of ergosterol in single kernel and ground grain by gas chromatography–mass spectrometry. Journal of Agricultural and Food Chemistry 54, 4121–4125. [DOI] [PubMed] [Google Scholar]
- EFSA CONTAM Panel 2013. Scientific Opinion on risks for animal and public health related to the presence of nivalenol in food and feed. EFSA Journal 11, 3262–3381. [Google Scholar]
- Food Safety Commission of Japan 2010. http://www.fsc.go.jp/english/evaluationreports/nm_toxins/rar_donniv_fs872_2010_nm.pdf http://www.fsc.go.jp/english/evaluationreports/nm_toxins/rar_donniv_fs872_2010_nm.pdf Risk assessment report deoxynivalenol and nivalenol (mycotoxin). Risk assessment report–mycotoxin FS/872/2010.
- Gale LR, Harrison SA, Ward TJ, O’Donnell K, Milus EA, Gale SW, Kistler HC. 2011. Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology 101, 124–134. [DOI] [PubMed] [Google Scholar]
- Gardiner SA, Boddu J, Berthiller F, Hametner C, Stupar RM, Adam G, Muehlbauer GJ. 2010. Transcriptome analysis of the barley–deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Molecular Plant-Microbe Interactions 23, 962–976. [DOI] [PubMed] [Google Scholar]
- Goswami RS, Kistler HC. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Molecular Plant Pathology 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Gottschalk C, Barthel J, Engelhardt G, Bauer J, Meyer K. 2009. Simultaneous determination of type A, B and D trichothecenes and their occurrence in cereals and cereal products. Food Additives and Contaminants 9, 1273–1289. [Google Scholar]
- Gratz SW, Dinesh R, Yoshinari T, Holtrop G, Richardson AJ, Duncan G, MacDonald S, Lloyd A, Tarbin J. 2016. Masked trichothecene and zearalenone mycotoxins withstand digestion and absorption in the upper GI tract but are efficiently hydrolyzed by human gut microbiota in vitro. Molecular Nutrition and Food Research (in press). [DOI] [PubMed] [Google Scholar]
- Hedman R, Pettersson H, Lindberg JE. 1997. Absorption and metabolism of nivalenol in pigs. Archives of Animal Nutrition 50, 13–24. [DOI] [PubMed] [Google Scholar]
- Jansen C, von Wettstein D, Schäfer W, Kogel KH, Felk A, Maier FJ. 2005. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proceedings of the National Academy of Sciences, USA 102, 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G-L, Dong Y, Lewis JM, Siler L, Ward RW. 2006. Characterization of resistance to Fusarium graminearum in a recombinant inbred line population of wheat. Crop Science 46, 2590–2597. [Google Scholar]
- Kazan K, Gardiner DM, Manners JM. 2012. On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Molecular Plant Pathology 13, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnem PR, Spolti P, Del Ponte EM, Cummings JA, Bergstrom GC. 2015. Trichothecene genotype composition of Fusarium graminearum not differentiated among isolates from maize stubble, maize ears, wheat spikes, and the atmosphere in New York. Phytopathology 105, 695–699. [DOI] [PubMed] [Google Scholar]
- Lee T, Han YK, Kim KH, Yun SH, Lee YW. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Applied and Environmental Microbiology 68, 2148–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens M, Scholz U, Berthiller F, et al. 2005. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Molecular Plant-Microbe Interactions 18, 1318–1324. [DOI] [PubMed] [Google Scholar]
- Li X, Shin S, Heinen S, Dill-Macky R, Berthiller F, Nersesian N, Clemente T, McCormick S, Muehlbauer GJ. 2015. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Molecular Plant-Microbe Interactions 28, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. 2004. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnology and Bioengineering 87, 623–631. [DOI] [PubMed] [Google Scholar]
- Lysøe E, Seong KY, Kistler HC. 2011. The transcriptome of Fusarium graminearum during the infection of wheat. Molecular Plant-Microbe Interactions 24, 995–1000. [DOI] [PubMed] [Google Scholar]
- Maier FJ, Miedaner T, Hadeler B, Felk A, Salomon S, Lemmens M, Kassner H, Schäfer W. 2006. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Molecular Plant Pathology 7, 449–461. [DOI] [PubMed] [Google Scholar]
- Maul R, Warth B, Schebb NH, Krska R, Koch M, Sulyok M. 2015. In vitro glucuronidation kinetics of deoxynivalenol by human and animal microsomes and recombinant human UGT enzymes. Archives of Toxicology 89, 949–960. [DOI] [PubMed] [Google Scholar]
- McCormick SP, Stanley AM, Stover NA, Alexander NJ. 2011. Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesterhazy A. 1995. Types and components of resistance to fusarium head blight of wheat. Plant Breeding 114, 377–386. [Google Scholar]
- Michlmayr H, Malachová A, Varga E, Kleinová J, Lemmens M, Newmister S, Rayment I, Berthiller F, Adam G. 2015. Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-d-glucoside. Toxins 7, 2685–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl V, Woechtl B, Schwartz-Zimmermann HE, Hennig-Pauka I, Moll WD, Adam G, Berthiller F. 2014. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in pigs. Toxicology Letters 229, 190–197. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Ohmichi K, Sakamoto S, Sago Y, Kushiro M, Nagashima H, Yoshida M, Nakajima T. 2011. Detection of a new Fusarium masked mycotoxin in wheat grain by high-resolution LC-Orbitrap™ MS. Food Additives and Contaminants. Part A 28, 1447–1456. [DOI] [PubMed] [Google Scholar]
- Nathanail AV, Syvähuoko J, Malachová A, Jestoi M, Varga E, Michlmayr H, Adam G, Sieviläinen E, Berthiller F, Peltonen K. 2015. Simultaneous determination of major type A and B trichothecenes, zearalenone and certain modified metabolites in Finnish cereal grains with a novel liquid chromatography-tandem mass spectrometric method. Analytical and Bioanalytical Chemistry 407, 4745–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara P, Blechl A, McCormick S, Alexander N, Dill-Macky R, Hohn T. 2002. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene. Theoretical and Applied Genetics 106, 74–83. [DOI] [PubMed] [Google Scholar]
- Pasquali M, Beyer M, Logrieco A, et al. 2016. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Frontiers in Microbiology 7, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquet JC, Changenet V, Macadré C, et al. 2016. A Brachypodium UDP-glycosyltransferase confers root tolerance to deoxynivalenol and resistance to Fusarium infection. Plant Physiology 172, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron A, Mimoun S, Murate LS, et al. 2016. Intestinal toxicity of the masked mycotoxin deoxynivalenol-3-β-d-glucoside. Archives of Toxicology 90, 2037–2046. [DOI] [PubMed] [Google Scholar]
- Poapolathep A, Sugita-Konishi Y, Doi K, Kumagai S. 2003. The fates of trichothecene mycotoxins, nivalenol and fusarenon-X, in mice. Toxicon 41, 1047–1054. [DOI] [PubMed] [Google Scholar]
- Poppenberger B. 2003. Molecular mechanisms of resistance to Fusarium mycotoxins in plants. PhD thesis, BOKU-Universität für Bodenkultur. [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G. 2003. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. Journal of Biological Chemistry 278, 47905–47914. [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Soeno K, et al. 2005. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proceedings of the National Academy of Sciences, USA 102, 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM, McCormick SP. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Molecular Plant-Microbe Interactions 8, 593–601. [DOI] [PubMed] [Google Scholar]
- Qiu J, Shi J. 2014. Genetic relationships, carbendazim sensitivity and mycotoxin production of the Fusarium graminearum populations from maize, wheat and rice in eastern China. Toxins 6, 2291–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ. 2001. Higher plant glycosyltransferases. Genome Biology 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Ohtsubo K, Izumiyama N, Nakamura K, Tanaka T, Yamamura H, Ueno Y. 1988. The acute and chronic toxicities of nivalenol in mice. Fundamental and Applied Toxicology 11, 38–47. [DOI] [PubMed] [Google Scholar]
- Sampietro DA, Ficoseco ME, Jimenez CM, Vattuone MA, Catalán CA. 2012. Trichothecene genotypes and chemotypes in Fusarium graminearum complex strains isolated from maize fields of northwest Argentina. International Journal of Food Microbiology 153, 229–233. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger W, Boddu J, Shin S, Poppenberger B, Berthiller F, Lemmens M, Muehlbauer GJ, Adam G. 2010. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Molecular Plant-Microbe Interactions 23, 977–986. [DOI] [PubMed] [Google Scholar]
- Schweiger W, Pasquet JC, Nussbaumer T, et al. 2013. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Molecular Plant-Microbe Interactions 26, 781–792. [DOI] [PubMed] [Google Scholar]
- Shimada T, Otani M. 1990. Effects of Fusarium mycotoxins on the growth of shots and roots at germination in some Japanese wheat cultivars. Cereal Research Communications 18, 229–232. [Google Scholar]
- Shin S, Torres-Acosta JA, Heinen SJ, McCormick S, Lemmens M, Paris MP, Berthiller F, Adam G, Muehlbauer GJ. 2012. Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. Journal of Experimental Botany 63, 4731–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit E, Schatzmayr G, Tassis P, et al. 2012. Current situation of mycotoxin contamination and co-occurrence in animal feed—focus on Europe. Toxins 4, 788–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Food and Drug Administration 2010. Guidance for industry and FDA: advisory levels for deoxynivalenol (DON) in finished wheat products for human consumption and grains and grain by-products used for animal feed. Silver Spring, MD: US Food and Drug Administration. [Google Scholar]
- van der Lee T, Zhang H, van Diepeningen A, Waalwijk C. 2015. Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Additives and Contaminants. Part A 32, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga E, Wiesenberger G, Hametner C, et al. 2015. New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environmental Microbiology 17, 2588–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proceedings of the National Academy of Sciences, USA 99, 9278–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TJ, Clear RM, Rooney AP, O’Donnell K, Gaba D, Patrick S, Starkey DE, Gilbert J, Geiser DM, Nowicki TW. 2008. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genetics and Biology 45, 473–484. [DOI] [PubMed] [Google Scholar]
- Wetterhorn KM, Newmister SA, Caniza RK, Busman M, McCormick SP, Berthiller F, Adam G, Rayment I. 2016. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: a UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry 55, 6175–6186. [DOI] [PubMed] [Google Scholar]
- Yoshinari T, Sakuda S, Furihata K, Furusawa H, Ohnishi T, Sugita-Konishi Y, Ishizaki N, Terajima J. 2014. Structural determination of a nivalenol glucoside and development of an analytical method for the simultaneous determination of nivalenol and deoxynivalenol, and their glucosides, in wheat. Journal of Agricultural and Food Chemistry 62, 1174–1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.