Highlight
StAmy23, StBAM1 and StBAM9 play distinct roles in potato cold-induced sweetening by preferentially acting on soluble phytoglycogen, soluble starch and starch granules, respectively, in different subcellular locations.
Keywords: α-Amylase, β-amylase, cold-induced sweetening, potato, reducing sugar, starch degradation, tuber.
Abstract
Cold-induced sweetening (CIS) in potato is detrimental to the quality of processed products. Conversion of starch to reducing sugars (RS) by amylases is considered one of the main pathways in CIS but is not well studied. The amylase genes StAmy23, StBAM1, and StBAM9 were studied for their functions in potato CIS. StAmy23 is localized in the cytoplasm, whereas StBAM1 and StBAM9 are targeted to the plastid stroma and starch granules, respectively. Genetic transformation of these amylases in potatoes by RNA interference showed that β-amylase activity could be decreased in cold-stored tubers by silencing of StBAM1 and collective silencing of StBAM1 and StBAM9. However, StBAM9 silencing did not decrease β-amylase activity. Silencing StBAM1 and StBAM9 caused starch accumulation and lower RS, which was more evident in simultaneously silenced lines, suggesting functional redundancy. Soluble starch content increased in RNAi-StBAM1 lines but decreased in RNAi-StBAM9 lines, suggesting that StBAM1 may regulate CIS by hydrolysing soluble starch and StBAM9 by directly acting on starch granules. Moreover, StBAM9 interacted with StBAM1 on the starch granules. StAmy23 silencing resulted in higher phytoglycogen and lower RS accumulation in cold-stored tubers, implying that StAmy23 regulates CIS by degrading cytosolic phytoglycogen. Our findings suggest that StAmy23, StBAM1, and StBAM9 function in potato CIS with varying levels of impact.
Introduction
Potato (Solanum tuberosum L.) is the most important non-grain food crop in the world. Significant amounts of potatoes are used for making crisps, French fries and other products. For a continuous supply of raw material, potato tubers are often stored at low temperature to reduce sprouting, water loss and pathogenesis. However, cold storage (normally less than 10 °C) often leads to accumulation of reducing sugar (RS) in tubers, which is known as cold-induced sweetening (CIS). RS reacts with α-amino acid groups of nitrogenous compounds during frying resulting in a Maillard browning of the products accompanied by harmful acrylamide accumulation (Tareke et al., 2002; Shepherd et al., 2010). Acrylamide formation correlated well with the concentrations of RS (Amrein et al., 2003), and there was a positive correlation between acrylamide content of potato crisps and colour (Pedreschi et al., 2005)—the higher the acrylamide accumulated, the darker the crisp colour will be. Therefore, CIS poses a significant challenge to the potato industry and raises a worldwide food safety concern (Xin and Browse, 2000; Mottram et al., 2002; Halford et al., 2012).
Sucrose hydrolysis by invertase and starch degradation have been reported to be the main pathways involved in potato CIS (Blenkinsop et al., 2003; Bhaskar et al., 2010; Zhang et al., 2014b; Lin et al., 2015). The invertase activity is considered critical for sucrose cleavage (Bhaskar et al., 2010), and a protein complex, StvacINV1–StInvInh2B–SbSnRK1, is implicated in the regulation of the enzyme activity in cold-stored tubers (Lin et al., 2015). Nevertheless, specific amylase genes responsible for potato CIS remain to be elucidated. Starch degradation is either hydrolytic (via amylases) or phosphorolytic (via starch phosphorylases). In both cases, the semi-crystalline structures of starch granules must be solubilized. Reversible glucan phosphorylation at the granule surface is essential for complete starch breakdown (Silver et al., 2014). The semi-crystalline matrix of the starch granule surface is disrupted by phosphorylation mediated by glucan, water dikinase (GWD) and phosphoglucan, water dikinase (Baunsgaard et al., 2005; Kötting et al., 2005; Xu et al., 2016). In potato leaves, the phosphorylation level of the starch granule surface is increased during starch breakdown (Ritte et al., 2004). In potato tubers, approximately 0.5% of the glucose residues are phosphorylated, which is considered highly phosphorylated (Mikkelsen et al., 2006). The analysis of transgenic potato plants with decreased GWD expression revealed that starch degradation was largely reduced in cold stored potato tubers, which was accompanied by a reduction in RS accumulation (Lorberth et al., 1998).
The hydrolytic pathway of starch degradation involves α-amylase (AMY) and β-amylase (BAM). AMY is an endoamylolytic enzyme that specifically hydrolyses α-1,4-glucan bonds to yield various linear and branched malto-oligosaccharides. Multiple genes encode different amylase isoforms that may have different roles depending on plant tissues and species. For example, suppressing rice α-amylase I-1 resulted in increased starch accumulation in young leaves under a sugar-supplemented condition (Asatsuma et al., 2005; Kitajima et al., 2009). In contrast, in Arabidopsis, all AtAMY single, double and triple knockout mutants displayed normal starch breakdown (Yu et al., 2005; Glaring et al., 2011). A recent study revealed that two α-amylase genes (StAmy1 and StAmy23) were expressed in potato tubers, but only StAmy23 transcripts could be induced by low temperature (Zhang et al., 2014a). The cold-responsive nature of this amylase gene was also reported in apple, where the expression of Amy8, which shows the highest identity to potato Amy23, was transiently upregulated at 0.5 °C in fruit (Wegrzyn et al., 2000). These observations imply that Amy23 may function under low-temperature conditions.
BAM belongs to the glycosyl hydrolase 14 family and hydrolyses α-1,4-linked glucan chains from the non-reducing end and catalyses the release of β-maltose (Weise et al., 2005). It is believed that the major pathway of starch degradation occurs via BAMs in Arabidopsis and other organisms (Scheidig et al., 2002; Fulton et al., 2008; Valerio et al., 2011; Monroe et al., 2014). The Arabidopsis genome codes for nine BAMs with varying functions in starch hydrolysis (Fulton et al., 2008; Li et al., 2009; Reinhold et al., 2011; Valerio et al., 2011; Monroe et al., 2014; Soyk et al., 2014). Some isoforms of BAM are considered cold-responsive. BAM3 contributes to leaf starch degradation in mesophyll cells at night and under cold stress (Kaplan and Guy, 2005; Fulton et al., 2008; Monroe et al., 2014). RNAi-BMY8 lines of Arabidopsis exhibited less maltose accumulation in response to cold stress (Kaplan and Guy, 2004, 2005). Cold-treated bam5/1 plants revealed an increase in BAM3 transcripts and reducing sugars (Monroe et al., 2014). Similar results were reported from other species. Overexpression of PtrBAM1 from Poncirus trifoliata in tobacco caused an increase in BAM activity and starch degradation, which was accompanied by a greater accumulation of maltose and soluble sugars at room temperature or under cold stress (Peng et al., 2014). The β-amylase activity and reducing sugar content increased sharply in five potato cultivars during the first week of storage at 4 °C (Cottrell et al., 1993). When the storage temperature was reduced from 20 °C to 5 or 3 °C, the β-amylase activity of potato tubers was enhanced by 4- to 5-fold over a 10-day period, together with maltose accumulation (Nielsen et al., 1997). Recent research demonstrated a substantial increase in β-amylase expression and an abundant accumulation of reducing sugars in potato tubers cooled to 3–5 °C (Wiberley-Bradford et al., 2016). These results suggest that β-amylase may play a significant role in potato CIS.
Seven BAMs (StBAM1, StBAM3, StBAM4, StBAM5, StBAM7, StBAM8, and StBAM9) were identified from the potato genome. Moreover, StBAM1 and StBAM9 showed higher transcripts in tubers than the others and were strongly induced by low temperature (Zhang et al., 2014a). Tomato Affymetrix GeneChip analysis showed significant up-regulation of StBMY7 and PCT-BMY1 in potato tubers exposed to low temperature (Bagnaresi et al., 2008). By modulating the expression of amylase inhibitor gene SbAI through potato transformation, the obvious changes in β-amylase activity resulted in a decrease in RS accumulation in cold-stored over-expressing tubers, and protein–protein interactions occurred between SbAI and StAmy23, and StBAM1 and StBAM9 (Zhang et al., 2014b). Thus, it is important to study the functional mechanism of the CIS-associated amylase genes for a systematic dissection and efficient control of potato cold-induced sweetening.
Here, we clarified the phylogeny of the α-amylase and β-amylase gene family in plants and provide the first report on the locations of α-amylase StAmy23 and β-amylases StBAM1 and StBAM9 in potato, together with the interaction between StBAM1 and StBAM9. Moreover, their roles in starch degradation and potato CIS were elucidated by individual gene silencing (StAmy23, StBAM1, and StBAM9) and collective silencing of both StBAM1 and StBAM9 in CIS-sensitive potato genotype and metabolomics analysis of the transgenic tubers.
Materials and methods
Phylogenetic analysis and alignment of plant amylases
The amylase sequences from different species were obtained from public databases (Supplementary Table S1 at JXB online). Phylogenetic analysis of glucosyl hydrolase domains of β-amylase was performed using Phylogeny.fr (Dereeper et al., 2008), and the phylogram was displayed with iTOL online software (Letunic and Bork, 2016). Exon–intron structures were analysed using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Phylogenetic analysis of α-amylase was performed by MUSCLE. Multiple alignments of the glucosyl hydrolase domains of β-amylase were performed using the ClustalX program with default settings and displayed by Jalview (http://www.jalview.org/). The crystal structure of the soybean β-amylase GmBMY1 (PDB ID 1BYB) was used for alignment analysis.
Gene cloning and subcellular localization
Based on the Potato Genome Sequencing Consortium (PGSC) database, StAmy23, StBAM1, and StBAM9 were cloned from CIS-sensitive cultivar E-potato 3 (E3) cDNA with specific primers shown in Supplementary Table S2. To analyse the locations of StAmy23, StBAM1, and StBAM9, the N-terminal regions of three proteins were analysed for the presence of possible chloroplast transit peptides or signal peptides using ChloroP (http://www.cbs.dtu.dk/services/ChloroP/), TargetP (http://www.cbs.dtu.dk/services/TargetP/) and iPSORT (http://ipsort.hgc.jp/index.html). The open reading frames (ORFs) of these genes without a termination codon were amplified with specific primers modified to contain the Gateway (Invitrogen) attB recombination sites. PCR products were purified and recombined into pDONR221 (Invitrogen) to generate entry clones via BP reactions. C-terminal green fluorescent protein (GFP) fusions of StAmy23–GFP, StBAM1–GFP and StBAM9–GFP were performed by recombining the entry clones with pK7FWG2 driven by the 35S silencing promoter using LR clonase (Invitrogen). For a starch granule marker, full-length granule-bound starch synthase I (StGBSS) without a stop codon was amplified from E3 cDNA with specific primers and cloned into pJCV55 obtained by restriction with BglII using Exnase II (Vazyme). Primer sequences are shown in Supplementary Table S2. Agrobacterium tumefaciens (GV3101) containing StAmy23–GFP, StBAM1–GFP, StBAM9–GFP and StGBSS–red fluorescent protein (RFP) were pressure infiltrated into the leaves of 4-week-old Nicotiana benthamiana plants. A. tumefaciens was resuspended in agroinfiltration medium at a final concentration of OD600=0.1. For coexpression, Agrobacterium cultures carrying the appropriate vectors were mixed before infiltration. Two days after infiltration, cells expressing fluorescent protein fusions were observed using a Carl Zeiss AXIO Observer A1 inverted fluorescence microscope.
Western blotting
Leaf discs (200 mg) expressing StAmy23–GFP, StBAM1–GFP, StBAM9–GFP, StGBSS–RFP, GFP and RFP were harvested at 2.5 dpi, ground in liquid nitrogen, suspended in 800 μl of protein extraction buffer (25 mM Tris–HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 10 mM DTT, 2% PVPP, 1% (v/v) protease inhibitor cocktail (Sigma-Aldrich; P0044), 0.1% Tween-20) on ice for 30 min, and centrifuged for 15 min at 2000 g at 4 °C. The supernatant was incubated by addition of 5×SDS-PAGE loading buffer, heated to 99 °C for 10 min, and analysed by 10% SDS-PAGE. Following blotting with blocking buffer (5% skimmed milk in phosphate buffered saline with Tween 20), hybridization was performed using GFP antibody (anti-GFP pAb-HRP-DirecT) (MBL; 598-7) and RFP antibody (anti-RFP pAb-HRP-DirecT) (MBL; PM005-7) diluted by the hybridization buffer (1% skimmed milk in phosphate-buffered saline) according to the manufacturer’s protocols.
Vector construction and potato transformation
To construct the StBAM1 and StBAM9 RNA interference vector, a 373-bp fragment from 1254 bp downstream of the start codon was amplified from the StBAM1 cDNA with specific primers (Supplementary Table S2, StBAM1-C1254 and StBAM1-C1626). A 281-bp fragment containing the ORF and the 3′-untranslated region was obtained from StBAM9 cDNA with specific primers (Supplementary Table S2, StBAM9-T1620 and StBAM9-T1900). The corresponding fragment was subcloned into the entry vector pDONR221 and cloned into the pHellsGate8 vector using Gateway technology (Invitrogen). To construct the RNAi vector of simultaneous repression of both StBAM1 and StBAM9, a 239-bp fragment starting from 751 bp downstream of the StBAM1 start codon was amplified with primers [Supplementary Table S2, (StBAM1+StBAM9)-1-C751 and (StBAM1+StBAM9)-1-C989], a 218-bp fragment from 976 bp downstream of StBAM9 start codon was amplified with primers [Supplementary Table S2, (StBAM1+StBAM9)-9-C976 and (StBAM1+StBAM9)-9-C1193]. The amplified PCR products were mixed and treated as templates, a chimeric fragment was obtained by overlap PCR with primers [Supplementary Table S2, (StBAM1+StBAM9)-1-C751 and (StBAM1+StBAM9)-9-C1193] and cloned into the pHellsGate8 vector. Primer sequences are shown in Supplementary Table S2. These constructs were driven by the 35S CaMV promoter, introduced into A. tumefaciens strain LBA4404 and transformed into the CIS-sensitive potato cv E3 as previously described (Si et al., 2003). Moreover, plants with RNA interference of StAmy23 in S. tuberosum L. cv. Solara have been previously obtained (Ferreira, 2011).
Plant material and sampling
The plants were grown at 18–25 °C in 24 cm-diameter plastic pots in the greenhouse (light intensity ranged from 400 to 1000 µmol m–2 s–1) at the National Centre for Vegetable Improvement (Central China), Huazhong Agricultural University (Wuhan, China). The morphology of plants was observed 8 weeks after planting. When the leaves had senesced naturally, the mature tubers were harvested, and the tuber yield per plant and starch granule size were determined. Harvested tubers were treated and sampled as described by Lin et al. (2015).
RNA isolation and quantitative RT-PCR
The frozen tuber and leaf samples were ground to a fine powder in liquid nitrogen for RNA isolation as described previously (Liu et al., 2013). Quantitative RT-PCR was performed with the Bio-Rad CFX ConnectTM Real-Time System (Bio-Rad, USA). Potato gene ef1a was used as a reference (Nicot et al., 2005), and the primers for amylase family genes were used as described before (Zhang et al., 2014a). Gene expression levels were calculated by the 2–∆∆Cq method as described by Bio-Rad (http://www.bio-rad.com/zh-cn/applications-technologies/real-time-pcr-experimental-design).
Determination of enzyme activity, starch, soluble starch, sugar, and crisp colour
For amylase extraction, 1 ml of 0.1 M citric acid buffer, pH 5.6, was added to 100 mg of powdered tuber tissue and extracted for 20 min on ice. The extract was centrifuged for 10 min at 4 °C and 2000 g, and the amylase activity in the supernatant was determined using assay kits from Megazyme (Bray, Ireland) as previously described (Zhang et al., 2014b). Protein quantification in the extraction was performed with a bicinchoninic acid kit (PPLYGEN; P1511). Starch, glucose, fructose and sucrose contents were determined as described previously (Müller-Röber et al., 1992). The soluble starch content was determined by measuring the amount of glucose released by treatment with amyloglucosidase (Smith and Zeeman, 2006). The fry test was carried out, and crisp colour was determined according to Liu et al. (2011) with modifications. Briefly, each sampled tuber was peeled and cut in half longitudinally. One part was used for the fry test; the tubers were cut into about 1 mm slices and fried at 170 °C for 3 min or until the cessation of bubbles in a Frymaster (USA) H14 electric fryer. The crisp colour was visually determined by using the Color Standards Reference Chart for Potato Chips from scale 1 (light) to 10 (dark) (Snack Food Association, USA). The crisp colour index (CCI) was calculated for each line with 15 crisps from three tubers by applying the colour scale value (C) to the formula CCI = [∑(Ci×Ni)]/N, where Ci is colour scale i, Ni is number of crisps of Ci and N is the total number of the crisps tested. For the statistical analyses, biological triplicates were used for each measurement, and all the data are presented as means±SD. Significance was determined by Student’s t test and LSD test with the software SPSS Statistics v. 20.0 for Windows.
Bimolecular fluorescence complementation
The ORFs of StBAM1 and StBAM9 without termination codon were amplified with specific primers (see Supplementary Table S2) and were separately cloned into NYFP and CYFP vector by restriction with BamHI and SalI using Exnase II (Vazyme). Transient expression by agroinfiltration and fluorescence detection were performed as described above (Gene cloning and subcellular localization).
Yeast two-hybrid assays
For vector construction, the full-length StBAM1, StBAM9, StGBSS, StGWD, StLSF1 StLSF2, and StBAM9 without transit peptides were amplified with specific primers (see Supplementary Table S2) and separately cloned into a pGBKT7 vector by restriction with EcoRI and SalI using Exnase II (Vazyme). The vectors pGADT7-StAmy23, pGADT7-StBAM1 and pGADT7-StBAM9 were obtained previously (Zhang et al., 2014b). Afterwards, pairwise vectors between pGADT7-StAmy23, pGADT7-StBAM1, pGADT7-StBAM9 and pGBKT7-StBAM1, pGBKT7-StBAM9, pGBKT7-StGBSS, pGBKT7-StGWD, pGBKT7-StLSF1, and pGBKT7-StLSF2 were transformed into yeast strain AH109 with BD Matchmaker Screening Kit according to the manufacturer’s protocols. A positive control was used as previously described (Lin et al., 2013).
Results
Multiple genes encode different amylase isoforms in potato
Together with Gene ID, alternative name, chloroplast transit peptide or signal peptide information and confirmed localization, seven β-amylases (StBAM1, StBAM3, StBAM4, StBAM5, StBAM7, StBAM8, and StBAM9) and two α-amylases (StAmy1 and StAmy23) were identified in the potato genome sequence consortium database (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml) and listed in Table 1.
Table 1.
General information for potato α-amylases and β-amylases
| Gene name | PGSC namea | Alternative name | No. of amino acids | ChloroP/TargetP | TargetP | Demonstrated localization |
|---|---|---|---|---|---|---|
| (chloroplast transit peptide) | (signal peptide) | |||||
| StBAM1 | PGSC0003DMG400001549 | StBAM1 b | 579 | Yes/Yes | Plastid stroma (this work) | |
| StBAM3 | PGSC0003DMG402020509 | StBAM2 b, PCT-BMY1c | 541 | No/No | No | Chloroplast stromac |
| StBAM4 | PGSC0003DMG400012129 | StBAM3 b | 541 | Yes/Yes | ||
| StBAM5 | PGSC0003DMG400026199 | StBAM4 b | 587 | Yes/Yes | ||
| StBAM7 | PGSC0003DMG400000169 | StBAM5 b | 554 | No/No | No | |
| StBAM8 | PGSC0003DMG400024145 | StBAM6 b | 856 | No/Yes | ||
| StBAM9 | PGSC0003DMG400010664 | StBAM7 b | 535 | Yes/Yes | Starch granule (this work) | |
| StAmy1 | PGSC0003DMG400020603 | 441 | No/No | Yes | ||
| StAmy23 | PGSC0003DMG400009891 | 407 | No/No | No | Cytoplasm (this work) |
a PGSC names are from the Potato Genome Sequence Consortium database.
b Alternative names are from Zhang et al. (2014a).
c Alternative name and demonstrated localization are from Scheidig et al. (2002).
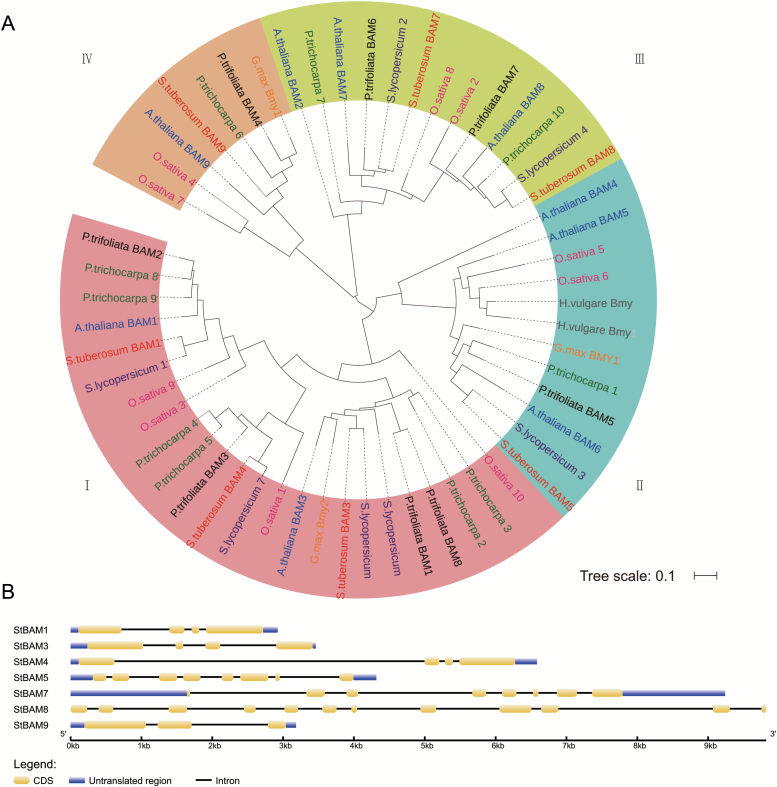
A phylogenetic analysis was performed using the conserved glucosyl hydrolase domains of 56 β-amylase proteins from potato (Solanum tuberosum), tomato (Solanum lycopersicum), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), soybean (Glycine max), barley (Hordeum vulgare), trifoliate orange (Poncirus trifoliate), and poplar (Populus trichocarpa) (see Supplementary Table S1), suggesting that β-amylases may be evolutionarily conserved among different species. The analysis displayed four major subfamilies (Fig. 1A). Potato β-amylases StBAM1, StBAM3, and StBAM4 were assigned to subfamily I, StBAM5 to subfamily II, StBAM7 and StBAM8 to subfamily III, while StBAM9 was clustered in subfamily IV. In most cases, the β-amylases, such as BAM1, BAM3, BAM4, BAM5, BAM7, and BAM8 from Solanaceae (potato and tomato) were similar. However, no β-amylase from subfamily IV was found in tomato. Moreover, the intron/exon structures of the StBAM gene family were consistent with the phylogenetic analysis. StBAMs in the same subfamily possessed similar gene structures except for subfamily III, in which StBAM7 and StBAM8 differed in the structure of exons at the 5′ and 3′ ends (Fig. 1B). The structure of StBAM9, which had only three exons, was different from other StBAM genes, suggesting a distinct function.
Fig. 1.
Phylogenetic analysis of β-amylases and gene structures of StBAM1–9. (A) A phylogram of 56 β-amylases from eight plant species. The glucosyl hydrolase domains of 56 β-amylases were aligned by MUSCLE and were used to construct a maximum-likelihood tree. The robustness of the tree is derived from 100 bootstrap replicates. The scale bar refers to 0.1 amino acid substitution per site. The phylogram displayed with Tree Of Life software (Letunic and Bork, 2016) showed that plant β-amylases were clustered into four subfamilies. Potato proteins are shown in red, tomato proteins in purple, Arabidopsis proteins in blue, rice proteins in rose red, P. trifoliate proteins in black, poplar proteins in dark green, soybean proteins in orange, and barley proteins in grey. (B) Gene structures for potato StBAMs based on the number and position of CDS (yellow), intron (solid lines) and untranslated region (blue).
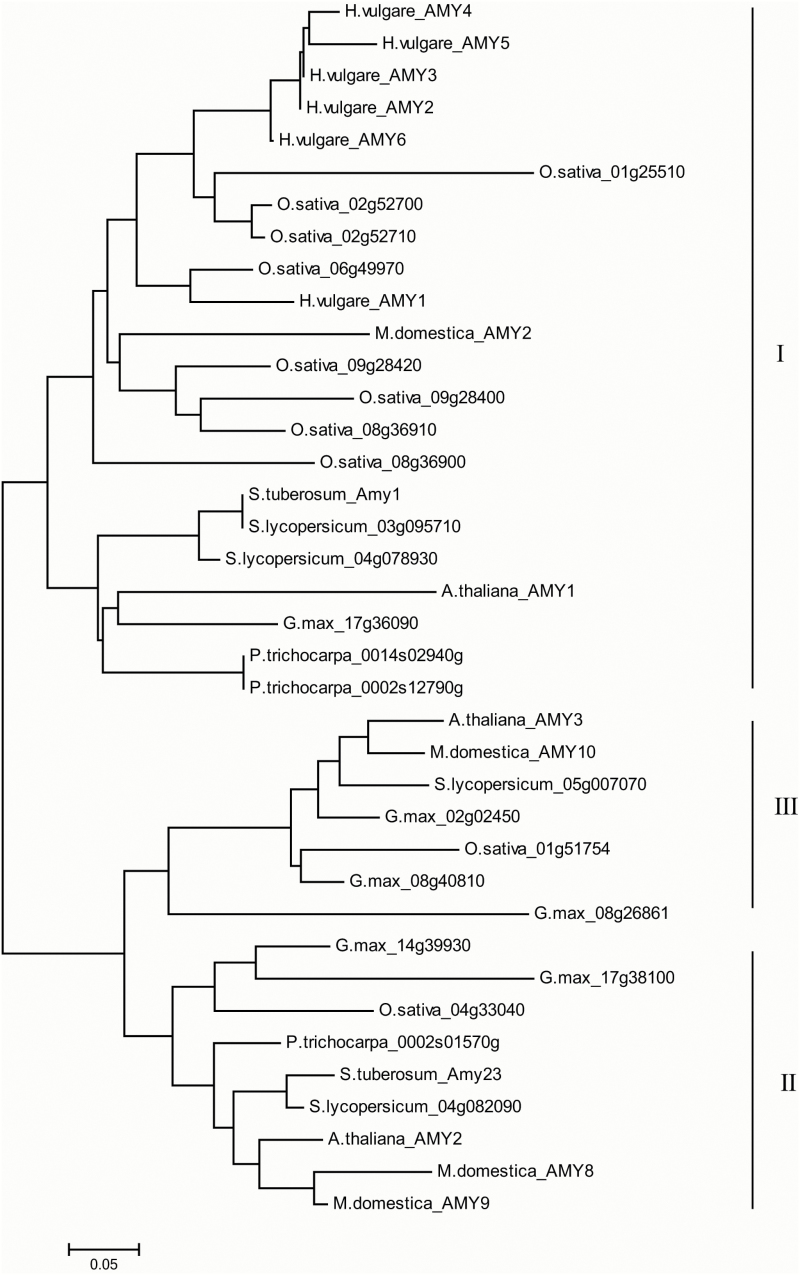
We also performed a phylogenetic analysis of 38 plant α-amylases from potato, tomato, Arabidopsis, rice, soybean, barley, poplar, and apple (see Supplementary Table S1). As shown in Fig. 2, the AMYs were classified into three major subfamilies, represented by AtAMY1, AtAMY2, and AtAMY3. StAmy1 was clustered into subfamily I, which was obviously divided into monocot and dicot proteins except for MdAMY2 from apple. Analysis using iPSORT indicated that StAmy1 possessed a signal peptide (Table 1) and may be targeted to a secretory pathway by its function. StAmy23 belonged to the subfamily II and was most closely related to AtAMY2 from Arabidopsis.
Fig. 2.
The phylogenetic relations of 38 α-amylases from eight plant species. Thirty-eight α-amylases were aligned by MUSCLE with default settings and used to generate a neighbour-joining tree. The scale bar refers to amino acid substitution per site. The phylogram showed that plant α-amylases were clustered into three subfamilies. α-Amylases of tomato, rice, poplar, and soybean were named in terms of the corresponding locus names, and the names of α-amylase for potato, Arabidopsis, barley, and apple were derived from the reports.
The structure of StBAM9 predicts an inactive protein
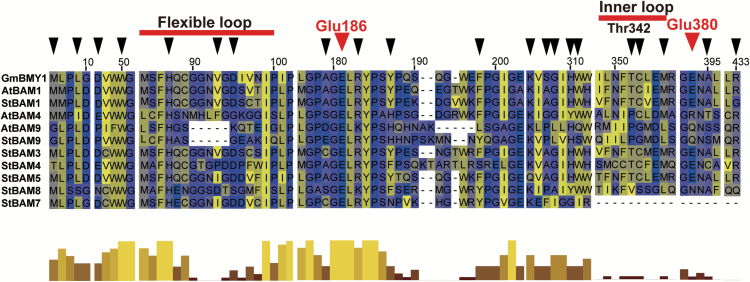
The conserved glucosyl hydrolase domains of the potato StBAM proteins, together with Arabidopsis AtBAM1, AtBAM4, and AtBAM9, were aligned with that of the soybean β-amylase GmBMY1 for which the crystal structure information was available (Mikami et al., 1994) (Fig. 3). Based on the structure of GmBMY1, 21 substrate binding residues (black arrowheads) and two active sites (red arrowheads) were identified. Most StBAMs had high sequence similarity in subfamilies I and II, but subfamilies III and IV showed remarkable variations in the flexible outer loop, inner loop and conserved residues. It was noticeable for StBAM9 that its flexible outer loop sequence had five amino acid deletions. The inner loop residue Thr-342 was changed to Pro, and the catalytic residue Glu-380 was replaced with Gln. The results also showed that StBAM9 was similar to AtBAM9 and AtBAM4 in the lack of conserved residues necessary for catalysis (Fulton et al., 2008). It was suggested that the movement of the flexible outer and inner loop affected substrate binding, and the two conserved Glu residues (Glu-186 and Glu-380) controlled catalysis (Mikami et al., 1994). Thus, the structure of StBAM9 suggests that it could be an inactive β-amylase.
Fig. 3.
Alignment of the conserved glucosyl hydrolase domains from potato StBAMs, soybean GmBMY1, and Arabidopsis AtBAM1, AtBAM4, and AtBAM9. Most and same colour shading indicates identical residues and conservative substitutions for each column, whereas the less conserved residues are presented as a different colour. An overall picture of sequence conservation is displayed in the bar graph, together with tall yellow bars representing high sequence conservation and short brown bars representing low sequence conservation. Substrate binding sites and two catalytic residues are marked with black and larger red arrowheads, respectively. The deletion and substitution of the flexible loop and inner loop in StBAM9, AtBAM9, and AtBAM4 were indicated with red lines.
StAmy23, StBAM1 and StBAM9 are localized in the cytoplasm, plastid stroma and starch granules, respectively
We analysed the N-terminal regions of the encoded proteins for the presence of possible chloroplast transit peptides and signal peptides using ChloroP, TargetP, and iPSORT. Both, StBAM1 and StBAM9 possessed predicted chloroplast transit peptides, whereas StAmy23 had no targeting peptide (Table 1).
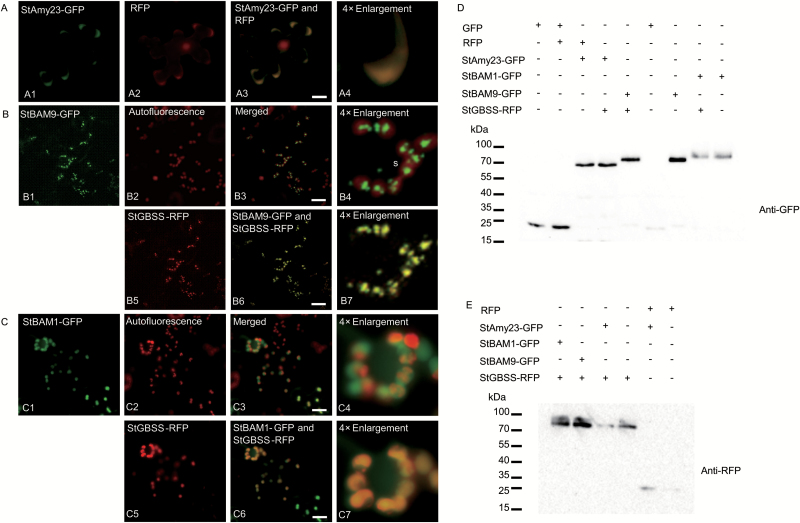
To discover whether these were chloroplast proteins, the coding regions of StAmy23, StBAM1, and StBAM9 were fused to a sequence encoding an enhanced green fluorescent protein (GFP). The constructs were then expressed transiently by infiltration of transformed Agrobacterium tumefaciens cells into Nicotiana benthamiana leaves. The construct encoding red fluorescent protein (RFP)-tagged granule-bound starch synthase I (StGBSS–RFP) was used as a positive control for starch granule localization (Szydlowski et al., 2009; Bahaji et al., 2011; Wang et al., 2013), while free GFP and RFP were used for cytosolic localization (Fulton et al., 2008). As expected, free GFP and RFP fluorescence could be observed in the cytosol and nuclei (see Supplementary Fig. S1A, B). Moreover, StGBSS–RFP was mainly localized to starch granules (restricted fluorescence on spherical or oval-shaped structures) rather than in the chloroplast stroma (diffuse fluorescence in the total chloroplast volume) (Supplementary Fig. S1C).
As shown in Fig. 4A, the fluorescence from StAmy23–GFP was completely co-localized with the cytosolic regions of free RFP (Fig. 4A1–4), suggesting that StAmy23 was exclusively located in the cytoplasm. In contrast, the non-uniform fluorescence from StBAM9–GFP (Fig. 4B1–4) and StBAM1–GFP (Fig. 4C1–4) coincided with the chlorophyll autofluorescence in most cases, indicating chloroplastic localization. However, the non-uniform distribution and restricted fluorescence rather than diffused fluorescence of StBAM9–GFP within the chloroplasts indicated that StBAM9 might be located on the surface of starch granules in the chloroplasts (Fig. 4B1–4). This speculation was confirmed by coexpression with the starch granule marker StGBSS–RFP. StBAM9–GFP showed full co-localization with StGBSS–RFP indicating that StBAM9 was localized to starch granules (Fig. 4B1, B5–7). Contrastingly, the diffuse fluorescence in the total chloroplast volume for StBAM1–GFP suggested that StBAM1 might be localized mainly in the chloroplast stroma rather than starch granules (Fig. 4C1–4). Interestingly, StBAM1–GFP could be co-localized with the diffused fraction of StGBSS–RFP fluorescence (Fig. 4C1, C5–7). The diffuse fluorescence implied that StGBSS might move to the chloroplast stroma from starch granules when coexpressed with StBAM1–GFP (Fig. 4C5), and the protein interaction may lead to the relocalization of StGBSS. Immunoblot analysis demonstrated that each GFP-fusion protein or RFP-fusion protein was intact (Fig. 4D, E), indicating that the fluorescence accurately reflects the localizations of StAmy23, StBAM1, StBAM9, and StGBSS in each case. Therefore, these results provide evidence that StAmy23 is a cytosolic enzyme, StBAM1 is localized in the plastid stroma, and StBAM9 is located on the starch granules. These results may also indicate the subcellular locations where these amylases function.
Fig. 4.
Subcellular localizations of StAmy23, StBAM1, and StBAM9 in Nicotiana benthamiana leaves. (A) StAmy23–GFP was coexpressed with cytosol marker RFP (A1–A4). (B) StBAM9–GFP was coexpressed with starch granule marker StGBSS–RFP (B1–B7). (C) StBAM1–GFP was coexpressed with StGBSS–RFP (C1–C7). The first column indicates GFP fluorescence (A1, B1, and C1), the second column indicates RFP fluorescence (A2, B5, and C5) or chlorophyll autofluorescence (B2 and C2), the third column shows merged images, and the last column shows ×4 enlargement of merged images (A4, B4, B7, C4 and C7). Bars: 10 μm. (D, E) Western blots probed with GFP antibody (E) and RFP antibody (F) showing stable protein fusions of potato StAmy23–GFP, StBAM1–GFP, StBAM9–GFP, StGBSS–RFP, and free GFP and RFP with expected size.
StAmy23, StBAM1 and StBAM9 silencing
To explore the roles of StAmy23, StBAM1, and StBAM9 in potato CIS, the expression vectors with StBAM1 and StBAM9 silenced individually and collectively were transformed into a CIS-sensitive cultivar, E-potato 3 (E3), using Agrobacterium-mediated RNA interference. The vector construction for these RNA interference vectors is shown in Supplementary Fig. S2. To check the specificity of silencing, all of the selected interference fragments were searched using blastn performed in the potato genome database (PGSC S. tuberosum group Phureja, DM1-3 Transcripts v3.4). RNAi-StAmy23 plants from the potato cultivar Solara obtained previously (Ferreira, 2011) were also employed for the functional test. Based on the relative transcript abundance of StBAM1 and StBAM9 in all of the transgenic plants (see Supplementary Fig. S3), three transgenic lines exhibiting a lower transcript abundance compared with the untransformed control were selected from each transformation for further function analysis. All of the transgenic lines displayed normal plant morphology and tuber development relative to the corresponding control under normal greenhouse conditions (Supplementary Fig. S4).
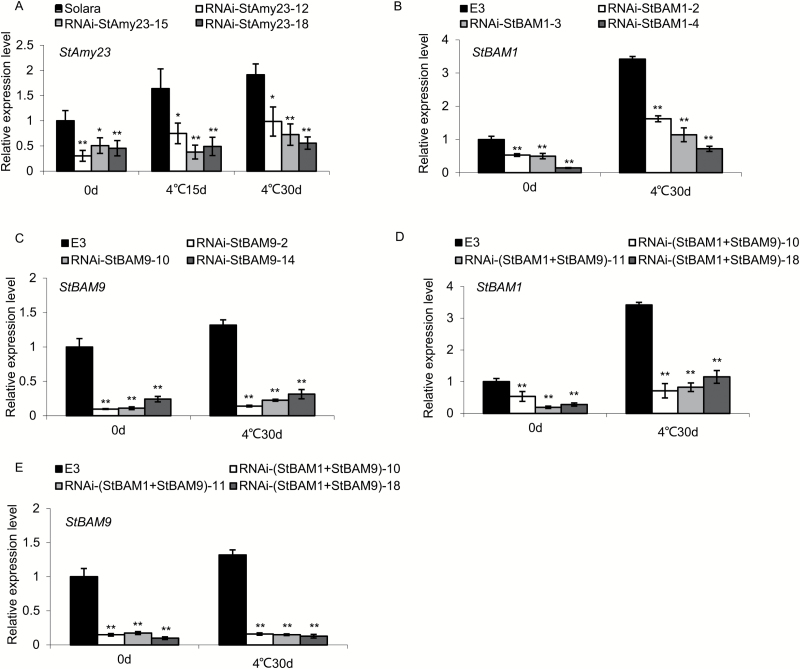
The transgenic tubers were stored at 4 °C for 0, 15 and 30 d to investigate the association between gene transcripts and sugar accumulation. Since both transgenic and untransformed control tubers had little variation in sugar content when stored at 20 °C for up to 30 d (data not shown), the tubers previously stored at 4 °C for 0 day were used for comparison. As shown in Fig. 5, the low temperature generally stimulated gene expression. Compared with the untransformed control, the transcripts of StAmy23, StBAM1, and StBAM9 were decreased in the RNAi-lines. For example, StAmy23 was suppressed by 52–73% in tubers stored at 4 °C for 30 d (Fig. 5A), StBAM1 by 57–82% (Fig. 5B), and StBAM9 by 76–89% (Fig. 5C). Obviously, the collective silence of StBAM1 and StBAM9 resulted in a dramatic decline in abundance of StBAM1 and StBAM9 mRNA showing 66–79% decline for StBAM1 (Fig. 5D) and 88–91% for StBAM9 (Fig. 5E). However, transcripts of other StBAM genes showed no remarkable decrease (see Supplementary Fig. S5), suggesting a specific silencing of the target genes in the present research.
Fig. 5.
Transcripts of StAmy23, StBAM1, and StBAM9 in transgenic tubers stored at 4 °C for 0, 15 and 30 d. (A) The relative expression of StAmy23 in RNAi-StAmy23 tubers. (B) The relative expression of StBAM1 in RNAi-StBAM1 tubers. (C) The relative expression of StBAM9 in RNAi-StBAM9 tubers. (D) The relative expression of StBAM1 in RNAi-(StBAM1+StBAM9) tubers. (E) The relative expression of StBAM9 in RNAi-(StBAM1+StBAM9) tubers. The columns represent the mean values of three biological replicates and the bars indicate the standard deviation. *P < 0.05, **P < 0.01 by Student’s t test.
StAmy23, StBAM1 and StBAM9 play different roles in potato CIS
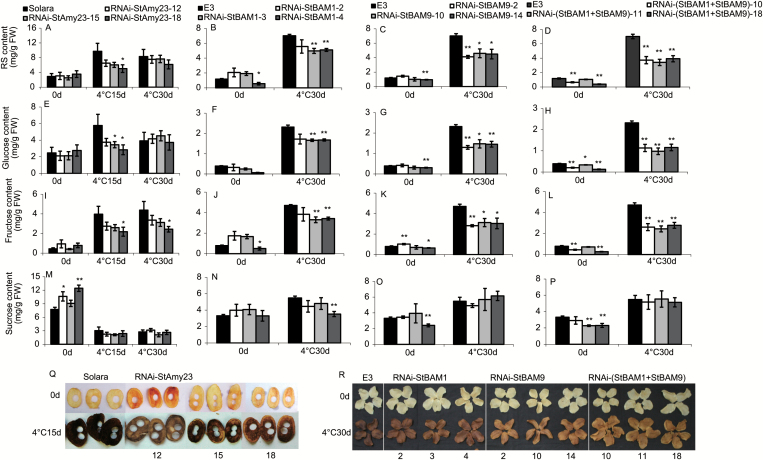
In addition to RNA transcripts, the contents of RS and sucrose were analysed in the same tubers (Fig. 6). It was obvious that the sucrose content was not affected by the transformation after cold storage (Fig. 6M–P). Looking into individual genes, silencing StAmy23 did not significantly alter RS, glucose and fructose, although RNAi-StAmy23-18 showed a significantly lower fructose content than control in the tubers stored at 4 °C for 30 d (Fig. 6I). These results revealed that StAmy23 might not be the primary factor influencing potato CIS.
Fig. 6.
The sugar content and crisp colour of RNAi-StAmy23, RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers stored at 4 °C for 0, 15 and 30 d. (A–D) Reducing sugar (RS) content. (E–H) Glucose content. (I–L) Fructose content. (M–P) Sucrose content. (Q) Colour of potato crisps from RNAi-StAmy23 tubers stored at 4 °C for 0 and 15 d. (R) Colour of potato crisps from RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers stored at 4 °C for 0 and 30 d. The columns represent the mean values of three biological replicates and the bars indicate the standard deviation. *P < 0.05, **P < 0.01 by Student’s t test.
In contrast, most of the StBAM1 (Fig. 6B, F, J) and StBAM9 (Fig. 6C, G, K) silencing lines accumulated less RS in the tubers stored at 4 °C for 30 d (in total and in glucose and fructose fractions) than in the control (E3). The reduction caused by single gene suppression was enhanced by collective suppression of both StBAM1 and StBAM9 (Fig. 6D, H, L; Supplementary Fig. S6). The changes in RS were also reflected by the colour of fried crisps from each of the transgenic tubers. RNAi-StBAM1 and RNAi-StBAM9 tubers exhibited visibly lighter crisp colour than the control (E3) after cold storage. A more obvious improvement was observed for RNAi-(StBAM1+StBAM9) tubers, while significant differences were not observed for RNAi-StAmy23 tubers (Fig. 6Q, R). Meanwhile, the crisp colour was visually determined using the Color Standards Reference Chart for Potato Chips. In comparison with the corresponding control, the crisp colour index decreased one to three levels in transgenic tubers (see Supplementary Fig. S7). Together, the results indicate that StBAM1 and StBAM9 are the main contributors to potato CIS. Moreover, we also found that silencing StBAM9 resulted in a lower RS and lighter crisp colour compared with StAmy23 or StBAM1 silenced lines (Supplementary Figs S6 and S7), indicating that StBAM9 may play a more crucial role in potato CIS.
StBAM1 and StBAM9 repression impair starch degradation in potato tubers
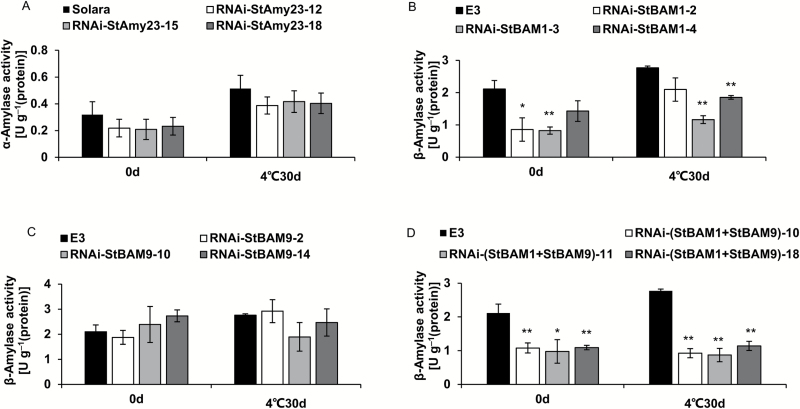
To understand how StAmy23, StBAM1, and StBAM9 function in potato CIS, the amylase activities and starch contents of tubers were measured before and after cold storage. The α-amylase activity showed no significant difference in RNAi-StAmy23 tubers (Fig. 7A), but total β-amylase activity was noticeably reduced in RNAi-StBAM1 and RNAi-(StBAM1+StBAM9) tubers and remained unchanged in RNAi-StBAM9 tubers (Fig. 7B–D). In accordance with structure analysis (Fig. 3), silencing StBAM9 did not impact the total β-amylase activity, supporting the functional prediction that StBAM9 is an inactive amylase.
Fig. 7.
The amylase activity of RNAi-StAmy23, RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers stored at 4 °C for 0 and 30 d. (A) α-Amylase activity of RNAi-StAmy23 tubers. (B) β-Amylase activity of RNAi-StBAM1 tubers. (C) β-Amylase activity of RNAi-StBAM9 tubers. (D) β-Amylase activity of RNAi-(StBAM1+StBAM9) tubers. The columns represent the mean values of three biological replicates and the bars indicate the standard deviation. *P < 0.05, **P < 0.01 by Student’s t test.
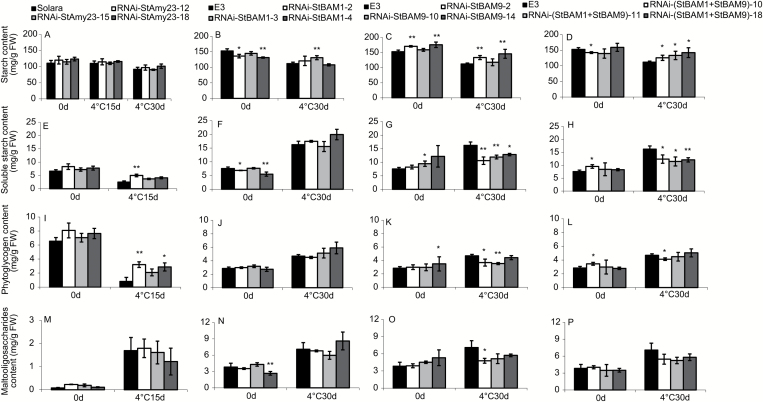
Moreover, compared with the untransformed control, the starch content showed no significant change in RNAi-StAmy23 tubers (Fig. 8A). However, the starch content of RNAi-StBAM1 tubers exhibited a remarkable increase in one of the three RNAi lines after cold storage (Fig. 8B). The starch content was elevated in RNAi-StBAM9 tubers before and after cold storage (Fig. 8C). There was no obvious difference in starch content in RNAi-(StBAM1+StBAM9) tubers before cold storage, but it was significantly higher than control (E3) after storage (Fig. 8D), indicating that both StBAM1 and StBAM9 are essential for starch degradation in potato tubers. Furthermore, their synergistic roles may be an underlying mechanism of potato CIS via the starch degradation pathway. Since StBAM9 has no catalytic activity, it may function in a way different from StBAM1.
Fig. 8.
The starch content of RNAi-StAmy23, RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers stored at 4 °C for 0, 15 and 30 d. (A–D) Starch content. (E–H) Soluble starch content. (I–L) Phytoglycogen content. (M–P) Malto-oligosaccharide content. The columns represent the mean values of three biological replicates and the bars indicate the standard deviation. *P < 0.05, **P < 0.01 by Student’s t test.
StAmy23 and StBAM1 preferentially operate on soluble starch, but StBAM9 acts on starch granules
To clarify the roles of these three amylases in starch breakdown, we measured the soluble starch and its fractions, phytoglycogen and low-molecular-mass malto-oligosaccharide (Fig. 8). Silencing StAmy23 mainly resulted in an increase in soluble phytoglycogen in cold stored tubers, but there was no obvious change in malto-oligosaccharide (Fig. 8I, M). Although the soluble starch and its fractions showed an increasing trend in StBAM1-silenced tubers after cold storage, the increase was not significant (Fig. 8F, J, N). In contrast, the soluble starch content of RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers dramatically decreased after cold storage (Fig. 8G, H), and both phytoglycogen and malto-oligosaccharide also declined (Fig. 8K, L, O, P). These observations are in accordance with the subcellular locations of StAmy23, StBAM1, and StBAM9 (Fig. 4), suggesting that they may play distinct roles in starch degradation, particularly under cold conditions. StAmy23 may act on soluble phytoglycogen that is mainly deposited in cytoplasm. StBAM1 may operate on soluble starch accumulated in the amyloplast stroma, while StBAM9, together with other active enzymes, may be involved in the solubilization of starch granules, which might be a prerequisite for the subsequent degradation of the soluble starch by StAmy23, StBAM1 or other enzymes associated with starch degradation.
StBAM9 and StBAM1 are shown to interact on the starch granules
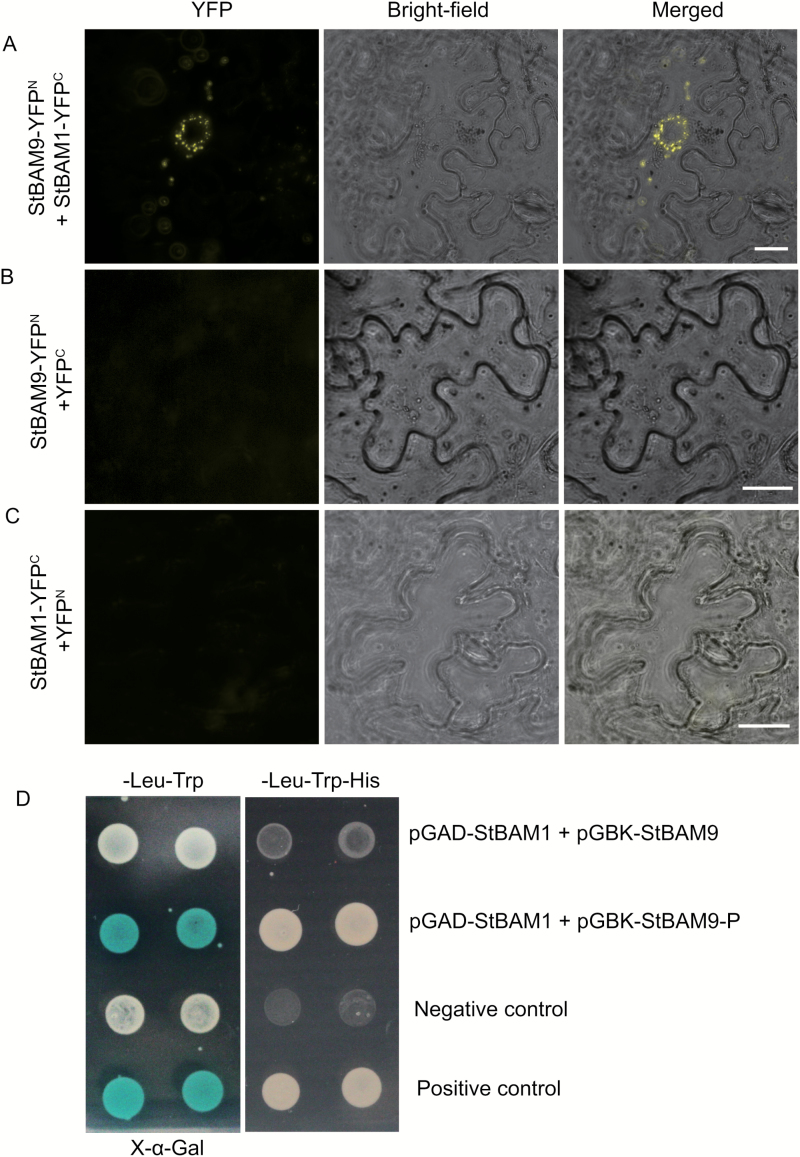
The interaction between StBAM9 and StBAM1 was accomplished using a bimolecular fluorescence complementation (BiFC) assay in which StBAM9-YFPN (StBAM9 fused with the N-terminal half of yellow fluorescent protein (YFP)) was coexpressed with StBAM1-YFPC (StBAM1 fused with the C-terminal half of YFP) in N. benthamiana. We observed the non-uniform distribution and restricted fluorescence pattern with StBAM9–YFPN and StBAM1–YFPC (Fig. 9A), but not with StBAM9–YFPN and YFPC (Fig. 9B), or StBAM1–YFPC and YFPN (Fig. 9C), suggesting that StBAM9 interacts with StBAM1 on the starch granules. The interaction was further confirmed by Gal4-based yeast two-hybrid (Y2H) assay and X-α-Gal assay (Fig. 9D). Interestingly, only StBAM9 without the transit peptide (1–63 aa) interacted with StBAM1, but not full-length StBAM9 in yeast.
Fig. 9.
The interaction between StBAM9 and StBAM1. (A–C) StBAM9 and StBAM1 interact on the starch granules in a BiFC assay. (A) Transient coexpression of StBAM9–YFPN and StBAM1–YFPC in N. benthamiana. (B) As a negative control, StBAM1–YFPC and YFPN were coexpressed. (C) StBAM9–YFPN and YFPC were coexpressed. Bars: 10 μm. (D) StBAM9 and StBAM1 interact in a yeast two-hybrid assay. Yeast coexpressing StBAM9 and StBAM9-P (StBAM9 without transit peptide) with StBAM1 grows on the double dropout (–Leu–Trp) medium by addition of X-α-Gal and triple dropout (–Leu–Trp–His) medium, respectively. The colour of the clones on SD/–Leu–Trp/X-a-Gal and the survival on SD/–Leu–Trp–His selecting plates represent the interaction.
Discussion
The potato genome encodes seven BAM and two AMY proteins relative to nine BAMs and three AMYs in Arabidopsis (Table 1). Consistent with the categories of Arabidopsis, trifoliate orange or apple (Stanley et al., 2002; Fulton et al., 2008; Peng et al., 2014), StBAMs (Fig. 1A) and StAMYs (Fig. 2) were classified into four and three major subfamilies, respectively. This phylogenetic analysis suggests that BAMs and AMYs are evolutionarily conserved in higher plants. Our results demonstrated that potato α-amylase StAmy23 and β-amylases StBAM1 and StBAM9 are involved in the starch degradation in potato CIS. The functional dissection, subcellular positioning and interaction assay led to the conclusion that these amylases have varied impact on potato CIS in different ways.
In general, all of the transgenic tubers at harvest, either by single silencing of StAmy23, StBAM1, and StBAM9 or by collective silence of StBAM1 and StBAM9, showed no obvious changes in starch yield per plant and starch granule size quantified by staining with iodine and viewing under a microscope (data not shown). Moreover, no obvious and consistent changes were observed in the content of different kinds of sugars (Fig. 6) and starch (Fig. 8) before cold storage. This result suggested that the observed effects on sugar and starch metabolism in the transgenic tubers after cold storage could be directly attributed to the contribution of StAmy23, StBAM1, and StBAM9.
StAmy23 may impact on potato CIS by breaking down soluble phytoglycogen in the cytoplasm
Plant AMYs are classified into three major subfamilies, and StAmy23 belongs to subfamily II (Fig. 2) and is localized in the cytoplasm (Fig. 4A), in accordance with the results of Stanley et al. (2002). To the best of our knowledge, StAmy23 is the first cytosolic α-amylase experimentally proven in planta. The expression of StAmy23 was strongly induced by low temperature in potato tubers (Zhang et al., 2014a). RNAi silencing resulted in the lower accumulation of RS in tubers stored at 4 °C for 15 d and improved crisp colour to some extent (Fig. 6A, E, I, Q), implying that StAmy23 is involved in potato CIS. Interestedly, silencing StAmy23 yielded a higher accumulation of phytoglycogen in cold stored tubers (Fig. 8I), while the starch degradation was not obviously affected (Fig. 8A), suggesting that StAmy23 might operate on soluble phytoglycogen. Therefore, these results suggest that StAmy23 might impact potato CIS by hydrolysing soluble phytoglycogen, which is a small fraction of potato starch and is mainly deposited in the cytoplasm.
StBAM1 may play a role in the potato CIS process by hydrolysing soluble starch in the amyloplast stroma
Based on the phylogenetic analysis (Fig. 1A) and protein alignment (Fig. 3), StBAM1 is most closely related to AtBAM1 with the highest sequence similarity, implying they may have a similar function. Previous research showed that bam1 single mutants showed no changes in starch content in Arabidopsis, but the mutants lacking both BAM1 and BAM3 accumulate more starch than the mutant lacking only BAM3, suggesting that BAM1 contributes to starch degradation in the absence of BAM3 (Fulton et al., 2008). Similar results were obtained by Monroe et al. (2014). Moreover, mutants lacking BAM1 accumulate more starch in leaf guard cell chloroplasts during the day in Arabidopsis (Valerio et al., 2011).
Our results demonstrated that StBAM1 repression decreased the total β-amylase activity in cold-stored tubers (Fig. 7B), accompanied by an increase in the starch content (Fig. 8B). As a consequence, the RS content was remarkably reduced (Fig. 6B) and the crisp colour was improved to a certain extent (Fig. 6R). Higher starch accumulation could be a result of a slight accumulation of soluble starch (Fig. 8F), suggesting that StBAM1 may directly break down the soluble starch to regulate potato CIS. This finding is supported by its plastid stromal localization (Fig. 4C) where the soluble starch was deposited. Therefore, it is reasonable to conclude that StBAM1 is the main β-amylase in starch breakdown and plays a major role in potato CIS by hydrolysing soluble starch in the amyloplast stroma.
StBAM9 plays vital and distinct roles in the starch degradation pathway of potato CIS by acting on starch granules
In the present study, StBAM9 was experimentally confirmed to be located on the starch granules (Fig. 4B). To our knowledge, this is the first report providing evidence for the existence of starch granule-localized β-amylase in potato. Suppressing StBAM9 resulted in an enhanced starch accumulation (Fig. 8C), together with the apparently decreased RS (Fig. 6C, G, K) and obviously lighter crisp colour (Fig. 6R), the reduction of RS content was more remarkable than silencing StAmy23 or StBAM1. These results indicate a dominant function of StBAM9 in potato CIS. In contrast to suppression of StAmy23 and StBAM1, StBAM9 repression resulted in a clear decline in soluble starch accumulation (Fig. 8G). Therefore, considering the association of StBAM9 with starch granules, we speculate that StBAM9 might contribute to the release of soluble glucan from the surface of starch granules. All of these findings strongly suggest that unlike StAmy23 or StBAM1, StBAM9 is the primary contributor to potato CIS by directly acting on starch granules.
StBAM9 may recruit StBAM1 to starch granule and thereby facilitate starch degradation
The gene structure of StBAM9 clustered in the most divergent subfamily IV is different from the other StBAM genes (Fig. 1), suggesting that StBAM9 may be a distinct protein in the β-amylase family. Furthermore, StBAM9 is an inactive enzyme (Fig. 3; Zhang et al., 2014b). Nevertheless, it is essential in regulating the process of starch metabolism and potato CIS on starch granules. Similarly, AtBAM9 and another β-amylase, AtBAM4, are also inactive, but AtBAM4 plays an important role in starch degradation (Fulton et al., 2008). Thus, this type of protein is proposed to participate in a potentially novel and complex pathway of starch metabolism. The functioning of StBAM9 is worth further investigation. It has been shown that sweet potato β-amylase is a tetramer of identical subunits (Cheong et al., 1995). Isoamylase isoforms ISA1 (catalytic subunits) and ISA2 (non-catalytic subunits) are associated with a multimeric enzyme and form a complex with each other in potato and Arabidopsis (Hussain et al., 2003; Delatte et al., 2005). Moreover, complexes containing class I and class II starch branching enzyme (BE) and complexes containing combinations of starch biosynthetic enzymes SSI, SSII, and class II BE exist in wheat (Tetlow et al., 2004, 2008). Therefore, we speculate that StBAM9 may facilitate starch breakdown by recruiting other proteins to form protein complexes. The paired interactions between amylases (StAmy23, StBAM1, and StBAM9) and StGWD and phosphatases StLSF1, StLSF2, and StGBSS in the Y2H system were investigated (see Supplementary Fig. S8). It identified that only StLSF2 might interact with StBAM9. LSF2 was localized in the chloroplast and specifically hydrolysed the phosphate bound to the C3-position during starch degradation in Arabidopsis (Santelia et al., 2011); complete starch degradation at the starch granule surface requires removal of phosphate (Silver et al., 2014). Therefore, we speculate that the StBAM9–StLSF2 protein complex might bind StLSF2 to the granule surface for further starch degradation. However, we found that the recombinant StBAM9 or StBAM1 had no effect on the dephosphorylation of StLSF2, and StLSF2 also did not affect the activity of StBAM9 or StBAM1 (data not shown). In addition, StBAM1 was captured by StBAM9 from the Y2H library by using StBAM9 without a transit peptide as bait (unpublished data). Moreover, the BiFC and Y2H assay showed that StBAM9 and StBAM1 interacted on the starch granules (Fig. 9); the StBAM9–StBAM1 protein complex may help bind StBAM1 to the starch granule for starch degradation. This is the first report of the existence of an interaction between β-amylases in plants, and provides a new direction for understanding starch metabolism. Furthermore, the activity of recombinant StBAM1 showed no change as the StBAM9 concentration subsequently increased in vitro where p-nitrophenyl-β-D-maltoheptaoside (the specific substrate for β-amylase) was used as the substrate (data not shown). This result was in accordance with those from the β-amylase activity of silenced StBAM9 lines (Fig. 7C). Therefore, we speculate that other proteins interacting with StBAM9 may also exist in vivo, but further work is needed to test this hypothesis.
In addition, although collective suppression of both StBAM1 and StBAM9 resulted in lower RS content than each single repression (Fig. 6D, R), the degree of improvement of CIS was lower than that of the amylase inhibitor SbAI overexpression tubers (Zhang et al., 2014b). These findings imply that functional redundancy possibly exists, and other amylase family members can compensate for their absence. Moreover, CIS may be regulated by a concerted action of multiple enzymes associated with starch degradation.
In conclusion, potato CIS is regulated by the concerted action of multiple amylases acting on different substrates in different subcellular locations. StBAM9 may release soluble glucan by directly attacking starch granules. The degradation of the soluble glucan was then completed with the participation of the amyloplast stromal StBAM1, cytosolic StAmy23 and other starch degrading enzymes. Moreover, StBAM9 and StBAM1 interact on the starch granules. To the best of our knowledge, this is the first report to demonstrate the interaction between amylases. Moreover, this study provides a novel approach to potato CIS improvement by modulating starch degradation associated with the amylases StAmy23, StBAM1, and StBAM9.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Subcellular localizations of free eGFP, RFP and starch granule marker StGBSS in Nicotiana benthamiana leaves.
Fig. S2. Constructions of RNA interference vectors.
Fig. S3. The relative expression level of StBAM1 and StBAM9 in all of the transgenic potato plants.
Fig. S4. The morphology of RNAi-StAmy23, RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) plants grown in pots.
Fig. S5. The relative expression level of other StBAM genes in RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers.
Fig. S6. The RS content of transgenic tubers containing the different constructs stored at 4 °C for 30 d.
Fig. S7. The crisp colour index of RNAi-StAmy23, RNAi-StBAM1, RNAi-StBAM9 and RNAi-(StBAM1+StBAM9) tubers after cold storage.
Fig. S8. Interactions between StAmy23, StBAM1, StBAM9 and StLSF2, StLSF1, StGWD, StGBSS proteins.
Table S1. Information for the AMYs and BAMs used to perform the phylogenetic analyses in Figs 1–3.
Table S2. Primers used in this research.
Supplementary Material
Acknowledgements
We thank Jörg Hofmann (Friedrich-Alexander-University Erlangen-Nuernberg) for assistance in the assay method of soluble starch content, Dr Lin Chen (Huazhong Agriculture University) for assistance with the methods of phylogenetic analysis. We also thank Dehuan Wang, Ying Yao, and Peng Li for their assistance with plant growth and sugar measurements, and Xiaojun Peng for assistance with protein purification. This research was supported by grants from the National Science Foundation of China (31671749 and 31401437) and the Earmarked Fund for Modern Agro-Industry Technology Research System of China (CARS-10-P06).
Glossary
Abbreviations:
- AMY
α-amylase
- BAM
β-amylase
- CIS
cold-induced sweetening;
- GWD
glucan water dikinase
- RS
reducing sugar.
References
- Amrein TM, Bachmann S, Noti A, et al. 2003. Potential of acrylamide formation, sugars, and free asparagine in potatoes: a comparison of cultivars and farming systems. Journal of Agricultural and Food Chemistry 51, 5556–5560. [DOI] [PubMed] [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T. 2005. Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant & Cell Physiology 46, 858–869. [DOI] [PubMed] [Google Scholar]
- Bagnaresi P, Moschella A, Beretta O, Vitulli F, Ranalli P, Perata P. 2008. Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahaji A, Li J, Ovecka M, et al. 2011. Arabidopsis thaliana mutants lacking ADP-glucose pyrophosphorylase accumulate starch and wild-type ADP-glucose content: further evidence for the occurrence of important sources, other than ADP-glucose pyrophosphorylase, of ADP-glucose linked to leaf starch biosynthesis. Plant & Cell Physiology 52, 1162–1176. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Lütken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. 2005. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. The Plant Journal 41, 595–605. [DOI] [PubMed] [Google Scholar]
- Bhaskar PB, Wu L, Busse JS, Whitty BR, Hamernik AJ, Jansky SH, Buell CR, Bethke PC, Jiang J. 2010. Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiology 154, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkinsop RW, Copp LJ, Yada RY, Marangoni AG. 2003. A proposed role for the anaerobic pathway during low‐temperature sweetening in tubers of Solanum tuberosum. Physiologia Plantarum 118, 206–212. [Google Scholar]
- Cheong CG, Eom SH, Chang C, et al. 1995. Crystallization, molecular replacement solution, and refinement of tetrameric β-amylase from sweet potato. Proteins 21, 105–117. [DOI] [PubMed] [Google Scholar]
- Cottrell J, Duffus C, Paterson L, Mackay G, Allison M, Bain H. 1993. The effect of storage temperature on reducing sugar concentration and the activities of three amylolytic enzymes in tubers of the cultivated potato, Solanum tuberosum L. Potato Research 36, 107–117. [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC. 2005. Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. The Plant Journal 41, 815–830. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira SJ. 2011. Transcriptome based analysis of starch metabolism in Solanum tuberosum. Dissertation, Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany. [Google Scholar]
- Fulton DC, Stettler M, Mettler T, et al. 2008. β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. The Plant Cell 20, 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaring MA, Baumann MJ, Abou Hachem M, et al. 2011. Starch-binding domains in the CBM45 family – low-affinity domains from glucan, water dikinase and α-amylase involved in plastidial starch metabolism. The FEBS Journal 278, 1175–1185. [DOI] [PubMed] [Google Scholar]
- Halford NG, Curtis TY, Muttucumaru N, Postles J, Elmore JS, Mottram DS. 2012. The acrylamide problem: a plant and agronomic science issue. Journal of Experimental Botany 63, 2841–2851. [DOI] [PubMed] [Google Scholar]
- Hussain H, Mant A, Seale R, et al. 2003. Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. The Plant Cell 15, 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. 2005. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiology 137, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. 2004. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiology 135, 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. 2005. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. The Plant Journal 44, 730–743. [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, et al. 2009. The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. The Plant Cell 21, 2844–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research 44, W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Francisco P, Zhou W, Edner C, Steup M, Ritte G, Bond CS, Smith SM. 2009. Catalytically-inactive β-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein. Archives of Biochemistry and Biophysics 489, 92–98. [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu J, Liu X, Ou Y, Li M, Zhang H, Song B, Xie C. 2013. Interaction proteins of invertase and invertase inhibitor in cold-stored potato tubers suggested a protein complex underlying post-translational regulation of invertase. Plant Physiology and Biochemistry 73, 237–244. [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu T, Liu J, et al. 2015. Subtle regulation of potato acid invertase activity by a protein complex of invertase, invertase inhibitor, and sucrose nonfermenting1-related protein kinase. Plant Physiology 168, 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng S, Liu J, Ou Y, Song B, Zhang C, Lin Y, Li X-Q, Xie C. 2013. The potato protease inhibitor gene, St-Inh, plays roles in the cold-induced sweetening of potato tubers by modulating invertase activity. Postharvest Biology and Technology 86, 265–271. [Google Scholar]
- Liu X, Zhang C, Ou Y, Lin Y, Song B, Xie C, Liu J, Li XQ. 2011. Systematic analysis of potato acid invertase genes reveals that a cold-responsive member, StvacINV1, regulates cold-induced sweetening of tubers. Molecular Genetics and Genomics 286, 109–118. [DOI] [PubMed] [Google Scholar]
- Lorberth R, Ritte G, Willmitzer L, Kossmann J. 1998. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nature Biotechnology 16, 473–477. [DOI] [PubMed] [Google Scholar]
- Mikami B, Degano M, Hehre EJ, Sacchettini JC. 1994. Crystal structures of soybean β-amylase reacted with β-maltose and maltal: active site components and their apparent roles in catalysis. Biochemistry 33, 7779–7787. [PubMed] [Google Scholar]
- Mikkelsen R, Suszkiewicz K, Blennow A. 2006. A novel type carbohydrate-binding module identified in α-glucan, water dikinases is specific for regulated plastidial starch metabolism. Biochemistry 45, 4674–4682. [DOI] [PubMed] [Google Scholar]
- Monroe JD, Storm AR, Badley EM, Lehman MD, Platt SM, Saunders LK, Schmitz JM, Torres CE. 2014. β-Amylase1 and β-amylase3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiology 166, 1748–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram DS, Wedzicha BL, Dodson AT. 2002. Food chemistry: Acrylamide is formed in the Maillard reaction. Nature 419, 448–449. [DOI] [PubMed] [Google Scholar]
- Müller-Röber B, Sonnewald U, Willmitzer L. 1992. Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. The EMBO Journal 11, 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Nielsen TH, Deiting U, Stitt M. 1997. A β-amylase in potato tubers is induced by storage at low temperature. Plant Physiology 113, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedreschi F, Moyano P, Kaack K, Granby K. 2005. Color changes and acrylamide formation in fried potato slices. Food Research International 38, 1–9. [Google Scholar]
- Peng T, Zhu X, Duan N, Liu JH. 2014. PtrBAM1, a β-amylase-coding gene of Poncirus trifoliata, is a CBF regulon member with function in cold tolerance by modulating soluble sugar levels. Plant, Cell & Environment 37, 2754–2767. [DOI] [PubMed] [Google Scholar]
- Reinhold H, Soyk S, Simková K, Hostettler C, Marafino J, Mainiero S, Vaughan CK, Monroe JD, Zeeman SC. 2011. β-Amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. The Plant Cell 23, 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Scharf A, Eckermann N, Haebel S, Steup M. 2004. Phosphorylation of transitory starch is increased during degradation. Plant Physiology 135, 2068–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Kötting O, Seung D, et al. 2011. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. The Plant Cell 23, 4096–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J. 2002. Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. The Plant Journal 30, 581–591. [DOI] [PubMed] [Google Scholar]
- Shepherd LVT, Bradshaw JE, Dale MFB, McNicol JW, Pont SDA, Mottram DS, Davies HV. 2010. Variation in acrylamide producing potential in potato: segregation of the trait in a breeding population. Food Chemistry 123, 568–573. [Google Scholar]
- Si H-J, Xie C-H, Liu J. 2003. An efficient protocol for Agrobacterium-mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agronomica Sinica 29, 801–805. [Google Scholar]
- Silver DM, Kötting O, Moorhead GB. 2014. Phosphoglucan phosphatase function sheds light on starch degradation. Trends in Plant Science 19, 471–478. [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. 2006. Quantification of starch in plant tissues. Nature Protocols 1, 1342–1345. [DOI] [PubMed] [Google Scholar]
- Soyk S, Simková K, Zürcher E, Luginbühl L, Brand LH, Vaughan CK, Wanke D, Zeeman SC. 2014. The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. The Plant Cell 26, 1746–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Fitzgerald AM, Farnden KJ, MacRae EA. 2002. Characterisation of putative α-amylases from apple (Malus domestica) and Arabidopsis thaliana. Biologia, Bratislava 57, 137–148. [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, et al. 2009. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. The Plant Cell 21, 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareke E, Rydberg P, Karlsson P, Eriksson S, Törnqvist M. 2002. Analysis of acrylamide, a carcinogen formed in heated foodstuffs. Journal of Agricultural and Food Chemistry 50, 4998–5006. [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ. 2008. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiology 146, 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ. 2004. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. The Plant Cell 16, 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, Sparla F. 2011. Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. Journal of Experimental Botany 62, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu B, Zhao J, et al. 2013. Autophagy contributes to leaf starch degradation. The Plant Cell 25, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn T, Reilly K, Cipriani G, Murphy P, Newcomb R, Gardner R, MacRae E. 2000. A novel α-amylase gene is transiently upregulated during low temperature exposure in apple fruit. European Journal of Biochemistry 267, 1313–1322. [DOI] [PubMed] [Google Scholar]
- Weise SE, Kim KS, Stewart RP, Sharkey TD. 2005. β-Maltose is the metabolically active anomer of maltose during transitory starch degradation. Plant Physiology 137, 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberley-Bradford AE, Busse JS, Bethke PC. 2016. Temperature-dependent regulation of sugar metabolism in wild-type and low-invertase transgenic chipping potatoes during and after cooling for low-temperature storage. Postharvest Biology and Technology 115, 60–71. [Google Scholar]
- Xin Z, Browse J. 2000. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell & Environment 23, 893–902. [Google Scholar]
- Xu X, Dees D, Dechesne A, Huang X-F, Visser RG, Trindade LM. 2016. Starch phosphorylation plays an important role in starch biosynthesis. Carbohydrate Polymers 157, 1628–1637. [DOI] [PubMed] [Google Scholar]
- Yu TS, Zeeman SC, Thorneycroft D, et al. 2005. α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. The Journal of Biological Chemistry 280, 9773–9779. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hou J, Liu J, Xie C, Song B. 2014a Amylase analysis in potato starch degradation during cold storage and sprouting. Potato Research 57, 47–58. [Google Scholar]
- Zhang H, Liu J, Hou J, Yao Y, Lin Y, Ou Y, Song B, Xie C. 2014b The potato amylase inhibitor gene SbAI regulates cold-induced sweetening in potato tubers by modulating amylase activity. Plant Biotechnology Journal 12, 984–993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.