Abstract
This study examined the potential pathogenicity of Shiga toxin-producing Escherichia coli (STEC) in feces of sika deer by PCR binary typing (P-BIT), using 24 selected STEC genes. A total of 31 STEC strains derived from sika deer in 6 prefectures of Japan were O-serotyped and found to be O93 (n=12), O146 (n=5), O176 (n=3), O130 (n=3), O5 (n=2), O7 (n=1), O96 (n=1), O116 (n=1), O141 (n=1), O157 (n=1) and O-untypable (n=1). Of the 31 STEC strains, 13 carried both stx1 and stx2, 5 carried only stx1, and 13 carried one or two variants of stx2. However, no Stx2 production was observed in 3 strains that carried only stx2: the other 28 strains produced the appropriate Stx. P-BIT analysis showed that the 5 O5 strains from two wild deer formed a cluster with human STEC strains, suggesting that the profiles of the presence of the 24 P-BIT genes in the deer strains were significantly similar to those in human strains. All of the other non-O157 STEC strains in this study were classified with strains from food, domestic animals and humans in another cluster. Good sanitary conditions should be used for deer meat processing to avoid STEC contamination, because STEC is prevalent in deer and deer may be a potential source of STEC causing human infections.
Keywords: deer, Shiga toxin-producing Escherichia coli (STEC), wild animal
Shiga toxin-producing Escherichia coli (STEC) causes diarrhea, hemorrhagic colitis and hemolytic uremic syndrome (HUS) in humans. STEC is prevalent in feces of ruminants, especially cattle, and therefore, ruminants are considered to be a reservoir and an important source of STEC infections in humans [4]. Although many STEC serotypes have been found in human patients, O157:H7 is the most prevalent and important serotype associated with outbreaks and sporadic cases in many countries [33]. However, non-O157 serotypes, mainly O26, O103, O111 and O145, also cause symptoms similar to those of O157:H7 in humans [29].
In Japan, the number of sika deer (Cervus nippon) has increased and their habitat has expanded during the last two decades. Therefore, herbivorous damage to agricultural products and forest vegetation by wild deer has become serious problems in many prefectures in Japan. To reduce the number of deer, some jurisdictions in Japan have supported deer hunting and deer meat is served in local restaurants and retail meat shops. Deer meat is also sold through the internet and consumed in individual homes.
Sporadic cases of STEC O157:H7 infection due to contaminated deer meat have been reported in Japan [28] and the United States [1, 35, 37], although the prevalence of this serotype was low (0.25–2.4%) in deer in the United States [9, 36]. In contrast, high prevalence of non-O157 STEC strains has been reported in deer carcasses, meat and feces [8, 25, 31, 38]. A recent study reported that genes encoding subtilase cytotoxin (subA and subB) are very common among STEC strains from deer and suggested that these strains may have more pathogenic potential for humans, although they do not carry genes encoding intimin (eae) or Shiga toxin (stx) subtypes that are associated with severe clinical outcomes [39]. However, the pathogenicity of deer STEC strains for humans remains to be elucidated.
To analyze the epidemiology of STEC infection, STEC strains have been typed by seropathotyping [22], pulsed-field gel electrophoresis (PFGE) [43], multiple locus variable-number tandem repeat analysis [19] and multilocus sequence typing [30]. These methods could be applied for analysis of the epidemiologic relationship of the strains. To evaluate the potential pathogenicity of the STEC strain, several mouse models have been developed to evaluate STEC pathogenicity [26]. Furthermore, several genetic markers which may be involved in the pathogenicity have been identified and used for the genetic assessment of STEC strains [24]. Cornelius et al. [7] originally developed a PCR binary typing (P-BIT) system and applied it to the molecular characterization of pathogenic Campylobacter jejuni. Brandt et al. [3] reported that the pathogenicity of both STEC O157 and STEC non-O157 strains could be evaluated by P-BIT analysis using 24 marker genes associated with virulence.
In the present study, we evaluated the potential pathogenicity of STEC strains derived from feces of sika deer in Japan based on serotyping, subtyping of stx genes, Shiga toxin (Stx) production, phylogenetic analysis of three DNA markers (chuA, yjaA and TspE4.C2) [6] and P-BIT analysis.
MATERIALS AND METHODS
STEC strains
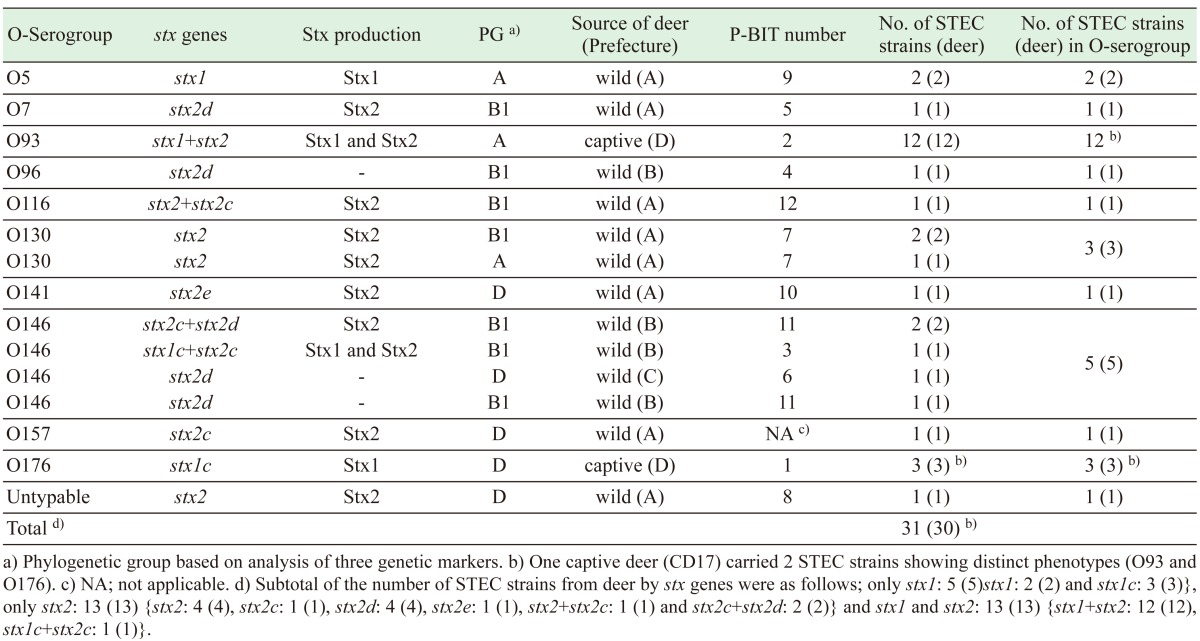
Thirty-one STEC strains had been isolated from 30 sika deer in 4 prefectures (A to D) in Japan from March 2010 to May 2013 (Table 1). The strains of STEC derived from feces of wild deer in Prefectures A (Tokai area), B (Kinki area) and C (Kitakanto/Koushin area) were obtained by using CHROMagarTM STEC plates (CHROMagar, Paris, France). The strains derived from feces of captive deer in Prefecture D (Tokai area) were obtained by using CHROMagarTM O157 plates (CHROMagar) or cefixime-tellurite-sorbitol-MacConkey agar plates (CT-SMAC) [i.e., Sorbitol MacConkey Agar (Eiken) and CT-supplement (0.05 µg/ml of cefixime and 2.5 µg/ml of tellurite; Merck, Darmstadt, Germany)]. The captive deer in Prefecture D were raised in the same facility (zoo). All the STEC strains from deer had been confirmed as oxidase production (−), indole production (+), acid production from glucose (+), fermentation of lactose/sucrose (+) and gas production from glucose (+) by biochemical analysis and were positive for stx1 and/or stx2 genes encoding Shiga toxin Stx1 and Stx2, respectively, by PCR [5]. The strains of E. coli with stx1 and/or stx2 genes were designated as STEC in this study.
Table 1. Properties of the STEC strains isolated from feces of sika deer in Japan.
Serotyping of STEC
Commercial antisera (Denka Seiken, Tokyo, Japan; and Statens Serum Institut, Copenhagen, Denmark) were used for O-serotyping. The O-serogroups of the STEC strains were determined by slide agglutination tests following the manufacturer’s instructions.
O93 was the most dominant O-serogroup (38.7%, 12/31) of the strains used in this study, followed by 5 O146 strains, 3 O130 strains, 3 O176 strains and 2 O5 strains (Table 1). There was each one strain of O7, O96, O116 and O141. In addition, a wild deer in Prefecture A was shown to be infected with an O157 strain. A strain could not be identified its O-serogroup (Untypable) in this study.
Production of Stx
Production of Stx1 and Stx2 in the culture supernatant by the STEC strains was examined by reverse passive latex agglutination tests using a VTEC-RPLA “Seiken” Kit (Denka Seiken) according to the manufacturer’s instructions.
Phylogenetic grouping
The phylogenetic groups of the STEC strains in this study were determined based on the presence (+) or absence (−) of three genetic markers (chuA, yjaA and TspE4.C2) by using multiplex PCR as described in the previous report [6].
PCR binary typing
To assess the potential pathogenicity of non-O157 STEC strains, the P-BIT typing system was used in this study as described by Brandt et al. [3]. The strains were tested for the presence or absence of 24 selected genes (ECs3737, pic, espC, iha, eae, nleB, nleC, nleG2-3, nleG, agn43EDL933, paa, ureC, ehxA, etpD, katP, espP, stx2, stx2c, lpfAO26, fyuA, cif, stx1, stx2d and iutA) by PCR. The results of these PCR experiments were analyzed using BioNumerics version 5.10 software (Applied Maths, Sint-Martens-Latam, Belgium). Inter-strain relationships were assessed by numerical analysis of the P-BIT data using a simple matching coefficient and Ward’s clustering. P-BIT data for non-O157 STEC (n=29) strains isolated in New Zealand from human and non-human sources were also included in this analysis [3].
Bayesian cluster analysis
Bayesian cluster analysis was carried out to confirm the clustering of the tested strains by Ward’s method. The analysis was done using STRUCTURE Ver. 2.3.4 software [34]. The setting options were as follows: length of burn-in period was 100,000, number of MCMC was 100,000, ancestry model was admixture model, and frequency model was allele frequency correlated. The number of iterations was 10 and the K values were 1 to 5. The number of clusters (K) was determined by the highest value of the log-likelihood and the delta K value, calculated by analyzing the results using STRUCTURE Harvester Ver. 0.6.93 software [10]. The assignment of tested strains to clusters was carried out using posterior probability values of tested strains to the clusters. When the posterior probability value (Q value) of a tested strain to a cluster was more than 0.9, the strain was assigned to the cluster [24].
RESULTS
Properties of STEC strains
Both stx1 and stx2 genes were detected in 13 (41.9%) of the 31 strains in this study, only stx1 was detected in 5 (16.1%) strains and only stx2 was detected in 13 (41.9%) strains (Table 1). All 18 STEC strains carrying stx1 gene produced Stx1, and 23 of the 26 strains carrying stx2 gene produced Stx2.
The 31 STEC strains formed three phylogenetic groups (PGs) based on analysis of three DNA markers (i.e., chuA, yjaA and TspE4.C2). The three PGs were designated A (n=15), B1 (n=9) and D (n=7) (Table 1).
P-BIT analysis of the STEC strains
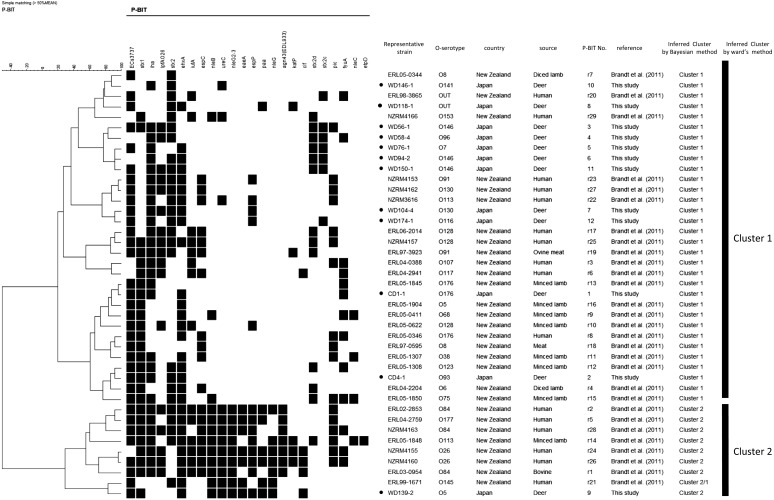
The non-O157 STEC deer strains (n=30) and 29 reference strains were typed by P-BIT analysis. The non-O157 STEC deer strains were classified into 12 types designated P-BIT 1 to P-BIT 12 (Table 1). A dendrogram of the P-BIT data using a simple matching coefficient and Ward’s clustering is shown in Fig. 1. All of the strains examined were grouped into two clusters.
Fig. 1.
Dendrogram of P-BIT analysis using a simple matching coefficient and Ward’s clustering. Deer STEC strains (n=30) and reference STEC strains (n=29) were compared based on the presence or absence of 24 target genes. The presence or absence of these genes is indicated by black and white blocks, respectively. Only the representative strain was included in this dendrogram, if several strains showed the same characteristics, including O-type and P-BIT pattern. Filled black circles in the column of “Representative strain” represent the reference STEC strains derived from deer in Japan.
Bayesian cluster analysis also showed that the highest log-likelihood and delta K values were obtained when K was assumed to be 2 (Table 2). Delta K was originally used to determine the K value when the standard deviation of the log-likelihood value was high [15], although the value in this study was quite low (Table 2). All 12 P-BIT patterns (P-BIT 1 to P-BIT 12) of the deer strains had Q values higher than 0.9 (Table 3). Based on these results, the tested strains were assigned to two clusters, designated clusters 1 and 2 (Table 3).
Table 2. Mean log-likelihood and delta K values for 1−5 clusters.
| K | Mean LnP (K) | STDEV LnP (K) | Delta K |
|---|---|---|---|
| 1 | −539.57 | 0.1947 | n/a |
| 2 | −429.72 | 0.8728 | 126.86772 |
| 3 | −430.6 | 8.5097 | 3.901415 |
| 4 | −464.68 | 21.1851 | 1.1999 |
| 5 | −524.18 | 25.4607 | n/a |
Table 3. Posterior probability values (Q values) of the STEC representative strains for the P-BIT patterns and their inferred clusters a).
| Strain No. | Q values |
Inferred cluster a) | P-BIT No. | |
|---|---|---|---|---|
| Cluster 1 | Cluster 2 | |||
| ERL03-0954 | 0.017 | 0.983 | 2 | r1 |
| ERL02-2853 | 0.007 | 0.993 | 2 | r2 |
| ERL04-0388 | 0.985 | 0.015 | 1 | r3 |
| ERL04-2204 | 0.991 | 0.009 | 1 | r4 |
| ERL04-2759 | 0.013 | 0.987 | 2 | r5 |
| ERL04-2941 | 0.916 | 0.084 | 1 | r6 |
| ERL05-0344 | 0.994 | 0.006 | 1 | r7 |
| ERL05-0346 | 0.986 | 0.014 | 1 | r8 |
| ERL05-0411 | 0.971 | 0.029 | 1 | r9 |
| ERL05-0622 | 0.97 | 0.03 | 1 | r10 |
| ERL05-1307 | 0.981 | 0.019 | 1 | r11 |
| ERL05-1308 | 0.993 | 0.007 | 1 | r12 |
| ERL05-1845 | 0.993 | 0.007 | 1 | r13 |
| ERL05-1848 | 0.032 | 0.968 | 2 | r14 |
| ERL05-1850 | 0.967 | 0.033 | 1 | r15 |
| ERL05-1904 | 0.992 | 0.008 | 1 | r16 |
| ERL06-2014 | 0.988 | 0.012 | 1 | r17 |
| ERL97-0595 | 0.99 | 0.01 | 1 | r18 |
| ERL97-3923 | 0.967 | 0.033 | 1 | r19 |
| ERL98-3865 | 0.994 | 0.006 | 1 | r20 |
| ERL99-1671 | 0.313 | 0.687 | 2/1 | r21 |
| NZRM3616 | 0.916 | 0.084 | 1 | r22 |
| NZRM4153 | 0.973 | 0.027 | 2 | r23 |
| NZRM4155 | 0.006 | 0.994 | 2 | r24 |
| NZRM4157 | 0.98 | 0.02 | 1 | r25 |
| NZRM4160 | 0.005 | 0.995 | 2 | r26 |
| NZRM4162 | 0.989 | 0.011 | 1 | r27 |
| NZRM4163 | 0.028 | 0.972 | 2 | r28 |
| NZRM4166 | 0.94 | 0.06 | 1 | r29 |
| CD1_1 | 0.991 | 0.009 | 1 | 1 |
| CD4_1 | 0.992 | 0.008 | 1 | 2 |
| WD56_1 | 0.993 | 0.007 | 1 | 3 |
| WD58_4 | 0.995 | 0.005 | 1 | 4 |
| WD76_1 | 0.994 | 0.006 | 1 | 5 |
| WD94_2 | 0.995 | 0.005 | 1 | 6 |
| WD104_4 | 0.986 | 0.014 | 1 | 7 |
| WD118_1 | 0.937 | 0.063 | 1 | 8 |
| WD139_2 | 0.073 | 0.927 | 2 | 9 |
| WD146_1 | 0.985 | 0.015 | 1 | 10 |
| WD150_1 | 0.994 | 0.006 | 1 | 11 |
| WD174_1 | 0.989 | 0.011 | 1 | 12 |
a) Clusters inferred by Bayesian cluster analysis.
Two non-O157 strains (the representative strain is WD139-2), which were derived from 2 feces of wild deer in Prefecture A, were typed as P-BIT 9. These strains formed a cluster (cluster 2) with 6 strains from human patients and 2 strains from the animals including lamb and ovine in New Zealand (Table 1 and Fig. 1). The other non-O157 strains (n=28) derived from deer were grouped in cluster 1 with 11 strains from the animals including lamb and ovine and 10 strains from patients in New Zealand. All of the deer STEC strains, and the human and other animal strains including lamb and ovine in cluster 1 did not carry the eae gene, but the 2 deer strains in P-BIT 9 tested positive for the eae gene.
Fifteen strains from captive deer in Prefecture D were typed and designated P-BIT 1 or 2, with 3 strains in P-BIT 1 and 12 strains in P-BIT 2 (Table 1).
DISCUSSION
STEC in O-serogroups, O5, O7, O93, O96, O116, O130, O141, O146, O157 and O176, derived from feces of sika deer in Japan were used in this study. Although, O146 is not the major serogroup of human STEC strains in Japan, it is a frequent STEC serotype identified in confirmed cases of STEC infection in the European Union in 2010 and 2011 [14] and has been diagnosed as a cause of HUS [41]. Furthermore, the strains of the serogroup have been isolated from both deer and human patients in Europe [25, 38]. O-serogroups of O5, O93 and O96, which was identified in the deer strains used in this study had also been found in the STEC isolates from healthy adults in Japan [27]. STEC strains of O93 and O96 were also reported to be found in deer in Japan in 1997 [2]. Further studies are needed to understand the ecology of the STEC with O-serogroups of O5, O93, O96 and O146 in Japan including the possibility of the transmission of the STEC between human and deer in Japan. In contrast, O130 and O141 were common O-serogroups in the deer strains used in this study and in deer strains isolated in Germany [11] and Spain [8], respectively. Therefore, the strains of these serogroups may unevenly distribute in deer.
STEC strains have been classified in four phylogenetic groups: A, B1, B2 and D [6, 18]. Non-pathogenic STEC and other E. coli strains tend to be grouped in phylogenetic group A, but most extra-intestinal E. coli strains are in groups B2 and D [13]. In the present study, STEC strains from Japanese sika deer were found to be in phylogenetic groups A, B1 and D. Most (9/16) strains isolated from wild deer in Japan were in phylogenetic group B1, as were the STEC strains in deer isolated in the United States [20]. Majority of the STEC strains has been known to belong to phylogenetic group B1 [13]. These results suggest that the genes involving in the development of STEC virulence were conserved in E. coli strains with specific evolutionary origins [13].
The present study is the first report using P-BIT analysis to assess the potential pathogenicity of STEC strains from deer. Brandt et al. [3] reported that PCR binary typing was a useful tool to evaluate the possible human health risk due to non-O157 STEC strains. However, the P-BIT cluster results could be affected by the number of strains analyzed. Therefore, Bayesian cluster analysis was also carried out to confirm the clustering of STEC strains by Ward’s method [23]. Both Ward’s and Bayesian cluster analysis showed that two deer strains belonging to P-BIT 9 formed a cluster (cluster 2) with 6 strains from human patients, including a sporadic HUS case [12]. These data suggested that the STEC strains from deer in cluster 2 may be potential human pathogens.
The other non-O157 strains derived from deer in this study belonging to P-BIT1–8 and 10–13 were grouped in cluster 1. Tsukamoto et al. [44] reported that deer may be a negligible source of human STEC infections, because the majority of serotypes and stx2 genotypes in non-O157 STEC strains from deer were different from those in human patients. The result that deer strains in cluster 1 do not carry the eae gene, encoding the intimin adherence protein, may support the suggestion that deer strains may not be a source of human STEC infections. However, recent studies have reported that STEC not carrying the eae gene have been isolated from human patients [21]. Deer STEC strains in 10 of the 12 P-BIT types carry the ihA gene, encoding the IrgA homologue adhesin [42]. In addition, the ehxA enterohemolysin and ureC urease genes, which have been shown to be associated with HUS caused by non-O157 STEC that do not carry the eae gene, were found in deer strains typed in P-BIT 9 and P-BIT 2, respectively [17]. Therefore, the non-O157 STEC strains in P-BIT cluster 2 need to be reassessed as potential human pathogens.
The number of sika deer has recently increased in Japan. Many deer are now found in the pastures of cattle ranches and therefore have close contact with animals known to be a major reservoir of STEC. In fact, STEC have been reported to be transmitted between various animals, including deer and domestic animals [8, 16, 25, 40]. Therefore, deer may carry pathogenic STEC acquired from cattle in ranch pastures. Furthermore, this study showed that STEC strains found in wild deer in Japan carried a variety of pathogenic genes. Since lateral gene transfer has been involved in bacterial evolution, STEC in wild deer play a role as a source of pathogenic genes for STEC and other enteric bacteria in wild deer and domestic animals [32].
Our study indicated that the STEC strains derived from deer in Japan included the strains which are potentially pathogenic to humans by a molecular analysis, P-BIT using multiple pathogenic genes. The opportunity to eat deer meat is increasing and sporadic cases of STEC infections caused by consumption of contaminated deer meat have been reported in several countries [1, 37]. Therefore, it is necessary to control STEC contamination in the processing of deer meat.
Acknowledgments
The authors appreciate Dr. Kosaku Soma and Dr. Takayoshi Masuko, Tokyo University of Agriculture for kindly providing deer feces samples for the study. This work was partially supported by a Grant-in-Aid for Scientific Research (No.26450412) and Academic frontier project from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Academic Frontier Project “Surveillance and Control for Zoonoses” from The Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Ahn C. K., Russo A. J., Howell K. R., Holt N. J., Sellenriek P. L., Rothbaum R. J., Beck A. M., Luebbering L. J., Tarr P. I.2009. Deer sausage: a newly identified vehicle of transmission of Escherichia coli O157:H7. J. Pediatr. 155: 587–589. doi: 10.1016/j.jpeds.2009.02.051 [DOI] [PubMed] [Google Scholar]
- 2.Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K.1998. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 42: 815–822. doi: 10.1111/j.1348-0421.1998.tb02356.x [DOI] [PubMed] [Google Scholar]
- 3.Brandt S. M., King N., Cornelius A. J., Premaratne A., Besser T. E., On S. L.2011. Molecular risk assessment and epidemiological typing of Shiga toxin-producing Escherichia coli by using a novel PCR binary typing system. Appl. Environ. Microbiol. 77: 2458–2470. doi: 10.1128/AEM.02322-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caprioli A., Morabito S., Brugère H., Oswald E.2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36: 289–311. doi: 10.1051/vetres:2005002 [DOI] [PubMed] [Google Scholar]
- 5.Cebula T. A., Payne W. L., Feng P.1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33: 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clermont O., Bonacorsi S., Bingen E.2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66: 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelius A. J., Gilpin B., Carter P., Nicol C., On S. L.2010. Comparison of PCR binary typing (P-BIT), a new approach to epidemiological subtyping of Campylobacter jejuni, with serotyping, pulsed-field gel electrophoresis, and multilocus sequence typing methods. Appl. Environ. Microbiol. 76: 1533–1544. doi: 10.1128/AEM.02215-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díaz-Sánchez S., Sánchez S., Herrera-León S., Porrero C., Blanco J., Dahbi G., Blanco J. E., Mora A., Mateo R., Hanning I., Vidal D.2013. Prevalence of Shiga toxin-producing Escherichia coli, Salmonella spp. and Campylobacter spp. in large game animals intended for consumption: relationship with management practices and livestock influence. Vet. Microbiol. 163: 274–281. doi: 10.1016/j.vetmic.2012.12.026 [DOI] [PubMed] [Google Scholar]
- 9.Dunn J. R., Keen J. E., Moreland D., Alex T.2004. Prevalence of Escherichia coli O157:H7 in white-tailed deer from Louisiana. J. Wildl. Dis. 40: 361–365. doi: 10.7589/0090-3558-40.2.361 [DOI] [PubMed] [Google Scholar]
- 10.Earl D. A., vonHoldt B. M.2012. Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resorces 4: 359–361. doi: 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- 11.Eggert M., Stüber E., Heurich M., Fredriksson-Ahomaa M., Burgos Y., Beutin L., Märtlbauer E.2013. Detection and characterization of Shiga toxin-producing Escherichia coli in faeces and lymphatic tissue of free-ranging deer. Epidemiol. Infect. 141: 251–259. doi: 10.1017/S0950268812000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott E. J., Robins-Browne R. M., O’Loughlin E. V., Bennett-Wood V., Bourke J., Henning P., Hogg G. G., Knight J., Powell H., Redmond D., Contributors to the Australian Paediatric Surveillance Unit. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85: 125–131. doi: 10.1136/adc.85.2.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escobar-Páramo P., Clermont O., Blanc-Potard A. B., Bui H., Le Bouguénec C., Denamur E.2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21: 1085–1094. doi: 10.1093/molbev/msh118 [DOI] [PubMed] [Google Scholar]
- 14.European Food Safety Authority. 2007. Scientific opinion of the panel on biological hazards on a request from EFSA on monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic types. EFSA Journal 579: 1–61. [Google Scholar]
- 15.Evanno G., Regnaut S., Goudet J.2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 16.Fischer J. R., Zhao T., Doyle M. P., Goldberg M. R., Brown C. A., Sewell C. T., Kavanaugh D. M., Bauman C. D.2001. Experimental and field studies of Escherichia coli O157:H7 in white-tailed deer. Appl. Environ. Microbiol. 67: 1218–1224. doi: 10.1128/AEM.67.3.1218-1224.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz E., van Hoek A. H., Wuite M., van der Wal F. J., de Boer A. G., Bouw E. I., Aarts H. J.2015. Molecular hazard identification of non-O157 Shiga toxin-producing Escherichia coli (STEC). PLoS One 10: e0120353. doi: 10.1371/journal.pone.0120353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girardeau J. P., Dalmasso A., Bertin Y., Ducrot C., Bord S., Livrelli V., Vernozy-Rozand C., Martin C.2005. Association of virulence genotype with phylogenetic background in comparison to different seropathotypes of Shiga toxin-producing Escherichia coli isolates. J. Clin. Microbiol. 43: 6098–6107. doi: 10.1128/JCM.43.12.6098-6107.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyytiä-Trees E., Smole S. C., Fields P. A., Swaminathan B., Ribot E. M.2006. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3: 118–131. doi: 10.1089/fpd.2006.3.118 [DOI] [PubMed] [Google Scholar]
- 20.Ishii S., Meyer K. P., Sadowsky M. J.2007. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl. Environ. Microbiol. 73: 5703–5710. doi: 10.1128/AEM.00275-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Käppeli U., Hächler H., Giezendanner N., Beutin L., Stephan R.2011. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000−2009. Emerg. Infect. Dis. 17: 180–185. doi: 10.3201/eid1702.100909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karmali M. A., Mascarenhas M., Shen S., Ziebell K., Johnson S., Reid-Smith R., Isaac-Renton J., Clark C., Rahn K., Kaper J. B.2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41: 4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi N., Lee K., Yamazaki A., Saito S., Furukawa I., Kono T., Maeda E., Isobe J., Sugita-Konishi Y., Hara-Kudo Y.2013. Virulence gene profiles and population genetic analysis for exploration of pathogenic serogroups of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 51: 4022–4028. doi: 10.1128/JCM.01598-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manel S., Berthier P., Luikart G.2002. Detecting wildlife poaching: Identifying the origin of individuals with bayesian assignment tests and multilocus genotypes. Conserv. Biol. 16: 650–659. doi: 10.1046/j.1523-1739.2002.00576.x [DOI] [Google Scholar]
- 25.Miko A., Pries K., Haby S., Steege K., Albrecht N., Krause G., Beutin L.2009. Assessment of Shiga toxin-producing Escherichia coli isolates from wildlife meat as potential pathogens for humans. Appl. Environ. Microbiol. 75: 6462–6470. doi: 10.1128/AEM.00904-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohawk K. L., O’Brien A. D.2011. Mouse models of Escherichia coli O157:H7 infection and shiga toxin injection. J. Biomed. Biotechnol. 2011: 258185. doi: 10.1155/2011/258185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita-Ishihara T., Iyoda S., Iguchi A., Ohnishi M.2016. Secondary Shiga Toxin-Producing Escherichia coli Infection, Japan, 2010−2012. Emerg. Infect. Dis. 22: 2181–2184. doi: 10.3201/eid2212.160783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano H., Hirochi T., Fujita K., Wakamori Y., Takeshi K., Yano S.2004. Phenotypic and genotypic characterization of beta-D-glucuronidase-positive Shiga toxin-producing Escherichia coli O157:H7 isolates from deer. J. Med. Microbiol. 53: 1037–1043. doi: 10.1099/jmm.0.05381-0 [DOI] [PubMed] [Google Scholar]
- 29.Nataro J. P., Kaper J. B.1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noller A. C., McEllistrem M. C., Stine O. C., Morris J. G., Jr., Boxrud D. J., Dixon B., Harrison L. H.2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41: 675–679. doi: 10.1128/JCM.41.2.675-679.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obwegeser T., Stephan R., Hofer E., Zweifel C.2012. Shedding of foodborne pathogens and microbial carcass contamination of hunted wild ruminants. Vet. Microbiol. 159: 149–154. doi: 10.1016/j.vetmic.2012.03.031 [DOI] [PubMed] [Google Scholar]
- 32.Ogura Y., Ooka T., Asadulghani, Terajima J., Nougayrède J. P., Kurokawa K., Tashiro K., Tobe T., Nakayama K., Kuhara S., Oswald E., Watanabe H., Hayashi T.2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8: R138. doi: 10.1186/gb-2007-8-7-r138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paton J. C., Paton A. W.1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11: 450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard J. K., Stephens M., Donnelly P.2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabatsky-Ehr T., Dingman D., Marcus R., Howard R., Kinney A., Mshar P.2002. Deer meat as the source for a sporadic case of Escherichia coli O157:H7 infection, Connecticut. Emerg. Infect. Dis. 8: 525–527. doi: 10.3201/eid0805.010373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renter D. G., Sargeant J. M., Hygnstorm S. E., Hoffman J. D., Gillespie J. R.2001. Escherichia coli O157:H7 in free-ranging deer in Nebraska. J. Wildl. Dis. 37: 755–760. doi: 10.7589/0090-3558-37.4.755 [DOI] [PubMed] [Google Scholar]
- 37.Rounds J. M., Rigdon C. E., Muhl L. J., Forstner M., Danzeisen G. T., Koziol B. S., Taylor C., Shaw B. T., Short G. L., Smith K. E.2012. Non-O157 Shiga toxin-producing Escherichia coli associated with venison. Emerg. Infect. Dis. 18: 279–282. doi: 10.3201/eid1802.110855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez S., García-Sánchez A., Martínez R., Blanco J., Blanco J. E., Blanco M., Dahbi G., Mora A., Hermoso de Mendoza J., Alonso J. M., Rey J.2009. Detection and characterisation of Shiga toxin-producing Escherichia coli other than Escherichia coli O157:H7 in wild ruminants. Vet. J. 180: 384–388. doi: 10.1016/j.tvjl.2008.01.011 [DOI] [PubMed] [Google Scholar]
- 39.Sánchez S., Beristain X., Martínez R., García A., Martín C., Vidal D., Díaz-Sánchez S., Rey J., Alonso J. M., Herrera-León S.2012. Subtilase cytotoxin encoding genes are present in human, sheep and deer intimin-negative, Shiga toxin-producing Escherichia coli O128:H2. Vet. Microbiol. 159: 531–535. doi: 10.1016/j.vetmic.2012.04.036 [DOI] [PubMed] [Google Scholar]
- 40.Singh P., Sha Q., Lacher D. W., Del Valle J., Mosci R. E., Moore J. A., Scribner K. T., Manning S. D.2015. Characterization of enteropathogenic and Shiga toxin-producing Escherichia coli in cattle and deer in a shared agroecosystem. Front. Cell. Infect. Microbiol. 5: 29. doi: 10.3389/fcimb.2015.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stritt A., Tschumi S., Kottanattu L., Bucher B. S., Steinmann M., von Steiger N., Stephan R., Hächler H., Simonetti G. D.2013. Neonatal hemolytic uremic syndrome after mother-to-child transmission of a low-pathogenic stx2b harboring shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 56: 114–116. doi: 10.1093/cid/cis851 [DOI] [PubMed] [Google Scholar]
- 42.Tarr P. I., Bilge S. S., Vary J. C., Jr., Jelacic S., Habeeb R. L., Ward T. R., Baylor M. R., Besser T. E.2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68: 1400–1407. doi: 10.1128/IAI.68.3.1400-1407.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terajima J., Izumiya H., Iyoda S., Mitobe J., Miura M., Watanabe H.2006. Effectiveness of pulsed-field gel electrophoresis for the early detection of diffuse outbreaks due to Shiga toxin-producing Escherichia coli in Japan. Foodborne Pathog. Dis. 3: 68–73. doi: 10.1089/fpd.2006.3.68 [DOI] [PubMed] [Google Scholar]
- 44.Tsukamoto T., Yamasaki S., Makino S., Asakura H., Takeda Y.2002. [The serotype and Shiga toxin type of Shiga toxin-producing Escherichia coli from humans and various animals]. Kansenshogaku Zasshi 76: 167–173. doi: 10.11150/kansenshogakuzasshi1970.76.167 [DOI] [PubMed] [Google Scholar]