Abstract
Hemoplasmas belong to Mycoplasmataceae (Mollicutes: Mycoplasmatales) and are able to infect a broad range of mammalian species. We investigated prevalence of hemotropic mycoplasma species in pig farms in the region of Zhejiang by a PCR scheme using universal primers targeting 16S rRNA and RNase P RNA gene (rnpB). Representative positive samples from different farms were selected for sequencing of 16S rRNA and the 219bp rnpB gene fragments for phylogenetic analysis. Sequencing analysis of PCR products from first samples identified a novel hemoplasma species present in several pig farms in the region with highest nucleotide identity of 92% to Candidatus Mycoplasma turicensis. A duplex PCR assay was then designed for differential detection of the novel hemoplasma from Mycoplasma parvum/M. suis in field samples. Of 324 blood samples from clinically healthy pigs, 26.5% was positive for this novel hemoplasma species and 50% positive for M. suis/M. parvum, indicating that the novel hemotropic mycoplasma species were of considerably high prevalence in Zhejiang province, China.
Keywords: hemoplasma, Mycoplasma suis, Mycoplasma parvum, novel hemotropic Mycoplasma hemosuis
Hemoplasmas or hemotropic mycoplasmas belong to Mycoplasmataceae (Mollicutes: Mycoplasmatales) and are able to infect a broad range of mammalian species. The organisms lack cell wall and are not cultivable. They adhere to the surfaces of host erythrocytes and cause their deformity or even damage, leading to hemolytic anemia and icterus. Moreover, they can persist for years in latently infected animals without causing clinical symptoms [1,2,3, 6]. Early studies proposed the two hemoplasma species that could infect pigs as Eperythrozoon suis and Eperythrozoon parvum by morphological features [7]. Further analysis of the 16S rRNA and rnpB gene sequences reclassified these two species as swine hemoplasma species Mycoplasma suis and Mycoplasma parvum [7].
Here, we report a novel hemoplasma agent that caused latent infection in pigs in Zhejiang, China. The 16S rRNA-based phylogenetic analysis suggests that this agent is closely related to Candidatus Mycoplasma turicensis isolated from feline [12, 13]. Duplex PCR of pig blood samples revealed that about one fourth of the tested pigs were subclinically infected with the novel hemotropic mycoplasma species.
MATERIALS AND METHODS
Blood samples and bacterial strains
The first blood sample positive by Giemsa stained smear as a putative novel hemoplasma species was collected from a 20-day old three-breed cross (Landrace, Yorkshire and Duroc) growing piglet (ZJSX1101) with clinical symptoms of eperythrozoonosis in Shangyu county of Zhejiang province. A total of 324 blood samples were then obtained for PCR analysis from clinically healthy pigs of 13 different farms in Zhejiang, China from 2009 to 2013, including 86 two-breed cross (Landrace-Yorkshire) sows and 238 three-breed cross growing pigs at 30–120 days of age. Strains of Hemophilus parasuis, Erysipelothrix rhusiopathiae, Mycoplasma hyosynoviae, Mycoplasma hyorhinis and Toxoplasma gondii for PCR specificity tests were kindly provided by Dr. Y.C. Wang at the swine disease group, Zhejiang Academy of Agricultural Sciences, Hangzhou, China. The M. wenyonii strain was kindly provided by Dr. X. M. Song at Zhejiang Academy of Agricultural Sciences, Hangzhou, China, and that of M. ovis by Dr X. X. Wang at Henan Agricultural University, Zhengzhou, China.
DNA extraction and PCR analysis
The hemoplasma DNA templates were extracted from 400 µl of blood samples using the Blood DNA Mini Kit (Simgen Biotech. Co., Ltd., Hangzhou, China). The 16S rRNA universal primers fHf1 and rHf2 as well as rnpB universal primers 80F1 and 290R1 of the hemoplasma speces were used for respective amplification of the 16S rRNA gene and the rnpB fragments of the putative novel strain and M. suis or M. parvum [4, 5] (Table 1). The 25 µl reaction mixture contained 2.5 µl of 10x PCR buffer containing MgCl2, 100 µM of each dNTP, 250 nM of each primer, 1U Taq DNA polymerase (TaKaRa Biotech. Co., Ltd., Dalian, China) and 5 µl DNA template. The thermal program for 16s RNA consisted of 95°C for 5 min, 35 cycles of 95°C for 1 min, 50°C for 1 min and 72°C for 1.5 min, and final extension at 72°C for 10 min on the PTC-200 Thermo Cycler (MJ Research, Co., Ltd., Waltham, MA, U.S.A.). The amplification parameters for rnpB were 95°C for 10 min; 35 cycles of 95°C for 30 sec, 45°C for 30 sec, and 72°C for 30 sec and final extension at 72°C for 10 min.
Table 1. PCR primers used in this study.
| Species | Gene name | Primers | Sequence (5′–3′) | Location | Amplification Length (bp) |
|---|---|---|---|---|---|
| Haemoplasma | 16S rRNA | fHf1 rHf2 | ACGCGTCGACAGAGTTTGATCCTGGCT CGCGGATCCGCTACCTTGTTACGACTT | 1,445/1,490 | |
| Haemoplasma | rnp B | 80F1 290R1 | GAGGAAAGTCCRYGCTWGCAC TCCCYTACCRAAATTTRGGTTTCT | 219–232 | |
| Novel Haemoplasma species | 16S rRNA | cmsf2 cmsr2 | AAACTCTGATGGTACCTCCTGAATAAGTGA CCTTCGCTGGGGATGTCAAACCT | 441–972 | 532 |
| Mycoplasma suis/M. parvum | 16S rRNA | msf2 msr2 | TAAATTAAAGGAGGCTGCCGMAAGGTG TACGCCCAATAAATCCGGATAATGCTC | 199–582 | 384 |
Sequence analysis of the 16S rRNA and rnpB gene fragments
The amplified gene fragments were cloned into pMD18-T vector (TaKaRa Biotech) and transformed into E. coli DH5α. Positive clones were sequenced using the M13 forward and reverse primers with the ABI Prism BigDye Terminator Cycle Sequencing kit on the ABI-PRISM3730 DNA Analyzer at Shanghai Sangon (Sangon Biotech Co., Ltd., Shanghai, China). Phylogenetic trees were conducted on the MEGA version 5.1 [8] using the neighbor-joining method. The data sets were resampled 1,000 times to generate bootstrap percentage values.
Development of a duplex PCR assay for detection of putative novel hemoplasma species and M. suis /M. parvum in blood samples
The primer pair cmsf2/cmsr2 were designed based on the 16S rRNA sequences of the hemoplasma species, and the primers msf2/msr2 were designed from the 16S rRNA sequences of M. suis and M. parvum (Table 1). The 25 µl reaction mixture contained 2.5 µl of 10x PCR buffer containing MgCl2, 200 nM of each primer, 100 µM of each dNTP, 1 U Taq DNA polymerase and 5µl templete DNA. The cycling parameters were: 95°C for 5 min; 35 cycles of 95°C for 30 sec, 61.5°C for 30 sec and 72°C for 30 sec; and final extension at 72°C for 10 min. Specificity of the duplex PCR was tested against M. ovis, M. wenyonii, H. parasuis, E. rhusiopathiae, M. hyosynoviae, M. hyorhinis, T. gondii and healthy pig blood DNA in individual PCRs. The PCR sensitivity was tested for the lowest concentration of detectable DNA templates on the pMD18-T backbone. The concentration of the recombinant pMD18-T containing the 16S rRNA fragment was adjusted to 100 ng/µl, as determined by spectrophotometry. Ten-fold serial dilutions (1 × 10−2 to 1 × 10−9ng/µl) were then made for PCR using the primer pair msf2/msr2 or cmhf2/cmhr2, as previously described [4, 15].
Occurrence of M. suis/M. parvum and the putative novel Hemoplasma species in pigs
To investigate the prevalence of hemoplasma infections in pigs in Zhejiang province, 324 blood samples from 13 pig farms were detected by the duplex PCR assay as described above. At least one PCR product from each verified hemoplasma positive pig farm was sequenced for phylogenetic analyses.
Nucleotide sequence accession numbers
The GenBank accession numbers of partial 16S rRNA gene and rnpB gene sequences derived from this study are JX489599 to JX489604, KC907396 and KC907397.
RESULTS
Molecular characterization of the putative novel Hemoplasma species
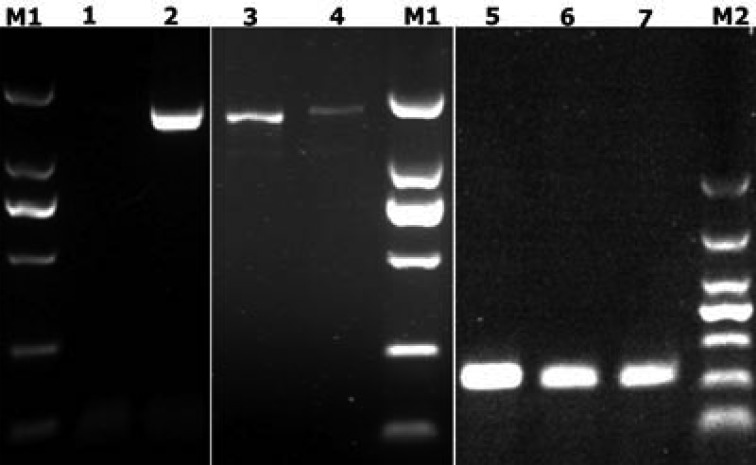
Initial microscopic examination showed that the blood smears obtained from the first infected pig ZJSX1101 had about 0.1% of red blood cells attached with hemoplasma-like organism with round and ellipse shapes (Fig. 1). The PCR products of the putative novel Hemoplasma species for both 16S rRNA and rnpB gene fragments were close to those of the M. suis and M. parvum (Fig. 2). Sequencing analysis revealed that the 16S rRNA PCR amplicon was 1,445 bp for the putative novel Hemoplasma species, while that of M. suis and M. parvum was 1,490 bp. The 1,445 bp 16S rRNA fragment of the putative hemoplasma species (accession no. JX489601) had 92% (1,295/1,401) nucleotide identity to Candidatus Mycoplasma turicensis and 81% (1,212/1,495) to M. wenyonii (Table 2). The 1,490bp fragment of the sample collected from a pig farm in Jiaxing, Zhejiang province (accession no. JX489599) had 99% (1,464/1,469) identity to M. parvum and 95% (1,402/1,470) to M. suis, while the 1,490 bp fragment of the sample collected from a pig farm in Ningbo, Zhejiang province (accession no. KC907396), had 99% (1,464/1,469) identity to M. suis and 95% (1,405/1,470) to M. parvum. These results suggest that pigs from these two farms had different infections, one with M. parvum and the other with M. suis.
Fig. 1.
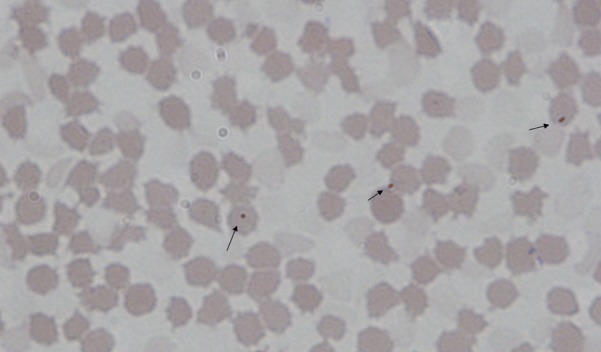
Giemas stained smear of peripheral blood from a pig infected with a putative novel hemoplasma species. Approximately 0.1% of red blood cells were infected.
Fig. 2.
Amplification of 16S rRNA and rnpB gene fragments from putative novel Hemoplasma, M. suis and M. parvum in pig blood samples. M1: DNA marker DL2000. Lanes: 1, negative control; 2, 16S rRNA of M. suis; 3, 16S rRNA of putative novel hemoplasma sp; 4, 16S rRNA of M. parvum; 5, rnpB gene of M. suis; 6, rnpB of putative novel hemoplasma sp.; 7, rnpB of M. parvum; M2 DNA Marker DL1000.
Table 2. Identity of the novel Haemoplasma species and other Haemoplasma species based on 16S rRNA and rnpB gene.
| 16s rRNA |
rnpB |
||||
|---|---|---|---|---|---|
| Species | Source/GenBank | Identity (%) | Species | Source/GenBank | Identity (%) |
| Candidatus Mycoplasma turicensis | feline/ DQ464423 | 92.0 | Candidatus M. haemohominis | Homo sapiens/ GU562825 | 81.0 |
| M. coccoides | murine/AY171918 | 91.0 | Candidatus M. turicensis | feline/ EF212003 | 76.0 |
| Candidatus Mycoplasma haemohominis | Homo sapiens/ GU562823 | 91.0 | M. parvum | pig/JX489602 | 74.0 |
| Mycoplasma sp. | buffalo/EF424082 | 89.8 | M. coccoides | murine/EU078619 | 73.0 |
| M. muris | murine/U82963 | 89.6 | M. suis | pig/KC907397 | 71.0 |
| M. haemocanis | canis/AY529641 | 88.4 | |||
| M. haemofelis | feline/EU145745 | 88.2 | |||
| M. parvum | pig/JX489599 | 82.0 | |||
| M. suis | pig/KC907396 | 82.0 | |||
| M. wenyonii | bovine/AY946266 | 81.0 | |||
The rnpB amplicon from the putative novel hemoplasma was 219 bp (accession no. JX489602), while M. suis and M. parvum had amplicons of 232 bp (accession no. KC907397) and 228 bp (accession no. JX489601), respectively. The sequenced rnpB fragment of the novel hemoplasma species was 79% similar to Candidatus M. hemohominis (Homo sapiens, Plymouth, GU562825) and 71% to M. suis (KC907397) (Table 2).
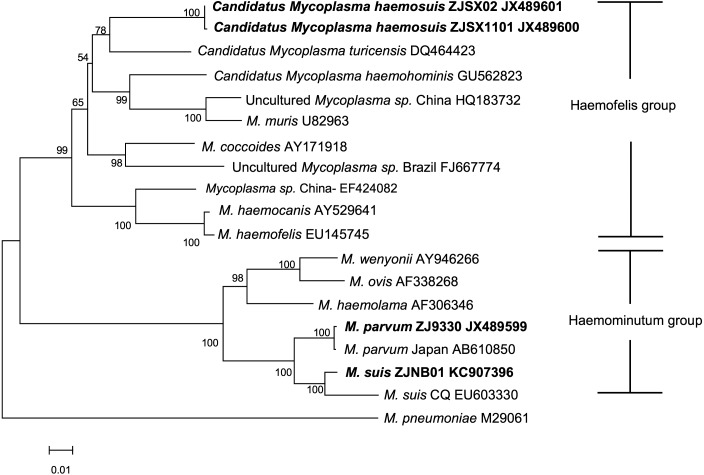
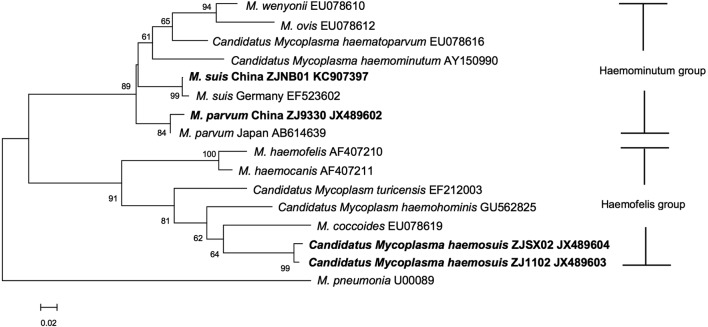
The putative novel porcine hemoplasma was phylogenetically closer to Candidatus M. turicensis, clustered together with M. hemocanis and M. hemofelis, and belonged to the hemofelis group by their 16S rRNA (Fig. 3). They apparently branched away from the hemominutum group including M. suis and M. parvum as previously described [5, 9]. Phylogenetic analysis of the rnpB gene sequences also showed results similar to the 16S rRNA (Fig. 4).
Fig. 3.
Phylogenetic tree displaying the relationship of the novel Hemoplasma strains to other relevant species based on nearly complete 16S rRNA sequences by the neighbor-joining method. The numbers at the nodes were generated from 1,000 bootstrap resamplings. Values of <50 are not shown. Evolutionary distances are to the scale shown. GenBank accession numbers are shown. The species names in bold were from this study.
Fig. 4.
Phylogenetic tree displaying the relationship of the novel Hemoplasma strains to other relevant species based on the rnpB fragments by the neighbor-joining method. The numbers at the nodes were generated from 1,000 bootstrap resamplings. Values of <50 are not shown. Evolutionary distances are to the scale shown. GenBank accession numbers are shown. The species names in bold were from this study.
Differential detection of M. parvum/M. suis and putative novel Hemoplasma species by duplex PCR
The duplex PCR assay was specific for detection of the novel porcine Hemoplasma species (532 bp), M. parvum and M. suis (384 bp) (Fig. 5). There were no amplicons from other tested bacterial species (Fig. S1). The lower detection limit was between 280 and 290 copies of the recombinant plasmids containing the 16S rRNA gene inserts (Fig. S2).
Fig. 5.
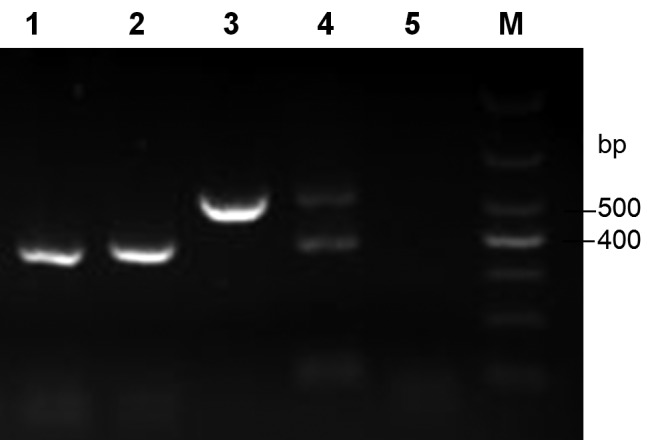
Differential detection of the novel Haemplasma species from M. suis/M. parvum by duplex PCR. M: DNA marker DL1000. Lanes: 1,384 bp product from blood infected by M. suis; 2,384 bp product from blood infected by M. parvum; 3,532 bp product from blood infected by new hemoplasma; 4, positive controls 384 bp and 532 bp of 16S rRNA inserts in pMD18-T; 5, negative control.
Occurrence of M. suis, M. parvum and the putative novel Hemoplasma species in pigs
To investigate the prevalence of the putative novel Hemoplasma species and M. suis/M. parvum in Zhejiang, blood samples from 13 pig farms were detected by PCR. Only one pig farm was negative for hemoplasma. Twelve farms had different levels of infection with hemoplasma species. Of the 324 samples tested, prevalence of the putative novel Hemoplasma species was 26.5% on average, while that of M. suis/M. parvum was 50%. Sows had higher infection rate with hemoplasma (36%) than growing pigs (24.1%). For M. suis/M. parvum, the infection rate was much higher in sows (70.9%) than in growing pigs (42.6%). The rate of mixed infections of putative novel Hemoplasma species and M. suis/M. parvum was 17.3% (Table 3).
Table 3. Frequency distribution of Haemoplasma species in the examined swine farms assessed using PCR assay.
| Organism(s) | Positive No/No. pigs tested (%) |
||
|---|---|---|---|
| Sows | Growing pigs | All pigs | |
| No. of pigs tested | 86 | 238 | 324 |
| M. suis/M. parvum | 61 (70.9) | 101 (42.4) | 162 (50) |
| M. suis/M. parvum only | 38 (44.2) | 68 (28.6) | 106 (32.7) |
| Novel Haemoplasma species | 31 (36) | 55 (23.1) | 86 (26.5) |
| Novel Haemoplasma species only | 8 (9.3) | 22 (9.2) | 30 (9.3) |
| Mixed infections with M. suis/M. parvum and novel Haemoplasma species | 23 (26.7) | 33 (13.9) | 56 (17.3) |
| Total | 69 (80.2) | 123 (51.7) | 192 (59.3) |
Of the 37 sequences representing 12 verified hemoplasma positive pig farms, 21 belonged to putative novel Hemoplasma species in samples from 12 farms, 13 M. parvum from 11 farms and 3 M. suis from 2 farms. One sample had mixed infection with all three Hemoplasma species, and one had dual infection with putative novel Hemoplasma and M. parvum. The 384 bp PCR fragments of M. suis/M. parvum had an overlapping region of about 120 bp with the 532 bp PCR fragments of the novel Hemoplasma. Phylogenic analysis of these short overlapping sequences shows that all putative novel Hemoplasma strains fell into the same group but different from M. suis and M. parvum. There appeared to be divergence also within the M. suis and M. parvum isolates from different farms in Zhejiang province.
DISCUSSION
The present study is the first report of a novel Hemoplasma species naturally infecting pigs in China. Sequence analysis of the 16S rRNA gene indicates that the novel Hemoplasma has close relationship with Candidatus Mycoplasma turicensis, a hemoplasma species isolated from feline [12, 14]. Phylogenetic tree confirmed that the new Hemoplasma species is different from M. suis and M. parvum.
We developed a duplex PCR method for differential detection of the novel Hemoplasma species and M. suis/M. parvum infections in pigs. Because there is high similarity of the 16S rRNA gene sequences between M. suis and M. parvum [11], it is difficult to design primers to differentiate M. suis from M. parvum by PCR. With the duplex PCR method, we found that infection with the novel Hemoplasma species in pigs from surveyed farms in Zhejiang was 26.5%, about half of that with M. suis/M. parvum. There were co-infections of M. suis/M. parvum and the novel Hemoplasma species that may contribute to pathogenicity in a synergistic manner. Sequencing results of 37 PCR positive samples from 12 pig farms show that M. parvum and the novel Hemoplasma species seemed to be prevalent in Zhejiang province. Sows had higher infection, possibly because of persistent exposure with increasing age [10].
In summary, we have identified the novel hemotropic mycoplasma species as a potential pathogen to pigs. The duplex PCR method could differentiate the novel Hemoplasma species from M. suis/M. parvum in blood samples. Although the overall carrier status was high with majority (nearly 60%) of surveyed pigs carrying hemoplasmal species, absence of apparent clinical manifestations suggests their low pathogenicity. They might be opportunistic pathogens. Their pathogenic potentials await further investigation, though it might be difficult because of their non-culturable nature [1].
CONFLICT OF INTEREST
There is no conflict of interest.
Supplementary
Acknowledgments
This work was supported by the Scientific and Technological Innovation Team of Zhejiang Province (2010R50027-26) and the Innovation Project of Zhejiang Academy of Agricultural Sciences, Hangzhou, China.
REFERENCES
- 1.Hoelzle L. E.2008. Haemotrophic mycoplasmas: recent advances in Mycoplasma suis. Vet. Microbiol. 130: 215–226. doi: 10.1016/j.vetmic.2007.12.023 [DOI] [PubMed] [Google Scholar]
- 2.Meli M. L., Willi B., Dreher U. M., Cattori V., Knubben-Schweizer G., Nuss K., Braun U., Lutz H., Hofmann-Lehmann R.2010. Identification, molecular characterization, and occurrence of two bovine hemoplasma species in Swiss cattle and development of real-time TaqMan quantitative PCR assays for diagnosis of bovine hemoplasma infections. J. Clin. Microbiol. 48: 3563–3568. doi: 10.1128/JCM.02224-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messick J. B.2004. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet. Clin. Pathol. 33: 2–13. doi: 10.1111/j.1939-165X.2004.tb00342.x [DOI] [PubMed] [Google Scholar]
- 4.Messick J. B., Cooper S. K., Huntley M.1999. Development and evaluation of a polymerase chain reaction assay using the 16S rRNA gene for detection of Eperythrozoon suis infection. J. Vet. Diagn. Invest. 11: 229–236. doi: 10.1177/104063879901100304 [DOI] [PubMed] [Google Scholar]
- 5.Peters I. R., Helps C. R., McAuliffe L., Neimark H., Lappin M. R., Gruffydd-Jones T. J., Day M. J., Hoelzle L. E., Willi B., Meli M., Hofmann-Lehmann R., Tasker S.2008. RNase P RNA gene (rnpB) phylogeny of Hemoplasmas and other Mycoplasma species. J. Clin. Microbiol. 46: 1873–1877. doi: 10.1128/JCM.01859-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Splitter E. J.1950. Eperythrozoon suis, the etiologic agent of ictero-anemia or an anaplasmosis-like disease in swine. Am. J. Vet. Res. 11: 324–330. [PubMed] [Google Scholar]
- 7.Splitter E. J.1950. Eperythrozoon suis n. sp. and Eperythrozoon parvum n. sp., 2 new blood parasites of swine. Science 111: 513–514. doi: 10.1126/science.111.2889.513 [DOI] [PubMed] [Google Scholar]
- 8.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tasker S., Peters I. R., Mumford A. D., Day M. J., Gruffydd-Jones T. J., Day S., Pretorius A. M., Birtles R. J., Helps C. R., Neimark H.2010. Investigation of human haemotropic Mycoplasma infections using a novel generic haemoplasma qPCR assay on blood samples and blood smears. J. Med. Microbiol. 59: 1285–1292. doi: 10.1099/jmm.0.021691-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasker S., Binns S. H., Day M. J., Gruffydd-Jones T. J., Harbour D. A., Helps C. R., Jensen W. A., Olver C. S., Lappin M. R.2003. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom. Vet. Rec. 152: 193–198. doi: 10.1136/vr.152.7.193 [DOI] [PubMed] [Google Scholar]
- 11.Watanabe Y., Fujihara M., Obara H., Nagai K., Harasawa R.2011. Two genetic clusters in swine hemoplasmas revealed by analyses of the 16S rRNA and RNase P RNA genes. J. Vet. Med. Sci. 73: 1657–1661. doi: 10.1292/jvms.11-0293 [DOI] [PubMed] [Google Scholar]
- 12.Willi B., Boretti F. S., Cattori V., Tasker S., Meli M. L., Reusch C., Lutz H., Hofmann-Lehmann R.2005. Identification, molecular characterization, and experimental transmission of a new hemoplasma isolate from a cat with hemolytic anemia in Switzerland. J. Clin. Microbiol. 43: 2581–2585. doi: 10.1128/JCM.43.6.2581-2585.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willi B., Tasker S., Boretti F. S., Doherr M. G., Cattori V., Meli M. L., Lobetti R. G., Malik R., Reusch C. E., Lutz H., Hofmann-Lehmann R.2006. Phylogenetic analysis of “Candidatus Mycoplasma turicensis” isolates from pet cats in the United Kingdom, Australia, and South Africa, with analysis of risk factors for infection. J. Clin. Microbiol. 44: 4430–4435. doi: 10.1128/JCM.00987-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willi B., Museux K., Novacco M., Schraner E. M., Wild P., Groebel K., Ziegler U., Wolf-Jäckel G. A., Kessler Y., Geret C., Tasker S., Lutz H., Hofmann-Lehmann R.2011. First morphological characterization of ‘Candidatus Mycoplasma turicensis’ using electron microscopy. Vet. Microbiol. 149: 367–373. doi: 10.1016/j.vetmic.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zachar V., Thomas R. A., Goustin A. S.1993. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 21: 2017–2018. doi: 10.1093/nar/21.8.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.