Abstract
In September and October 2015, suspected cases of bovine ephemeral fever (BEF) were reported in the mainland region of Kagoshima Prefecture and on Tanegashima Island. The genome of the BEF virus (BEFV) was detected in the diseased cows and the cows that had recovered. The serum obtained from the affected cows contained high titers of BEFV-neutralizing antibody. In total, 18 affected cows were demonstrated to be infected with BEFV during the outbreak. Our findings showed evidence that BEF occurred in mainland Japan after a 23-year absence. Phylogenetic analysis based on the surface glycoprotein (G) gene revealed that BEFVs detected in the affected cows were genetically distinct from previous Japanese BEFVs, but were close to BEFVs circulating in Taiwan and mainland China in recent years. Amino acid substitution in the neutralizing epitope domains of the G protein was limited between the detected viruses and the vaccine strain (YHL isolate), and high titers of the neutralizing antibody against the YHL isolate were induced in the infected cattle during the disease occurrences. Therefore, current BEF vaccines probably elicit protective immunity against the BEFVs detected in 2015, although their effectiveness should be assessed. Since the BEFV vaccination rates are estimated to be low, a BEF outbreak should be considered a possibility in mainland Japan.
Keywords: arbovirus, BEFV, bovine ephemeral fever, febrile illness, rhabdovirus
Bovine ephemeral fever (BEF) is an acute febrile illness in cattle and water buffalo caused by BEF virus (BEFV), which belongs to the genus Ephemerovirus in the family Rhabdoviridae [22]. The distribution of BEF ranges throughout the tropical, subtropical and temperate zones of Africa, the Middle East, Asia and Australia. BEF is characterized by sudden fever (40–42°C), anorexia, depression, rapid breathing, lacrimation, foamy salivation, arthritis, limping and ananastasia [16]. In most cases, diseased animals show mild or moderate symptoms and recover within a few days. Infected animals sometimes develop pneumonia with severe pulmonary emphysema. Abortion also occurs in a low percentage of infected pregnant cattle. In lactating cows, BEF causes the cessation of lactation and the reduction of milk production. Although the mortality rate is typically low (<1%), the economic impact of BEF is considerable due to the reduction of milk production in dairy cattle and the loss of conditions in beef cattle [22].
BEFV displays a bullet-shaped morphology and has a single-stranded, negative-sense RNA genome of 14.9 kb in size [22]. The virus particle consists of five structural proteins comprising a nucleoprotein, a polymerase-associated protein, a matrix protein, a large RNA-dependent RNA polymerase and a surface glycoprotein (G). The G protein is a class I transmembrane glycoprotein which forms surface spikes and elicits protective immunity in infected animals. Three major neutralizing epitopes (G1–G3) have been physically mapped on the G protein by competition binding assays using neutralizing monoclonal antibodies and cross-reactivity analysis of neutralization-resistant escape mutants [3, 4]. BEFV is thought to be an arthropod-borne virus (arbovirus). However, its principal vector remains unknown, although BEFV has been isolated from several species of field-collected mosquitoes and Culicoides biting midges [22].
In Japan, epidemics of BEF have occurred repeatedly since the 1950s [15]. During the largest outbreaks in 1949–1951, a total of 770,000 cases and over 10,000 deaths were reported [7]. The last occurrence of BEF in mainland Japan was recorded in 1992, and since then, the epidemics have been limited to the Yaeyama Islands and its neighboring Tarama Island, which are located at the southwestern end of Japan [1, 6, 13]. Inactivated BEF vaccines were developed on the basis of the YHL isolate that was obtained in Japan in 1966 [8] and have been used in the field.
In September 2015, a local veterinarian first identified several diseased cows with fever, foamy salivation and ananastasia in the southwestern part of the mainland region of Kagoshima Prefecture. In the following month, other cases with similar symptoms were reported on Tanegashima Island, which is located approximately 40 km south of the mainland region. On receiving the results of recent BEF outbreaks in southwestern end of Japan [6, 9, 13], we suspected that these cattle suffered from BEF based on their clinical symptoms. Herein, the detection of the causative agent of the disease and its epidemiological aspects are described.
MATERIALS AND METHODS
Study population
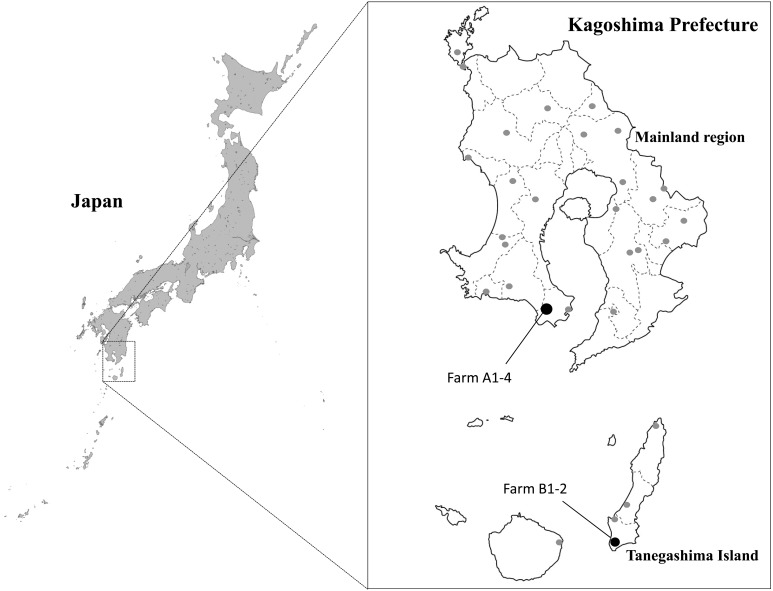
The 6 beef cattle herds (farms A1–A4, B1 and B2) included in this study contained 277 animals during the outbreak and located in the mainland region and Tanegashima Island (Fig. 1). Each held 5–35 cows and 2–63 calves under a zero grazing system. The vaccination against BEF had not been administered at all six farms before the disease was observed. Local veterinary practitioners checked animal conditions at the herds and reported clinical cases of suspected BEF to the prefectural livestock hygiene service centers. Serological tests and RT-PCR assay for BEFV were conducted at the diagnostic laboratory of Kagoshima Central Livestock Hygiene Service Center as described below.
Fig. 1.
Geographical distribution of herds (filled circles) affected by bovine ephemeral fever and sentinel herds (shaded circles) in Kagoshima Prefecture in 2015.
Blood samples
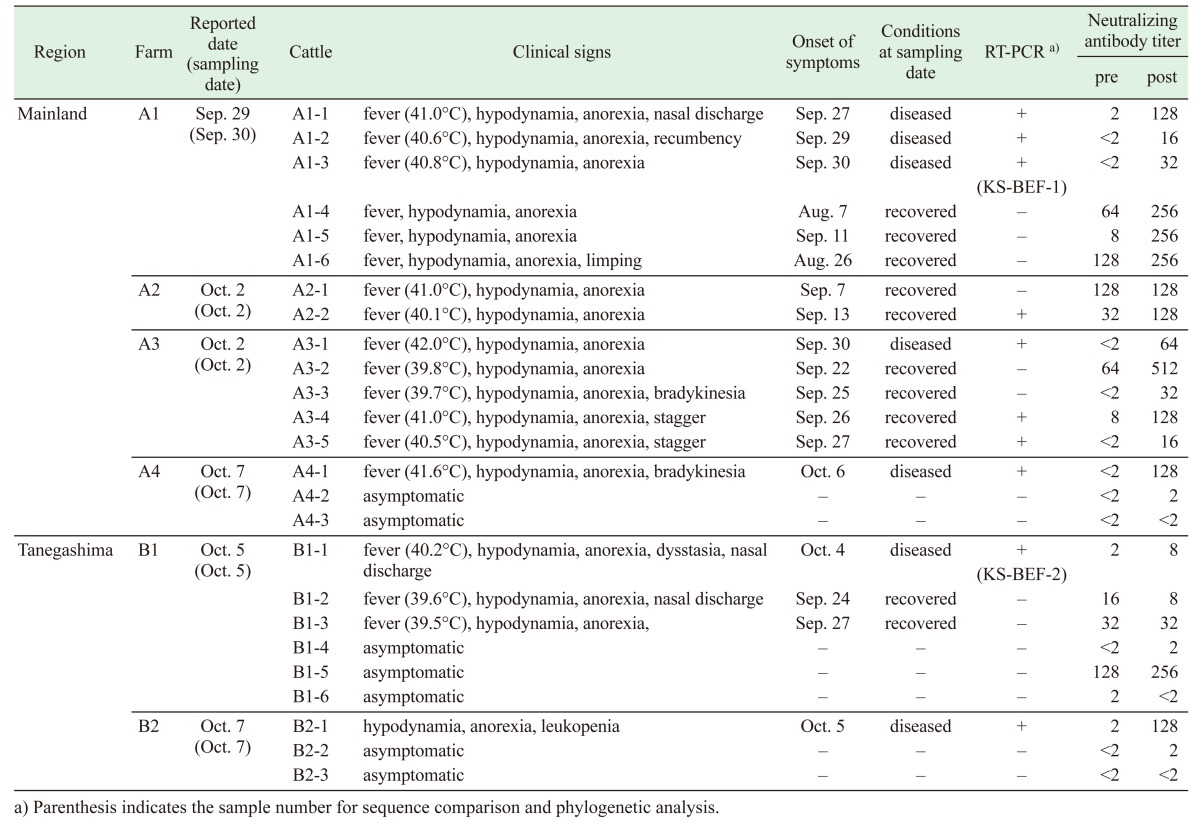
Heparinized blood samples were collected from 7 diseased cows and 11 cows that had recovered at six farms, on September 30 and October 2, 5 and 7 (Table 1). Blood samples were also drawn from 7 asymptomatic cattle raised at the same farms. The blood samples were separated into plasma and blood cells by centrifugation at 2,150 g for 10 min. The blood cells were washed three times with phosphate-buffered saline (PBS) and resuspended in PBS. Serum samples were obtained from the above-mentioned cattle at the same time and two weeks later.
Table 1. Clinical aspects, RT-PCR and virus neutralization tests for reported cases of bovine ephemeral fever in Kagoshima Prefecture in 2015.
Sentinel surveillance for bovine arboviruses was conducted with 120 calves that had not experienced summer as of June 2015 and were from non-affected herds scattered through Kagoshima Prefecture (Fig. 1). Serum samples were obtained from the calves in June, August, September, October and November 2015. In addition, sera were collected from 13 healthy cows kept at seven farms located within a 1-km radius of farms B1 and B2. The processed blood cells were kept at −80°C until virus isolation and viral genome detection. The serum samples were preserved at −20°C until they were used for the virus neutralization test.
Virus isolation
Hamster lung (HmLu-1), baby hamster kidney (BHK-21) and African green monkey kidney epithelial (Vero) cells were grown in Eagle’s minimum essential medium (MEM; Nissui, Tokyo, Japan) supplemented with 0.295% tryptose phosphate broth, 0.015% sodium bicarbonate and 5–10% fetal bovine serum at 37°C. Virus isolation was conducted with the processed blood cells as described previously [10]. In brief, tube-cultured HmLu-1, BHK-21 and Vero cells were washed three times with Earle’s solution, inoculated with the processed samples and incubated for 1 hr at 37°C. After the inocula were replaced with maintenance medium (MEM containing 0.295% tryptose phosphate broth and 0.015% sodium bicarbonate), the inoculated cultures were maintained with gentle rolling at 34°C and observed for cytopathic effect (CPE) over 7 days. Two more passages were conducted in the same manner until CPE was observed.
Virus neutralization test
The serum samples were heat-inactivated at 56°C for 30 min and serially two-fold diluted in MEM in flat-bottom 96-well plates. An equal volume of culture medium containing 100 TCID50 of BEFV YHL isolate was added, and the mixtures were maintained at 37°C for 1 hr. Subsequently, 100 µl of the suspension of HmLu-1 cells in serum-free medium (GIT; Wako Pure Chemical Industries, Osaka, Japan) was added to each well and incubated at 37°C for 7 days under a 5% CO2 atmosphere. The plates were microscopically observed for the presence of CPE, and the antibody titer was expressed as the reciprocal of the highest serum dilution at which CPE was inhibited.
RNA extraction and RT-PCR
Viral RNA was extracted from the blood cells with the High Pure Viral Nucleic Acid Kit (Roche, Mannheim, Germany) per the manufacturer’s instructions. For sensitive detection of the partial G gene of BEFV, reverse transcription polymerase chain reaction (RT-PCR) was conducted with the primer pair BEFV-AO-F (5′ GAATCATTATGGGATCGGATC 3′; at position 1140–1160)/BEFV-AO-R (5′ CCAACCTACAACAGCAGATAAAAC 3′; at position 1587–1564) using the OneStep RT-PCR Kit (QIAGEN, Hilden, Germany) under the conditions described previously [13]. The RT-PCR products which are expectedly 448 nucleotide (nt) in length were electrophoresed in 1.5% agarose and Tris-borate-EDTA (TBE) gel. The gels were stained with GelRed (Wako Pure Chemical Industries) and visualized with ultraviolet (UV) light. The primer pair gpF1 (5′-ATGTTCAAGGTCCTAATAATTACC-3′; at position 1–24)/gpR1872 (5′-TTAATGATCAAAGAATCTATC-3′: at position 1852–1872) [18] was used for amplification of the full-length BEFV G gene (1,872 nt in length). RT-PCR was performed with the PrimeScript™ One Step RT-PCR Kit Ver. 2 (TAKARA BIO, Kusatsu, Japan) under the following conditions: 1 cycle at 50°C for 30 min; 1 cycle at 94°C for 2 min; 45 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 2 min; and then maintenance at 4°C. The RT-PCR product of the expected size was identified by agarose gel electrophoresis.
Sequencing and phylogenetic analysis
The RT-PCR products of the expected length were processed with the High Pure PCR Cleanup Micro Kit (Roche) or the QIAquick PCR Purification Kit (QIAGEN) and sequenced directly using a BigDye Terminator v3.1 Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and ABI 3100-Avanti Genetic Analyzer (Thermo Fisher Scientific) according to the manufacturer’s recommendations. The nt sequence editing and the amino acid (aa) sequence deduction were performed with GENETYX ver. 10.1.3 (GENETYX, Tokyo, Japan). The sequences determined in this study were deposited in SAKURA of the DNA Data Bank of Japan (DDBJ) under accession numbers LC155957 and LC155958. The nt and aa sequence comparisons were carried out with homology search tools packaged within the software. A neighbor-joining (NJ) consensus tree was constructed by MEGA6 [17] from a CLUSTAL sequence alignment of the BEFV G gene nt sequences (1,823 bp; mapped to nts 25–1,847 of the gene). The reliability of the inferred consensus tree was tested by a boot-strap resampling of 1,000 pseudo-replicate data sets.
Insect collection
Insects were collected with suction light traps equipped with a 6W black light tube or UV light-emitting diodes [23, 25]. The traps were operated overnight in cowsheds. Surviving, non-engorged females were selected and sorted into species under the microscope based on the morphological keys. Some of the collected insects were kept at 25°C on 10% sucrose for three days until the engorged females digested blood meals, and then, the survivors were sorted into species. The other insects were pooled on the basis of place and species, and stored at −80°C until virus isolation. The collected insects were processed for virus isolation with BHK-21 cells as described previously [24].
RESULTS
Clinical cases
On September 29, 2015, it was reported that three cows developed a fever (over 40°C), hypodynamia, anorexia, respiratory manifestation, limping and recumbency at farm A1, located in the southern part of the mainland region of Kagoshima Prefecture (Fig. 1 and Table 1). Twenty of 61 Japanese black cattle in the same herd developed similar symptoms from August 7 to October 8. Diseased cows were also identified at three other farms (farms A2-4) located within a 1-km radius of farm A1 in early October. Each farm raised Japanese black cattle (range 45–92 animals per farm) at that time. After the first occurrence of disease, the sudden onset of fever, hypodynamia, anorexia, nasal discharge and dysstasia was observed in a total of 4 cows at two farms (farms B1 and B2) on Tanegashima Island between September 24 and October 5, 2015 (Fig. 1 and Table 1). The farms are in close proximity (approximately 300 m apart) and maintained 15 and 7 Japanese black cattle at the time, respectively. All of the diseased cows recovered after 2–3 days with treatment of their symptoms. No BEF cases were found in other areas of mainland Japan in 2015 [9].
RT-PCR detection and virus isolation
RT-PCR specific for the partial G gene showed positive results for 7 diseased (Cattle A1-1 to -3, A3-1, A4-1, B1-1 and B2-1) and 3 recovered cattle (A2-2, A3-4 and A3-5) (Table 1 ). The RT-PCR products that were selected from each affected farm were genetically identical to each other. The nt sequence of the RT-PCR products also showed high homology with the G gene of previously isolated BEFVs (87.5–100%). A total of 25 blood samples from the diseased, recovered and asymptomatic cattle kept at the affected farms were used for virus isolation in cultured cells. However, no virus was obtained from them.
Neutralization antibody test
Significant increases in the antibody titer to BEFV between paired sera were observed in all 7 diseased and in 6 of 11 recovered cows (Table 1). The other recovered cows already had the antibodies (ranging from 1:16 and 1:128) when the first blood sampling was conducted. All cattle which developed obvious clinical signs were seropositive against BEFV at the second blood sampling. A high antibody titer (1:256) to BEFV was also detected in a serum sample obtained from an asymptomatic cow (B1-5) at farm B1. The examined cattle with certain titers (≥1:16) at the first sampling tested negative by the RT-PCR assay. Serological surveillance at seven farms located within a 1-km radius of farms B1 and B2 could not detect the antibodies against BEFV. All sentinel cattle in Kagoshima Prefecture tested negative for BEFV antibodies in 2015.
Genetic comparison and phylogenetic analyses
The full-length BEFV G gene was successfully amplified from the extracted RNAs of two diseased cows (A1-3 and B1-1). We identified 1,827 nt of the G gene, excluding the PCR primer binding sequences. The RT-PCR products were designated as KS-BEF-1 and KS-BEF-2, respectively. No nt substitution was observed between these RT-PCR products. The sequence showed 96.3–96.8% nt and 98.7–99.3% aa identities to previous Japanese BEFVs isolated between 1988 and 2012 (Table 2 ). Relatively lower nt and aa identities (95.7 and 98.5%, respectively) were observed between the determined sequences and the corresponding sequence of the YHL isolate. The BEFV sequences detected in 2015 were most similar to Taiwanese isolates regarded as exotic viruses in 2013–2014 (99.7–99.8% nt and 99.7–99.8% aa identities). Recent Chinese BEFV isolates also shared high identities with the Japanese 2015 BEFVs (98.9–99.1% nt and 99.0–99.7% aa identities). BLAST searches of nt databases revealed that BEFVs isolated in Australia and the Middle East, except for several Turkish 2012 BEFVs, shared lower identities with the Japanese 2015 BEFVs (89.5–92.3% nt and 94.3–97.4% aa identities).
Table 2. Comparison of nucleotide (nt) and amino acid (aa) sequences of the surface glycoprotein (G) gene of Japanese 2015 bovine ephemeral fever virus (BEFV) with other East Asian BEFVs.
| nt sequence identity (%) | aa sequence identity (%) | |
|---|---|---|
| Taiwanese isolates (regarded as exotics) in 2013–2014 | 99.7–99.8 | 99.7–99.8 |
| Chinese isolates in 2011–2012 | 98.9–99.1 | 99.0–99.7 |
| Taiwanese isolates in 1984–2013 | 96.2–97.0 | 98.2–99.5 |
| Japanese isolates in 1988–2012 a) | 96.3–96.8 | 98.7–99.3 |
| Chinese isolate in 2002 | 96.7 | 99.2 |
| Chinese isolate in 1976 | 96.2 | 98.2 |
| YHL isolate a) | 95.7 | 98.5 |
a) Shorter length of the G gene, 1,821 nt and 607 deduced aa sequences, were used for the comparison.
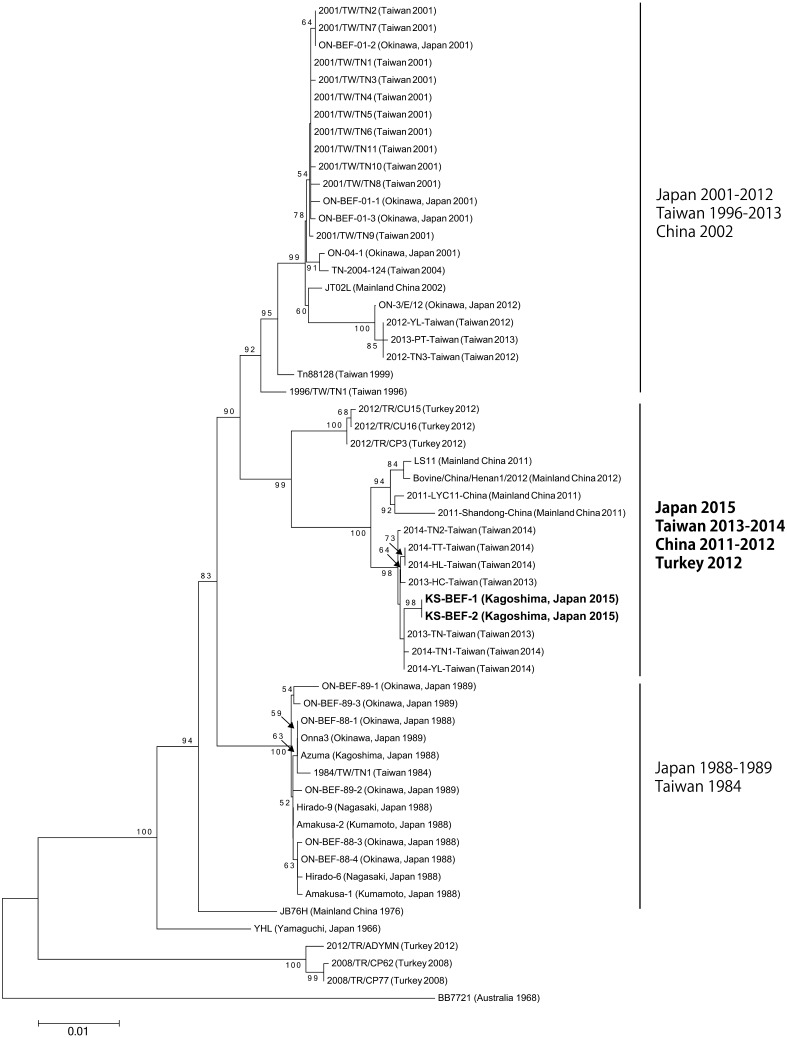
The phylogenetic tree using the G protein coding region showed that KS-BEF-1 and -2 are clustered with BEFVs from Taiwan in 2013–2014 and mainland China in 2011–2012 (Fig. 2). Three Turkish BEFVs, 2012/TR/CU15, 2012/TR/CU16 and 2012/TR/CP3, were also included in this clade. KS-BEF-1 and -2 were obviously separated from previous Japanese BEFVs in the phylogenetic analysis.
Fig. 2.
Neighbor-joining phylogenetic tree of nucleotide sequences of the surface glycoprotein (G) gene of bovine ephemeral fever virus isolates. The scale bar indicates 0.01 substitutions per site. Node support values (%) were generated from 1,000 bootstrap replicates (values <50% not shown).
Vector investigation
Virus isolation was conducted in 1,692 Culicoides biting midges and 2 Culicine mosquitoes that were collected at farms A1, A2 and A3. The most dominant species in the processed midges was Culicoides lungchiensis (1,486 midges). Eight additional Culicoides species, C. arakawae, C. cylindratus, C. jacobsoni, C. ohmorii, C. oxystoma, C. punctatus, C. sumatrae and C. tainanus, were included in the samples, but the numbers of these species were relatively low (1–168 midges). Species definition of Culicine mosquitoes was not available due to the unsuitable condition of the specimens. Virus isolation was unsuccessful in these insect samples.
DISCUSSION
Clinical observation of and serological and genetic tests on the affected cows in this study clearly demonstrated occurrences of BEF in Kagoshima Prefecture in 2015. While the occurrences were sporadic and small-scaled, our findings revealed a resurgence of BEF in mainland Japan after a 23-year absence. Because the clinical signs of the diseased cows were relatively mild during the occurrences, it may have taken several weeks before BEF was first identified. Abortion was also observed in two pregnant cows at farm A1, although no evidence supported the association of BEFV with these cases. For the early detection of BEF, it is important that clinical indices, such as the sudden onset of fever, anorexia, depression and recumbency, are not missed in the infected cattle.
Although previous BEF epizootics spread widely in mainland Japan [7, 12, 15], the areas affected in 2015 were quite limited. The affected area in the mainland region did not have a high density of cattle herds. In addition, insecticide treatment was immediately administered at the affected farms and in their neighboring cattle herds after the BEF outbreak. These factors may have buffered the spread of BEFV to a wider area.
The affected cows tested positive by the RT-PCR assay starting from 6 days after the onset of symptoms, indicating its efficacy on the BEF diagnosis. Although the results were likely consistent with the short viremia (2–8 days) of BEFV [26], a weak positive band in the RT-PCR assay was detected from a blood sample collected from a cow that had recovered (A2-2) 19 days after the symptoms developed. Fragments of BEFV RNA near the limit of detection might have been contained in the blood sample.
The partial BEFV G genes detected at all farms were identical to each other. This finding suggested that the origin of the BEFV was the same in two epizootic areas that were 100 km apart. The long-distance transport of infected vectors by wind has been considered for past BEF epizootics [2, 5, 6, 12, 15]. Estimation of the potential source of the infected vectors in the 2015 epizootic should be conducted by using atmospheric dispersal models in the future.
Phylogenetic studies based on the G gene have demonstrated that the currently analyzed BEFVs form three distinct lineages with different geographical origins: Australia, the Middle East and East Asia [20]. The East Asian lineage was further separated into three major clades in chronological order [13, 19]. The Japanese 2015 BEFVs were phylogenetically distinct from the previous Japanese isolates, but were included in the newest clade with BEFVs identified in Taiwan in 2013–2014, mainland China in 2011–2012 and Turkey in 2012. This clade first emerged in mainland China in 2011 and invaded Taiwan during 2013 and 2014 [19, 27]. The genotypic shift to the newest clade occurred rapidly across East Asia. Our phylogenetic analysis also provided evidence of an epidemiological relationship between the 2015 occurrence in Japan and the recent outbreaks in Taiwan and mainland China.
Vector control should be used as one of the preventive measures of BEFV spread. However, no principal vector species of BEFV has been identified so far [22]. Although mosquitoes are thought to be the most likely candidate for the BEFV vector, they were hardly collected at the farms studied in the mainland region. Due to this restricted abundance of mosquitoes, it is difficult to regard them as a principal vector. A large number of Culicoides biting midges were also captured at the same time, but no virus was isolated from them. The role of mosquitoes and Culicoides biting midges in BEFV transmission in the affected regions is still unclear. Experimental infections of field-caught Culicoides biting midges in previous studies revealed that they have very low or no susceptibility to BEFV [11, 21]. Furthermore, the low efficacy of BEFV isolation from field-caught hematophagous insects could be a major obstacle to vector identification [11].
Vaccination is considered to be the most effective measure to prevent BEF. Although aa substitutions between the YHL isolate and the currently circulating BEFVs in East Asia have been accumulated, their neutralization epitope domains were relatively conserved (data not shown). Only two aa residues located within the neutralization epitopes of the G protein were different between the YHL isolate and the Japanese 2015 BEFVs; 170N (asparagine) and 223D (aspartic acid) of the YHL isolate were substituted to threonine and glutamic acid in the 2015 BEFVs. In addition, the BEFV infection of cattle in 2015 still induced high antibody titers against the YHL isolate. Therefore, vaccines developed from the YHL isolate probably share antigenicity with the currently circulating BEFVs. In contrast, although an inactivated vaccine for BEF elicited a strong antibody response, it showed approximately 50% effectiveness in protection after three vaccinations during a BEF outbreak in Israel [2]. In addition, vaccinations unlikely elicit long-lasting immunity against BEFV, and the decreased immunity may not be sufficient to protect the disease [2, 18]. Effectiveness of current vaccines against the newly emerged BEFV strains is necessary to be assessed, and an efficient vaccination protocol should be found to keep protective immunity continuously to prevent BEF.
High mortality rates (>10%) were observed in recent large outbreaks associated with the newest clade of BEFVs in China and Turkey [14, 27]. The extensive spread of BEF may severely damage beef and dairy cattle in Japan. However, the vaccination rates for BEF were estimated to be low even in Kagoshima Prefecture in recent years (data not shown). We hope that the resurgence of BEF in mainland Japan in 2015 alerts cattle breeders and veterinarians to the danger of an outbreak occurring. Particular attention should be paid to vaccine promotion in order to minimize the economic loss caused by BEF. Moreover, informational exchange with neighboring nations/regions and sensitive monitoring in high-risk areas are essential for the prediction of and early warning regarding BEF outbreaks in Japan.
Acknowledgments
We thank all staff members at the Livestock Hygiene Service Center of Kagoshima Prefecture for their help in collecting and processing bovine blood samples. The authors send special thanks to Mr. Kanji Ushinohama for his kind cooperation in the field research. We are grateful to Ms. Mai Fujioka and Mr. Zenjiro Sakaguchi for their considerable support for the laboratory investigations and the preparation of this manuscript.
REFERENCES
- 1.Aizawa M., Takayoshi K., Kokuba T., Kato T., Yanase T., Yamakawa M., Tsuda T.2008. Molecular epidemiological analysis of bovine ephemeral fever virus isolated in Okinawa Prefecture. J. Jpn. Vet. Med. Assoc. 61: 363–366 (in Japanese with English summary). doi: 10.12935/jvma1951.61.363 [DOI] [Google Scholar]
- 2.Aziz-Boaron O., Klausner Z., Hasoksuz M., Shenkar J., Gafni O., Gelman B., David D., Klement E.2012. Circulation of bovine ephemeral fever in the Middle East—strong evidence for transmission by winds and animal transport. Vet. Microbiol. 158: 300–307. doi: 10.1016/j.vetmic.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 3.Cybinski D. H., Davis S. S., Zakrzewski H.1992. Antigenic variation of the bovine ephemeral fever virus glycoprotein. Arch. Virol. 124: 211–224. doi: 10.1007/BF01309803 [DOI] [PubMed] [Google Scholar]
- 4.Cybinski D. H., Walker P. J., Byrne K. A., Zakrzewski H.1990. Mapping of antigenic sites on the bovine ephemeral fever virus glycoprotein using monoclonal antibodies. J. Gen. Virol. 71: 2065–2072. doi: 10.1099/0022-1317-71-9-2065 [DOI] [PubMed] [Google Scholar]
- 5.Finlaison D. S., Read A. J., Kirkland P. D.2010. An epizootic of bovine ephemeral fever in New South Wales in 2008 associated with long-distance dispersal of vectors. Aust. Vet. J. 88: 301–306. doi: 10.1111/j.1751-0813.2010.00596.x [DOI] [PubMed] [Google Scholar]
- 6.Hayama Y., Moriguchi S., Yanase T., Suzuki M., Niwa T., Ikemiyagi K., Nitta Y., Yamamoto T., Kobayashi S., Murai K., Tsutsui T.2016. Epidemiological analysis of bovine ephemeral fever in 2012−2013 in the subtropical islands of Japan. BMC Vet. Res. 12: 47. doi: 10.1186/s12917-016-0673-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inaba Y.1971. Bovine ephemeral fever. Bull. Nat. Inst. Anim. Hlth. 62: 1–15 (in Japanese). [Google Scholar]
- 8.Inaba Y., Kurogi H., Sato K., Goto Y., Omori T., Matumoto M.1973. Formalin-inactivated, aluminum phosphate gel-adsorbed vaccine of bovine ephemeral fever virus. Arch. Gesamte Virusforsch. 42: 42–53. doi: 10.1007/BF01250506 [DOI] [PubMed] [Google Scholar]
- 9.MAFF2016. Animal Hygiene Weekly. Food Safety and Consumer Bureau, Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan 3399: 124. (in Japanese). [Google Scholar]
- 10.Miura Y., Goto Y., Kubo M., Kono Y.1988. Isolation of Chuzan virus, a new member of the Palyam subgroup of the genus Orbivirus, from cattle and Culicoides oxystoma in Japan. Am. J. Vet. Res. 49: 2022–2025. [PubMed] [Google Scholar]
- 11.Muller M. J., Standfast H.1993. Investigation of the vectors of bovine ephemeral fever virus in Australia. In: Proceedings of the 1st International Symposium on Bovine Ephemeral Fever and Related Rhabdoviruses, Beijing, PRC, 25–27 August 1992 (St. George, T. D., Uren, M. F., Young, P. L. and Hoffmann, D. eds.). Australian Centre for International Agricultural Research Proceedings 44: 29–32.
- 12.Nagano H., Hayashi K., Kubo M., Miura Y.1990. An outbreak of bovine ephemeral fever in Nagasaki Prefecture in 1988. Nihon Juigaku Zasshi 52: 307–314. doi: 10.1292/jvms1939.52.307 [DOI] [PubMed] [Google Scholar]
- 13.Niwa T., Shirafuji H., Ikemiyagi K., Nitta Y., Suzuki M., Kato T., Yanase T.2015. Occurrence of bovine ephemeral fever in Okinawa Prefecture, Japan, in 2012 and development of a reverse-transcription polymerase chain reaction assay to detect bovine ephemeral fever virus gene. J. Vet. Med. Sci. 77: 455–460. doi: 10.1292/jvms.14-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oğuzoğlu T. Ç., Ertürk A., Çizmeci Ş. G., Koç B. T., Akça Y.2015. A report on bovine ephemeral fever virus in Turkey: Antigenic variations of different strains of BEFV in the 1985 and 2012 outbreaks using partial glycoprotein gene sequences. Transbound. Emerg. Dis. 62: e66–e70. doi: 10.1111/tbed.12187 [DOI] [PubMed] [Google Scholar]
- 15.Shirakawa H., Ishibashi K., Ogawa T.1994. A comparison of the epidemiology of bovine ephemeral fever in South Korea and south-western Japan. Aust. Vet. J. 71: 50–52. doi: 10.1111/j.1751-0813.1994.tb06153.x [DOI] [PubMed] [Google Scholar]
- 16.St. George T. D.2004. Bovine ephemeral fever. pp. 1183–1193. In: Infectious Diseases of Livestock, 2nd ed. (Coetzer, J.A.W. and Tustin, R. C. eds.), Oxford University Press, Cape Town. [Google Scholar]
- 17.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ting L. J., Lee M. S., Lee S. H., Tsai H. J., Lee F.2014. Relationships of bovine ephemeral fever epizootics to population immunity and virus variation. Vet. Microbiol. 173: 241–248. doi: 10.1016/j.vetmic.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 19.Ting L. J., Lee M. S., Lin Y. L., Cheng M. C., Lee F.2016. Invasion of exotic bovine ephemeral fever virus into Taiwan in 2013−2014. Vet. Microbiol. 182: 15–17. doi: 10.1016/j.vetmic.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 20.Trinidad L., Blasdell K. R., Joubert D. A., Davis S. S., Melville L., Kirkland P. D., Coulibaly F., Holmes E. C., Walker P. J.2014. Evolution of bovine ephemeral fever virus in the Australian episystem. J. Virol. 88: 1525–1535. doi: 10.1128/JVI.02797-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venter G. J., Hamblin C., Paweska J. T.2003. Determination of the oral susceptibility of South African livestock-associated biting midges, Culicoides species, to bovine ephemeral fever virus. Med. Vet. Entomol. 17: 133–137. doi: 10.1046/j.1365-2915.2003.00414.x [DOI] [PubMed] [Google Scholar]
- 22.Walker P. J., Klement E.2015. Epidemiology and control of bovine ephemeral fever. Vet. Res. 46: 124. doi: 10.1186/s13567-015-0262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanase T., Hayama Y., Shirafuji H., Yamakawa M., Kato T., Horiwaki H., Tsutsui T., Terada Y.2014. Development of a light trap with a light-emitting diodes (LED) for the collection of Culicoides biting midges. Jpn. J. Appl. Entomol. Zool. 58: 127–132 (in Japanese with English summary). doi: 10.1303/jjaez.2014.127 [DOI] [Google Scholar]
- 24.Yanase T., Kato T., Kubo T., Yoshida K., Ohashi S., Yamakawa M., Miura Y., Tsuda T.2005. Isolation of bovine arboviruses from Culicoides biting midges (Diptera: Ceratopogonidae) in southern Japan: 1985–2002. J. Med. Entomol. 42: 63–67. doi: 10.1093/jmedent/42.1.63 [DOI] [PubMed] [Google Scholar]
- 25.Yanase T., Hirata M., Matsumori Y., Matsumura M., Kato T., Shirafuji H., Yamakawa M., Hayama Y., Tsutsui T.2011. Detection of Culicoides brevitarsis activity in Kyushu. J. Vet. Med. Sci. 73: 1649–1652. doi: 10.1292/jvms.11-0231 [DOI] [PubMed] [Google Scholar]
- 26.Zheng F. Y., Chen Q. W., Li Z., Gong X. W., Wang J. D., Yin H.2016. Experimental infection with bovine ephemeral fever virus and analysis of its antibody response cattle. Res. Vet. Sci. 104: 146–151. doi: 10.1016/j.rvsc.2015.12.018 [DOI] [PubMed] [Google Scholar]
- 27.Zheng F., Qiu C.2012. Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virol. J. 9: 268. doi: 10.1186/1743-422X-9-268 [DOI] [PMC free article] [PubMed] [Google Scholar]