Abstract
The gut microbiota has been implicated in the development of a number of chronic gastrointestinal and systemic diseases. These include inflammatory bowel diseases, irritable bowel syndrome, and metabolic (i.e. obesity, non-alcoholic fatty liver disease, and diabetes) and neurological diseases. The advanced understanding of host-microbe interactions has largely been due to new technologies such as 16S rRNA sequencing to identify previously unknown microbial communities and, more importantly, their functional characteristics through metagenomic sequencing and other multi-omic technologies, such as metatranscriptomics, metaproteomics, and metabolomics. Given the vast array of newly acquired knowledge in the field and technological advances, it is expected that mechanisms underlying several disease states involving the interactions between microbes, their metabolites, and the host will be discovered. The identification of these mechanisms will allow for the development of more precise therapies to prevent or manage chronic disease. This review discusses the functional characterization of the microbiome, highlighting the advances in identifying bioactive microbial metabolites that have been directly linked to gastrointestinal and peripheral diseases.
Keywords: cardiovascular disease, inflammatory bowel disease (IBD), metabolomics, metagenomics, neurological disease, meta-omics, metatranscriptomics, microbial metabolites, multi-omics
Major themes/advances in gut microbiota research
The gut microbiota has been implicated in the development of many chronic and systemic diseases, including inflammatory bowel diseases, metabolic disease, and neurological disorders. Over the past 15 years, the field of microbiome research has grown exponentially, in part because of new technologies, particularly 16S rRNA amplicon sequencing, which has led to the identification of previously unidentified members of the gut microbial community as well as an advanced understanding of their functional characteristics through shotgun metagenomic sequencing, and other multi-omic technologies such as metatranscriptomics, metaproteomics, and metabolomics. Although much has been uncovered about gut microbiota, whether or not it plays a causal role in the development of disease remains unclear. Given the vast array of newly gained knowledge in the field, it is likely that the mechanistic role of gut microbes and their microbial metabolites underlying several disease states will be discovered. With the identification of these mechanisms, development of personalized, precise therapies to improve or prevent chronic disease may become a reality. The goal of this review is to briefly discuss the functional capacity of the microbiome, challenges associated with multi-omic technologies, and recent advances in identifying microbial metabolites that have been directly linked to gastrointestinal and peripheral diseases.
Gut microbial ecosystem
The gut microbiota consists of over 10 trillion microbial cells and is a primary source of thousands of small molecules and other bioactive compounds that can trigger both host metabolic and immune pathways. The human gut microbiota also contains about 1000 different bacterial species with defined functions allowing them to thrive and create a niche in the midst of others with redundant or competing functions. Microbial ecosystems maintain homeostasis through a tight balance of cell-to-cell signaling and release of antimicrobial peptides to control neighboring bacterial clades allowing for their persistence in the confines of the human host. In addition to securing community dynamics with neighboring microbes, gut microbes also communicate with the human host in either a symbiotic or deleterious fashion, the latter contributing to the development of human disease.
Advances in metagenomics and metabolomics have led to the discovery of thousands of microbe-derived small molecules as well as the genes associated with their production. Although the advances in technology and wealth of big data sets have expanded our knowledge and appreciation for the contribution of gut microbes, the challenge remains in identifying small molecules that elicit a biological effect upon the host, the physiological levels necessary to do so, and how to assess the physiological impact of the targeted molecule (1). The following section will describe the basis of these technologies and discuss the strengths and challenges with these methods, particularly the challenge of incorporating and understanding these datasets simultaneously.
Functional characterization of the microbiome
Metagenomics
Many research studies that correlate the involvement of the microbiome with disease states in animal models and humans have relied heavily on the use of 16S rRNA marker gene amplicon-sequencing platforms. Although this technology has expanded our knowledge of gut microbial diversity and provides a relatively accurate fingerprint of phylogenetic community membership and structure, little can be learned about the functional properties of the community. In an attempt to predict function based on 16S rRNA sequences, tools have been designed to compare 16S marker gene sequences to reference gene databases (PICRUSt) (2). However, the reference genome database used is a limiting factor if it does not accurately reflect gene functions for the microbial community of interest, particularly in the case of rare members of the gut microbial community (3).
To gain more accurate insights into microbial community gene function, high throughput shotgun metagenomic sequencing has become an important tool, as it avoids many biases introduced by amplicon sequencing due to its untargeted nature. Shotgun metagenomic sequencing allows for an in-depth characterization of known microbial genes as well as identification of novel genetic microbial material. Following DNA shearing and template amplification, the short reads that are obtained can either be mapped to reference genomes or undergo de novo assembly, and functional annotation can be performed using specialized analyses platforms. For mapping purposes, off-line platforms such as HUMAnN (4) and on-line platforms, including MG-RAST (5) or JCVI Metagenomics Reports (METAPREP) (6), can be utilized. Assembly programs include khmer (7) and novel interfaces, such as A'nvio (8).
Although bias is avoided using this technique, if sequencing depth and coverage are insufficient, the reads cannot be properly assembled, and gene assignment based on known reference genomes cannot be completed. In addition, contaminating host DNA can overwhelm the sequencing output, inhibiting amplification of microbial sequences (3). In this case, samples such as this would require more sequencing depth and coverage, which can be cost-prohibitive, or require strategies to eliminate host DNA contamination and enrich for microbial DNA (3). Despite these limitations, the information gleaned from shotgun metagenomics has led to the identification of unique microbial strain-specific genes, particularly in the context of human disease (4).
Metatranscriptomics
Shotgun metagenomics provides a fingerprint of the gene content and functional capacity of the microbial community. However, it cannot be used to assess the activity of microbial gene expression. Combining shotgun metagenomic sequencing data with metatranscriptomic shotgun sequencing provides advantages for identification of the active microbial genome under differing conditions or disease states. Initially, total RNA from the microbial community is isolated and enriched for RNA (mRNA, lincRNA, and microRNA) followed by fragmentation. RNA is then converted to cDNA via reverse transcriptase with either random hexamers or oligo(dT) primers. Libraries are then constructed and sequenced (9).
Although this technology offers more precise characterization of the activity of the microbial whole genome, many technical issues can limit its utility. Collection and storage of gut microbial samples to maintain RNA integrity can be challenging, which can lead to an insufficient yield of quality microbial RNA for downstream purification strategies. For instance, in the process of enriching for mRNA, ribosomal RNA is eliminated, which constitutes nearly 90% of total RNA. Similar strategies to those used for shotgun metagenomics can be used for downstream data analysis with metatranscriptomic sequences. By applying these tools to the metatranscriptomic sequences, taxonomy can be assigned along with their actively expressed gene functions (9). This application can improve existing annotations, which remains a limiting factor of the utility of these technologies when inferring functionality of specific microbes.
By coupling shotgun metagenomic and metatranscriptomic sequencing, greater insights are possible with regard to functional genetic material and activity within the gut microbial community. Several bioinformatic tools can serve dual functions and integrate the two complementary strategies, including HUMAnN and Anvi'o (4, 8). Despite gaining an overall snapshot of the genetic composition and activity of specific genes within the gut microbial community, actual metabolic output remains unknown. Although some limitations exist, further exploration of microbially derived metabolites via metabolomics may provide the most valuable insights into gut microbial functional capacity and its impact on the host in health and disease.
Metabolomics
To determine the contribution of gut microbes to the host metabolism, one strategy has been to compare tissues of germ-free (GF) mice raised in the complete absence of microbiota to their conventionally raised or conventionalized counterparts. Although this tool has provided a great deal of insight, several limitations must first be recognized. First, it is acknowledged that GF mice exhibit several developmental abnormalities and altered structural features of the gastrointestinal tract, including increased transit time, enlarged cecum, shorter villi, and a thinner intestinal wall. Germ-free mice also display differences in other physiological features such as altered metabolism and reduced cardiac output (10–12). Nevertheless, discoveries using GF mice have allowed for opportunities to identify the contribution of gut microbes to the host metabolome as well as to investigate the potential of bacterial metabolites to drive host phenotypes and disease outcomes. For instance, several studies comparing GF mice to their conventionally raised counterparts showed that gut microbes impact thousands of metabolites in peripheral serum as well as in specific tissues, including the liver, brain, heart, kidney, and central nervous system (13–17).
Beyond the use of GF mice, recent technological advances in the discovery of metabolites (or small molecules) have helped to advance our understanding of the contribution of gut microbes to the host metabolome. It is well accepted that characterization of the human metabolome can provide much insight into determining states of health or disease. Thus far, identification of metabolites is achieved through the use of advanced targeted and untargeted analytical chemistry techniques, including nuclear magnetic resonance spectrometry (NMR), gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-mass spectrometry (LC-MS). These technologies, when used as complementary approaches, can yield vast amounts of information about the composition of both inorganic and organic compounds potentially derived from microbes, such as amino acids, lipids, sugars, biogenic amines, and organic acids, including volatile organic compounds (VOCs), ribosomally synthesized and post-translationally modified peptides, glycolipids, oligosaccharides, terpenoids or secondary bile acids, nonribosomal peptides, and polyketides (1).
VOCs, detected via GC-MS or selected ion flow tube-mass spectrometry (SIFT-MS), have been investigated for use as sensitive screening markers for inflammatory bowel disease (IBD). For example, VOCs were reported to distinguish IBD patients from healthy controls as well as Crohn's disease (CD) from ulcerative colitis (UC) patients. Specifically, dimethyl sulfide and hydrogen sulfide, produced by several classes of bacteria, were significantly different in CD versus UC patients (18). However, in this study, the authors were not able to determine the precise contribution of microbes alone, as the host can also produce these specific metabolites. In addition to this technology, further advances in matrix-assisted laser desorption ionization time-of-flight mass-spectrometry (MALDI-TOF) have allowed for the detection and imaging of specific metabolites in healthy and disease states (19). This technology has also been used in the clinical microbiology setting to identify specific bacteria in a high-throughput manner even in complex human fecal samples (20–22). Coupled with the ability to obtain pure cultures of specific microbes, the spectra peak output from techniques such as this can then be compared with existing databases, including MetaCyc (23), Human Metabolome Database (HMDB) (24), SetupX, and BinBase to aid in the identification and perhaps source of specific metabolites (25).
Although these technologies provide a great deal of information, computational tools to gain insights into large-scale datasets are prohibitive to discovery. To combat this, a large body of evidence as well as analytical tools and algorithms to identify biosynthetic gene clusters (BGCs) and the associated microbial metabolome have been developed and vetted by Medema and Fischbach (26). Another tool that may be especially useful in identifying microbe-derived bioactive compounds and coupling them to BGCs is the Integrated Microbial Genomes-Atlas of Microbial Gene Clusters (IMP-ABC), which is a publicly available database of biosynthetic gene clusters and predicted secondary metabolites that is based on a collection of thousands of published isolated genomes and metagenomes (27). This tool is expected to allow researchers to identify novel gene clusters from single isolates where whole genome sequencing has been performed or via metagenomes that may generate biologically active small molecules that impact host health.
Metabolomic research can also allow for better insight into the contribution of microbes to xenobiotic metabolism and the secondary metabolites they produce as a result that may have a significant impact on the host. Recent work has highlighted that the gut microbiota is a key contributor to regulating host drug metabolism because it is the first to interact with ingested xenobiotics prior to transport to the liver via portal circulation. Using RNA-seq to examine the community microbial metatranscriptome, Maurice et al. (28) showed that following short-term xenobiotic exposure, microbes exhibited altered gene expression pathways associated with tRNA biosynthesis, translation, vitamin biosynthesis, phosphate transport, the pentose phosphate pathway, and not surprisingly, xenobiotic metabolism/biodegradation using KEGG pathway abundance analysis coupled with HUMAnN and LEfSe. This xenobiotic metabolism can alter availability of specific drugs to the host. For instance, digoxin, a cardiac glycoside used to treat heart failure and arrhythmias, exhibits poor efficacy in some patients, due in part to gut microbial composition. Using several approaches, Haiser et al. (29) showed that specific strains of Eggerthella lenta could reduce digoxin, inhibiting its action in the host, and could be prevented by the presence of high levels of arginine. In another example, bioavailability of the Parkinson's therapy levidopa (l-dopa) was reduced in patients colonized with Helicobacter pylori in the stomach. Eradication of H. pylori led to significant increases in l-dopa levels in patient serum and improved efficacy (30). In addition, microbial metabolism of the colon cancer treatment, irinotecan, was found to induce gastrointestinal toxicity via β-glucuronidase activity. Blocking β-glucuronidase with an inhibitor greatly reduced the gastrointestinal side effects (31).
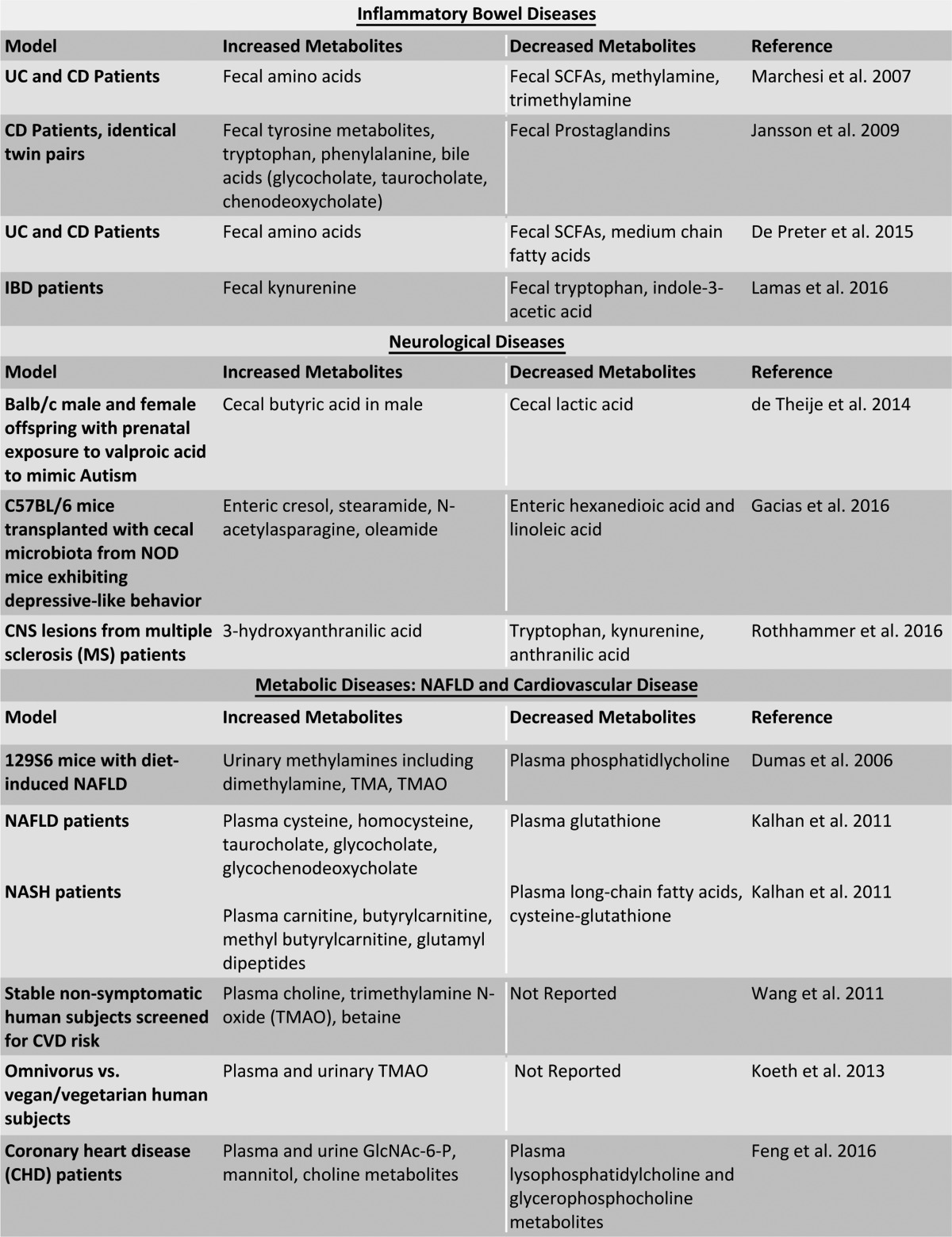
In the next section, bacterial metabolites will be examined through the lens of their biological significance related to gastrointestinal, neurological, and metabolic disease. Major metabolites related to these conditions will be highlighted, and a list of these metabolites can be found in Table 1.
TABLE 1.
Altered metabolites in gastrointestinal and systemic diseases
The following abbreviations are used: UC, ulcerative colitis; CD, Crohn's disease; IBD, inflammatory bowel diseases; SCFAs, short chain fatty acids; NOD, non-obese diabetic; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; CVD, cardiovascular disease.
The challenge in using multi-omic approaches is developing tools to identify the best candidate metabolites and microbes for further study. The best inferences may come from incorporating and simultaneously assessing results from metagenomic/metabolomics or metagenomic/metatranscriptomic datasets, for example. This approach will allow the overlay and prediction of which metabolites correspond to a bacterial gene/gene transcript, thus strengthening the ability to target a dynamic flow of events (32). These capabilities are in their infancy, and more advances in data processing are on the horizon.
Mechanisms underlying the link between microbe-derived metabolites and systemic disease
Inflammatory bowel diseases
IBD includes UC and CD. Ulcerative colitis is localized only to the colon and characterized by superficial ulcerative inflammation, whereas CD occurs sporadically along the length of the GI tract and is characterized by transmural inflammation penetrating through the epithelium. Both can be debilitating during flare-ups, persist throughout life, and may require surgical resection of regions of the GI tract if remission is not achieved using standard therapies. Understanding host-microbe interactions underlying the disease could lead to strategies to alleviate symptoms or prevent relapse for these individuals.
It is appreciated that gut microbes play an intimate role in the development of IBD. Indeed, the GI tract contains the majority of microbes in the body and is a major site of host-microbe interactions. UC and CD are associated with decreased microbial diversity based on phylogenetic measurements through 16S rRNA sequencing as well as metagenomic analysis (33). Another hallmark of IBD is microbial dysbiosis, which is characterized by the decreased abundance of commensals and increased abundance of pathogenic microbes, as well as reduced SCFA levels (34). Commensal gut microbes perform a variety of functions important to the host, including protection from pathogens through secretion of antimicrobial molecules and improved barrier function through the production of bioactive metabolites, including SCFAs. However, pathogenic microbes secrete detrimental metabolites such as hydrogen sulfide and bile acid derivatives that may exacerbate inflammatory states (35). Through metabolomic analysis, IBD patients exhibited dramatically reduced SCFA levels, methylamine, and trimethylamine (36). Decreases in the levels of these microbe-derived molecules can elicit negative consequences, as SCFAs aid in maintaining epithelial integrity, thereby protecting the host from bacterial invasion and infection. For instance, butyrate-mediated activation of G-protein-coupled receptor (GPR)43 improves barrier function via histone deacetylase (HDAC) inhibition, stabilization of hypoxia, and regulation of intestinal macrophages and regulatory T cells. GPR43 inhibition, however, increases inflammation in mice (37). Therefore, reduced levels of SCFA and lack of SCFA-mediated signaling leaves the epithelial barrier vulnerable to damage and bacterial penetrance.
Metabolomic analyses in patients with IBD also exhibit altered levels of tryptophan and tryptophan metabolites compared with control subjects (37, 38). Tryptophan is a precursor to several bacterial metabolites that have a functional impact on the host. Studies on bacterial metabolism of tryptophan date back to the 1950s, at which time it was understood that tryptophan metabolism may involve several different pathways (39). The tryptophan metabolite, indole, up-regulates the expression of tight junction proteins and reduces the expression of inflammatory genes (40, 41) presumably through interaction with the aryl hydrocarbon receptor (AHR) (42). Indeed, activation of AHR using tryptophan metabolites ameliorates dextran sulfate sodium-induced colitis in mice (43).
It was discovered that the host plays a significant role in the ability of the microbiota to metabolize tryptophan. For example, microbiota from mice lacking caspase recruitment domain family member 9 (CARD9) were unable to metabolize tryptophan, the products of which would otherwise activate AHR. Conventionalization of GF mice with CARD9−/− microbiota increased their susceptibility to develop colitis. Interestingly, intestinal inflammation in these mice was ameliorated with both the Lactobacillus strain and AHR agonist supplementation. Furthermore, microbiota obtained from IBD patients were less able to produce AHR ligands (44). This study nicely exemplifies the coordinated action of microbe and host to control inflammation via metabolite production in colitis. Metabolites that are regulated in IBD can be found in Table 1.
Neurological diseases
Mounting evidence shows that gut microbes contribute to neural development, as well as cognitive and behavioral health. Indeed, GF mice have increased blood-brain barrier (BBB) permeability from birth through adulthood, and conventionalization leads to improved BBB tight junction formation (45, 46). Correlations have been made between neurological disorders and gut microbial dysbiosis brought on by usage of antibiotics, resulting in the development of irritable bowel syndrome and depression (47). For example, Bercik et al. (48) found that antibiotic-treated mice displayed increased exploratory behavior and were less apprehensive. Furthermore, conventionalization of GF mice with BALB/c microbiota caused an increase in anxious and timid behavior compared with mice receiving NIH Swiss microbiota (48).
Metabolite-driven effects on the CNS may occur through a direct impact on BBB cells or by altering signaling through the enteric nervous system (47). SCFAs have been reported to activate sympathetic and autonomic nervous systems through both GPR41 and GPR43 activation in the gut and can directly cross the BBB. Animals exposed to an experimentally induced maternal microbial dysbiosis in utero, where SCFAs were significantly reduced, exhibited features of autism that mimicked human disease (47, 49). In a separate study using metabolomics analysis, transplantation of feces exhibiting elevated cresol levels induced depression in recipient mice (50). Furthermore, myelin expression was reduced in oligodendrocytes treated with cresol in vitro (50). In addition to their role in the gut and the down-regulation of inflammation in IBD, tryptophan metabolites have been shown to decrease inflammation in the brain. Conversion of tryptophan into AHR ligands by gut microbes has been shown to activate AHR in astrocytes, resulting in decreased CNS inflammation (51). Taken together, microbial metabolites can impact distal tissues, including the brain, contributing to neurological function and disease development. Further research in this area may lead to strategies for the prevention of neurological disorders.
Metabolic diseases: cardiovascular disease and non-alcoholic fatty liver disease (NAFLD)
Because the findings that bacterial metabolism of phosphatidylcholine and l-carnitine lead to cardiovascular disease (through production of trimethylamine and its subsequent conversion to trimethylamine N-oxide (TMAO) in the liver (52, 53)), a number of additional metabolites have been identified that are associated with cardiovascular as well as non-alcoholic fatty liver disease (NAFLD).2 Using metabolomic analysis, over 15 microbial metabolites were identified as predictors of coronary heart disease as well as their associated microbial source (54). These metabolites included GlcNAc-6-P, mannitol, and 15 unique choline metabolites. Integration of metagenomic and metabolomics analyses revealed Clostridium sp. HGF2 was associated with production of GlcNAc-6-P, and Clostridium sp. HGF2, Streptococcus sp. M143, and Streptococcus sp. M334 were associated with the production of mannitol (54). The exact mechanism for how these microbially derived metabolites drive coronary disease development remains to be explored.
Gut microbes play an important role in regulating choline and bile acid levels that are associated with fatty liver disease. For instance, using metabolomics analyses of plasma and blood, Dumas et al. (55) found that microbial metabolism of choline to methylamines mimicked choline deficiency in the host, resulting in NAFLD. In a separate human clinical trial, children presenting with NAFLD were given the probiotic VSL#3. Analysis of the urine metabolome following VSL#3 supplementation revealed that compared with healthy controls, children with NAFLD displayed decreased levels of tyrosine, valine, β-aminoisobutyric acid, pseudouridine, and methylguanidine, indicative of altered amino acid metabolism and RNA turnover (56). These changes were in conjunction with improved body mass index and a reduction in histological fatty liver previously published within the same subjects (57). Taken together, fatty liver disease appears to be associated with altered metabolites that are either produced or influenced by gut microbes.
Conclusion
Altogether, advances in the fields of metabolomics and microbiome analyses have resulted in the newly identified functional roles of several metabolites, such as SCFAs and tryptophan derivatives. These findings have shown great promise for the identification of novel therapeutic targets for several human diseases. Despite these advances, much remains to be discovered with regard to unidentified microbial metabolites and their implications in health and disease. To navigate the unknown microbial metabolome, advances in existing technologies will be required. However, with our current understanding, genetic modification of gut microbes to drive the production of specific metabolites may be a way in which physiological levels of these metabolites can be harnessed and formulated for medicinal use. This is not outside the realm of possibilities given the number of antimicrobial products derived from microbes that are in use today (1). Of course, the safe use of these products will require a thorough understanding on how they interact with the host, and this may be achieved by combining meta-omic approaches and functional testing utilizing GF mice and intestinal organoid technologies (58). Establishing new methods to integrate meta-omic datasets will also strengthen the ability to target potential bioactive microbial metabolites for further study. Given the wealth of data currently available and the current progress in the field, these achievements are on the forefront of microbiome research.
This work was supported by the GI Research Foundation, Chicago, IL and National Institutes of Health Grants DK042086, DK097268, and DK102872 from NIDDK (to E. B. C.). This is the second article in the Host-Microbiome metabolic interplay Minireview series. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NAFLD
- non-alcoholic fatty liver disease
- IBD
- inflammatory bowel disease
- UC
- ulcerative colitis
- CD
- Crohn's disease
- VOC
- volatile organic compound
- BGC
- biosynthetic gene cluster
- SCFA
- short chain fatty acid
- GF
- germ-free
- l-dopa
- levodopa
- GI
- gastrointestinal
- AHR
- aryl hydrocarbon receptor
- BBB
- blood-brain barrier.
References
- 1. Donia M. S., and Fischbach M. A. (2015) Human microbiota. Small molecules from the human microbiota. Science 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., Beiko R. G., and Huttenhower C. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharpton T. J. (2014) An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 5, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abubucker S., Segata N., Goll J., Schubert A. M., Izard J., Cantarel B. L., Rodriguez-Mueller B., Zucker J., Thiagarajan M., Henrissat B., White O., Kelley S. T., Methé B., Schloss P. D., Gevers D., et al. (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyer F., Paarmann D., D'Souza M., Olson R., Glass E. M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., and Edwards R. A. (2008) The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goll J., Rusch D. B., Tanenbaum D. M., Thiagarajan M., Li K., Methé B. A., and Yooseph S. (2010) METAREP: JCVI metagenomics reports–an open source tool for high-performance comparative metagenomics. Bioinformatics 26, 2631–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crusoe M. R., Alameldin H. F., Awad S., Boucher E., Caldwell A., Cartwright R., Charbonneau A., Constantinides B., Edvenson G., Fay S., Fenton J., Fenzl T., Fish J., Garcia-Gutierrez L., Garland P., et al. (2015) The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 4, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eren A. M., Esen Ö. C., Quince C., Vineis J. H., Morrison H. G., Sogin M. L., and Delmont T. O. (2015) Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 3, e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bashiardes S., Zilberman-Schapira G., and Elinav E. (2016) Use of metatranscriptomics in microbiome research. Bioinform. Biol. Insights 10, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coates M. E. (1975) Gnotobiotic animals in research: their uses and limitations. Lab. Anim. 9, 275–282 [DOI] [PubMed] [Google Scholar]
- 11. Al-Asmakh M., and Zadjali F. (2015) Use of germ-free animal models in microbiota-related research. J. Microbiol. Biotechnol. 25, 1583–1588 [DOI] [PubMed] [Google Scholar]
- 12. Nicklas W., Keubler L., and Bleich A. (2015) Maintaining and monitoring the defined microbiota status of gnotobiotic rodents. ILAR J. 56, 241–249 [DOI] [PubMed] [Google Scholar]
- 13. Martin F.-P., Dumas M.-E., Wang Y., Legido-Quigley C., Yap I. K., Tang H., Zirah S., Murphy G. M., Cloarec O., Lindon J. C., Sprenger N., Fay L. B., Kochhar S., van Bladeren P., Holmes E., and Nicholson J. K. (2007) A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wikoff W. R., Anfora A. T., Liu J., Schultz P. G., Lesley S. A., Peters E. C., and Siuzdak G. (2009) Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U.S.A. 106, 3698–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Claus S. P., Tsang T. M., Wang Y., Cloarec O., Skordi E., Martin F.-P., Rezzi S., Ross A., Kochhar S., Holmes E., and Nicholson J. K. (2008) Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsumoto M., Kibe R., Ooga T., Aiba Y., Kurihara S., Sawaki E., Koga Y., and Benno Y. (2012) Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsumoto M., Kibe R., Ooga T., Aiba Y., Sawaki E., Koga Y., and Benno Y. (2013) Cerebral low-molecular metabolites influenced by intestinal microbiota: a pilot study. Front. Syst. Neurosci. 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hicks L. C., Huang J., Kumar S., Powles S. T., Orchard T. R., Hanna G. B., and Williams H. R. (2015) Analysis of exhaled breath volatile organic compounds in inflammatory bowel disease: a pilot study. J. Crohns Colitis 9, 731–737 [DOI] [PubMed] [Google Scholar]
- 19. Singhal N., Kumar M., Kanaujia P. K., and Virdi J. S. (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stevenson L. G., Drake S. K., Shea Y. R., Zelazny A. M., and Murray P. R. (2010) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48, 3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seng P., Abat C., Rolain J. M., Colson P., Lagier J.-C., Gouriet F., Fournier P. E., Drancourt M., La Scola B., and Raoult D. (2013) Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51, 2182–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samb-Ba B., Mazenot C., Gassama-Sow A., Dubourg G., Richet H., Hugon P., Lagier J.-C., Raoult D., and Fenollar F. (2014) MALDI-TOF identification of the human gut microbiome in people with and without diarrhea in Senegal. PLoS ONE 9, e87419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caspi R., Foerster H., Fulcher C. A., Kaipa P., Krummenacker M., Latendresse M., Paley S., Rhee S. Y., Shearer A. G., Tissier C., Walk T. C., Zhang P., and Karp P. D. (2008) The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 36, D623–D631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., Bouatra S., Sinelnikov I., Arndt D., Xia J., Liu P., et al. (2013) HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wishart D. S. (2007) Current progress in computational metabolomics. Brief Bioinform. 8, 279–293 [DOI] [PubMed] [Google Scholar]
- 26. Medema M. H., and Fischbach M. A. (2015) Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadjithomas M., Chen I.-M., Chu K., Ratner A., Palaniappan K., Szeto E., Huang J., Reddy T. B., Cimermančič P., Fischbach M. A., Ivanova N. N., Markowitz V. M., Kyrpides N. C., and Pati A. (2015) IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio. 6, e00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maurice C. F., Haiser H. J., and Turnbaugh P. J. (2013) Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haiser H. J., Gootenberg D. B., Chatman K., Sirasani G., Balskus E. P., and Turnbaugh P. J. (2013) Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niehues M., and Hensel A. (2009) In vitro interaction of l-dopa with bacterial adhesins of Helicobacter pylori: an explanation for clinical differences in bioavailability? J. Pharm. Pharmacol. 61, 1303–1307 [DOI] [PubMed] [Google Scholar]
- 31. Carmody R. N., and Turnbaugh P. J. (2014) Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 124, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McHardy I. H., Goudarzi M., Tong M., Ruegger P. M., Schwager E., Weger J. R., Graeber T. G., Sonnenburg J. L., Horvath S., Huttenhower C., McGovern D. P., Fornace A. J. Jr., Borneman J., and Braun J. (2013) Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mondot S., and Lepage P. (2016) The human gut microbiome and its dysfunctions through the meta-omics prism. Ann. N.Y. Acad. Sci. 1372, 9–19 [DOI] [PubMed] [Google Scholar]
- 34. Leone V., Chang E. B., and Devkota S. (2013) Diet, microbes, and host genetics: the perfect storm in inflammatory bowel diseases. J. Gastroenterol. 48, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Devkota S., Wang Y., Musch M. W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D. A., Jabri B., and Chang E. B. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchesi J. R., Holmes E., Khan F., Kochhar S., Scanlan P., Shanahan F., Wilson I. D., and Wang Y. (2007) Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 6, 546–551 [DOI] [PubMed] [Google Scholar]
- 37. Zhang L. S., and Davies S. S. (2016) Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jansson J., Willing B., Lucio M., Fekete A., Dicksved J., Halfvarson J., Tysk C., and Schmitt-Kopplin P. (2009) Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS ONE 4, e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nozaki M., and Ishimura Y. (1972) Tryptophan metabolism in micro-organisms. Biochem. J. 128, 24P–25P [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bansal T., Alaniz R. C., Wood T. K., and Jayaraman A. (2010) The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimada Y., Kinoshita M., Harada K., Mizutani M., Masahata K., Kayama H., and Takeda K. (2013) Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 8, e80604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hubbard T. D., Murray I. A., Bisson W. H., Lahoti T. S., Gowda K., Amin S. G., Patterson A. D., and Perdew G. H. (2015) Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 5, 12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ji T., Xu C., Sun L., Yu M., Peng K., Qiu Y., Xiao W., and Yang H. (2015) Aryl hydrocarbon receptor activation down-regulates IL-7 and reduces inflammation in a mouse model of DSS-induced colitis. Dig. Dis. Sci. 60, 1958–1966 [DOI] [PubMed] [Google Scholar]
- 44. Lamas B., Richard M. L., Leducq V., Pham H.-P., Michel M.-L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., Brot L., Taleb S., Couturier-Maillard A., Nion-Larmurier I., Merabtene F., et al. (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M. L., Forssberg H., and Pettersson S. (2011) Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Tóth M., Korecka A., Bakocevic N., Ng L. G., Guan N. L., Kundu P., Gulyás B., Halldin C., Hultenby K., Nilsson H., et al. (2014) The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6, 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rogers G. B., Keating D. J., Young R. L., Wong M.-L., Licinio J., and Wesselingh S. (2016) From gut dysbiosis to altered brain function and mental illness: mechanisms and pathways. Mol. Psychiatry 21, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bercik P., Denou E., Collins J., Jackson W., Lu J., Jury J., Deng Y., Blennerhassett P., Macri J., McCoy K. D., Verdu E. F., and Collins S. M. (2011) The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609.e3 [DOI] [PubMed] [Google Scholar]
- 49. de Theije C. G., Wopereis H., Ramadan M., van Eijndthoven T., Lambert J., Knol J., Garssen J., Kraneveld A. D., and Oozeer R. (2014) Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav. Immun. 37, 197–206 [DOI] [PubMed] [Google Scholar]
- 50. Gacias M., Gaspari S., Santos P.-M., Tamburini S., Andrade M., Zhang F., Shen N., Tolstikov V., Kiebish M. A., Dupree J. L., Zachariou V., Clemente J. C., and Casaccia P. (2016) Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. Elife 5, e13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rothhammer V., Mascanfroni I. D., Bunse L., Takenaka M. C., Kenison J. E., Mayo L., Chao C.-C., Patel B., Yan R., Blain M., Alvarez J. I., Kébir H., Anandasabapathy N., Izquierdo G., Jung S., et al. (2016) Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y.-M., Wu Y., Schauer P., Smith J. D., Allayee H., Tang W. H., et al. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., Smith J. D., DiDonato J. A., Chen J., Li H., Wu G. D., et al. (2013) Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng Q., Liu Z., Zhong S., Li R., Xia H., Jie Z., Wen B., Chen X., Yan W., Fan Y., Guo Z., Meng N., Chen J., Yu X., Zhang Z., et al. (2016) Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci. Rep. 6, 22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dumas M.-E., Barton R. H., Toye A., Cloarec O., Blancher C., Rothwell A., Fearnside J., Tatoud R., Blanc V., Lindon J. C., Mitchell S. C., Holmes E., McCarthy M. I., Scott J., Gauguier D., and Nicholson J. K. (2006) Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U.S.A. 103, 12511–12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miccheli A., Capuani G., Marini F., Tomassini A., Praticò G., Ceccarelli S., Gnani D., Baviera G., Alisi A., Putignani L., and Nobili V. (2015) Urinary 1H-NMR-based metabolic profiling of children with NAFLD undergoing VSL#3 treatment. Int. J. Obes. 39, 1118–1125 [DOI] [PubMed] [Google Scholar]
- 57. Alisi A., Bedogni G., Baviera G., Giorgio V., Porro E., Paris C., Giammaria P., Reali L., Anania F., and Nobili V. (2014) Randomised clinical trial: the beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 39, 1276–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lukovac S., Belzer C., Pellis L., Keijser B. J., de Vos W. M., Montijn R. C., and Roeselers G. (2014) Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 5, e01438. [DOI] [PMC free article] [PubMed] [Google Scholar]