Abstract
Prions arise from proteins that have two possible conformations: properly folded and non-infectious or misfolded and infectious. The [PSI+] yeast prion, which is the misfolded and self-propagating form of the translation termination factor eRF3 (Sup35), can be cured of its infectious conformation by overexpression of Hsp104, which helps dissolve the prion seeds. This dissolution depends on the trimming activity of Hsp104, which reduces the size of the prion seeds without increasing their number. To further understand the relationship between trimming and curing, trimming was followed by measuring the loss of GFP-labeled Sup35 foci from both strong and weak [PSI+] variants; the former variant has more seeds and less soluble Sup35 than the latter. Overexpression of Saccharomyces cerevisiae Hsp104 (Sc-Hsp104) trimmed the weak [PSI+] variants much faster than the strong variants and cured the weak variants an order of magnitude faster than the strong variants. Overexpression of the fungal Hsp104 homologs from Schizosaccharomyces pombe (Sp-Hsp104) or Candida albicans (Ca-Hsp104) also trimmed and cured the weak variants, but interestingly, it neither trimmed nor cured the strong variants. These results show that, because Sc-Hsp104 has greater trimming activity than either Ca-Hsp104 or Sp-Hsp104, it cures both the weak and strong variants, whereas Ca-Hsp104 and Sp-Hsp104 only cure the weak variants. Therefore, curing by Hsp104 overexpression depends on both the trimming ability of the fungal Hsp104 homolog and the strength of the [PSI+] variant: the greater the trimming activity of the Hsp104 homolog and the weaker the variant, the greater the curing.
Keywords: amyloid, molecular chaperone, prion, protein folding, yeast, Hsp104
Introduction
The molecular chaperone, Hsp104, is a hexameric protein belonging to the triple-A ATPase family (1). Along with its roles in protein folding and conferring thermotolerance, this chaperone is essential for propagation of prions in budding yeast. Prions are infectious proteins that occur in two conformations, a properly folded non-infectious conformation and a misfolded infectious amyloid conformation (2). Yeast has more than a dozen prions, which pass from mother to daughter cells as prion seeds. The steady-state number of prion seeds is maintained by Hsp104, which severs the seeds; this severing reaction is dependent on the ATPase activity of Hsp104 and involves the presentation of the prion to Hsp104 by the molecular chaperones, Sis1 and Ssa1/Ssa2, followed by threading of the prions through the Hsp104 channel (3). Yeast prions are cured by inhibiting the ATPase activity of Hsp104; this inhibition can be caused by either overexpressing a dominant-negative Hsp104 mutant or by inactivating Hsp104 with guanidine. Curing is then caused by absence of prion seed severing, in combination with dilution of the seeds by cell division (4–6).
Surprisingly, one of the yeast prions, [PSI+], is not only cured by inactivation of Hsp104 but also by overexpression of Hsp104 (7). [PSI+] is the infectious form of Sup35, a protein that functions in translation termination (8, 9). In [PSI+] yeast, where most of the Sup35 is in the amyloid form and is therefore inactive, there is read-through of stop codons, which enables [PSI+] yeast to be readily distinguished from [psi−] yeast. A red/white colony assay is the standard method of detecting these two conformations. In this assay, yeast having nonsense mutations in the adenine synthesis pathway are plated on limiting adenine medium. Another method of detecting these two conformations is by fluorescence imaging using GFP-labeled Sup35. [PSI+] yeast have distinct fluorescent foci, whereas [psi−] yeast show diffuse fluorescence of the GFP-labeled Sup35 (10–12).
Using these methods, we previously showed that the curing of [PSI+] by overexpression of Saccharomyces cerevisiae Hsp104 (Sc-Hsp104) requires a new activity of Hsp104 called trimming (13). Like severing, trimming reduces the size of the prion seeds. However, unlike severing, trimming, which may occur through removal of Sup35 molecules from the ends of the prion fibers, is not accompanied by an increase in seed number. The dissolution of the prion seeds caused by overexpression of Sc-Hsp104 was shown to require the trimming activity of Hsp104. Mutants of Hsp104 that cannot trim also cannot cure [PSI+] prion by overexpression (14). For example, overexpression of the T160M point mutant of Sc-Hsp104 that lacks trimming activity but has severing activity comparable with wild-type Sc-Hsp104 does not cure [PSI+]. Consistent with our in vivo observations, in vitro evidence for dissolution of the prion seeds by Hsp104 has been obtained by Shorter and Lindquist (15). Other chaperones and cochaperones such as Hsp70, Sti1, Cpr7, Hsp90, and Sgt2 have been shown to affect curing of [PSI+] by Hsp104 overexpression, but it is not yet clear whether these chaperones function directly in the dissolution process (16–20).
Both the propagation and curing of the [PSI+] prion are dependent on the structure of Hsp104. The N-terminal domain of Hsp104 is important for curing by Hsp104 overexpression but not for propagating [PSI+] (21). Downstream of the N-terminal domain are two ATP-binding domains (NBD2 domains), and both prion propagation and curing are dependent on the hydrolysis of ATP by these domains (22). The M-domain, which bridges the two NBD domains, is important for the regulation of Hsp104 function by Ssa1/Ssa2 (23, 24). Finally, the highly acidic C-terminal domain has been implicated in the interaction of substrate with Hsp104 (25, 26); as yet, it is not known how this domain affects curing by Hsp104 overexpression. It has been shown that after its deletion from Hsp104, the resulting truncated fragment no longer assembles into hexamers in vitro and does not confer thermotolerance in vivo (27). Another factor that affects curing by Hsp104 overexpression is the fungal source of Hsp104. Similar to Sc-Hsp104, overexpression of Hsp104 from Candida albicans (Ca-Hsp104) has been reported to cure [PSI+] (28), whereas no curing was observed with overexpression of Hsp104 from Schizosaccharomyces pombe (Sp-Hsp104) (29, 30). Finally, curing by Hsp104 overexpression is dependent on the [PSI+] variant; weak [PSI+] variants, which have fewer seeds and a greater soluble pool of Sup35 than strong [PSI+] variants, are cured at a faster rate by overexpression of Sc-Hsp104 (31). The prion fibers differ in their physical properties and tertiary structure among the different [PSI+] variants (32–34). Fibers with greater thermostability result in a weaker [PSI+] variant because they are not fragmented as readily, so they produce fewer prion seeds that provide fewer ends to recruit and deplete soluble Sup35 (35, 36).
Our model predicts that a change in trimming causes a change in curing by Hsp104 overexpression. This prediction was tested by overexpressing different Hsp104 constructs and examining their ability both to cure different variants of [PSI+] and to trim the prion seeds. It was previously shown that overexpression of Sc-Hsp104 cures the weak [PSI+] variants faster than the strong [PSI+] variants (31), and we now show that Sc-Hsp104 trims the prion seeds of the weak [PSI+] variants faster than the prion seeds of the strong [PSI+] variants. Furthermore, we show that, in contrast to overexpression of Sc-Hsp104, overexpression of either Sp-Hsp104 or Ca-Hsp104 both trims and cures the weak [PSI+] variants, but overexpression of these Hsp104 homologs does not trim or cure the strong [PSI+] variants. Therefore, Sc-Hsp104 differs from the other fungal Hsp104 homologs in that its greater trimming activity enables it to cure both the strong and weak [PSI+] variants rather than just the weak [PSI+] variants. Therefore, the ability of Hsp104 overexpression to cure [PSI+] correlates with its trimming activity.
Results
Curing of different [PSI+] variants by overexpression of Sc-Hsp104
We previously showed that overexpression of Hsp104 cures [PSI+] by a dissolution mechanism involving the trimming of the prion seeds by Hsp104 (14). Because our data support a mechanism in which trimming is necessary for curing by Hsp104 overexpression, a change in trimming should be accompanied by a change in curing. To test this prediction, we looked at curing of [PSI+] by overexpression of Hsp104 using different Hsp104 constructs and different [PSI+] variants. Liebman and co-workers (31) previously established that weak [PSI+] variants cure faster than strong [PSI+] variants; they also showed that after overnight overexpression of Hsp104, there was 90% curing of a weak [PSI+] variant but only 50% curing of a strong [PSI+] variant. However, because this measurement was done at only a single time point, we extended their observation by determining the curing of different [PSI+] variants at several time points when Sc-Hsp104 was overexpressed from the GAL1 promoter. Furthermore, to make this study more comprehensive, curing was done in two different background strains (74D-694 and 779-6A), in which yeast either expressed Sup35 or GFP-labeled Sup35 (NGMC) from the Sup35 chromosomal locus. Using the GFP-labeled Sup35 enabled us to image by confocal microscopy the fluorescent changes of the prion as the yeasts were cured. The extent of curing was determined by the red/white colony assay after plating the yeast at ½ YPD medium.
The curing curves in Fig. 1A show the percent [PSI+] remaining versus generation time for the different [PSI+] variants. As expected, the weaker [PSI+] variants (L1758, L2888, and 1074[PSI+]DM) cured faster than the stronger [PSI+] variants (L1762, SY80, and 779-6A[PSI+]DM). However, we were surprised by how a large difference there was in the rates of curing of the weak and strong [PSI+] variants. For example, the weak L1758[PSI+] variant was more than 75% cured in two generations, whereas it took eight times longer to get comparable curing of the strong SY80[PSI+] variant. This very large difference in the rates of [PSI+] curing by Sc-Hsp104 overexpression predicts that there should be much faster trimming of the foci in the weak variants than in the strong variants. In our previous study, we defined trimming as the loss of detectable fluorescent foci in cells that are still [PSI+] (13). The foci then again became prominent in the trimmed cells after stress, whereas in [psi−] yeast, there was no change in the diffuse appearance of the GFP-Sup35 after stress (13).
Figure 1.
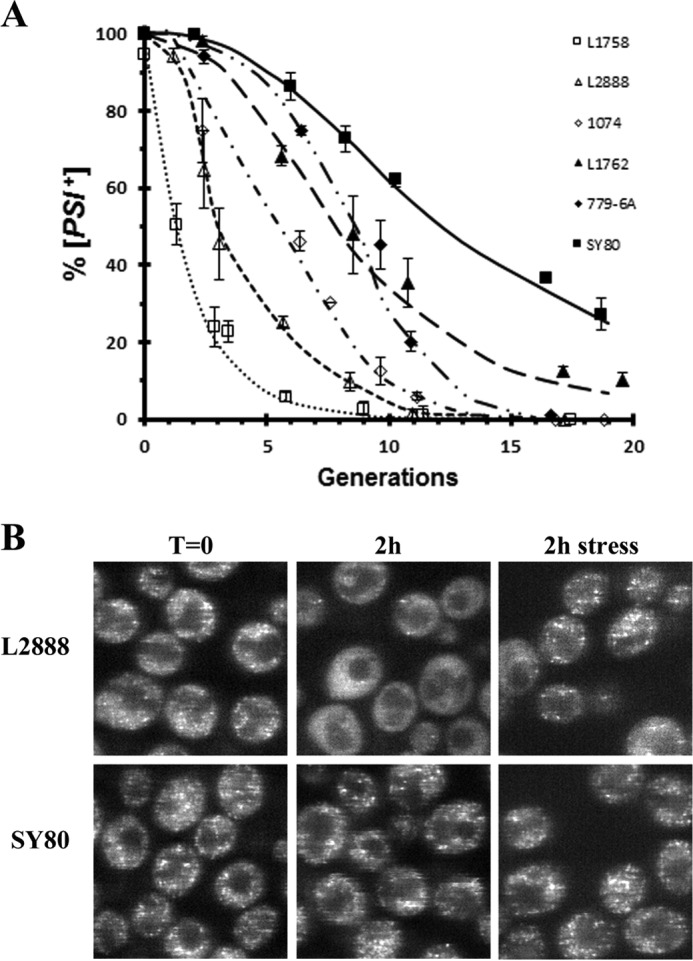
Rate of curing and trimming of prion seeds by Sc-Hsp104 is dependent on the [PSI+] variant. A, curing of [PSI+] by Sc-Hsp104 overexpression was measured in different [PSI+] variants. Sc-Hsp104 was expressed from the Gal1 promoter, and the extent of curing was measured at the indicated times by the red/white colony assay. The following variants were used: L1758[PSI+], L2888[PSI+], 1074[PSI+]DM, 779-6A[PSI+]DM, SY80[PSI+], and L1762[PSI+]. B, fluorescence imaging of yeast expressing GFP-labeled Sup35 (NGMC) in [PSI+] yeast overexpressing Sc-Hsp104. Images are of either the weak L2888[PSI+] or the strong SY80 [PSI+] variants. Images were taken after overnight incubation in raffinose medium (T = 0), 2 h after addition of galactose medium to induce Sc-Hsp104 expression, or 2 h in galactose medium and 1 h in 5 mm guanidine to stress the cells.
To test whether there is indeed faster trimming in the weak variants, we compared the rates of trimming of the weak L2888[PSI+] and the strong SY80[PSI+] variants; these variants have the same yeast background and express GFP-labeled Sup35. Following induction of Sc-Hsp104 expression with galactose, the cells were imaged and plated hourly for 3 h during which time the yeast had doubled in number. Z-stack confocal fluorescent images were obtained of both non-stressed and stressed cells. This enabled us to differentiate between cells with trimmed foci and cured [psi−] cells; trimmed foci reappear after stress, whereas stress has no effect on the cured cells. As expected, both weak and strong [PSI+] variants showed prominent foci in all cells prior to addition of galactose, while 2 h after galactose addition to induce Hsp104 overexpression, the appearance of the GFP-labeled Sup35 was very different in the weak and strong [PSI+] variants even though the plating assay showed they were 88 and 100% [PSI+], respectively. As shown in Fig. 1B, in the weak [PSI+] variant population, there were many cells with no apparent foci unless the cells were stressed, at which point there was a marked increase in the percent cells with detectable foci. In contrast, the strong [PSI+] variant cells had prominent foci even in the absence of stress, indicating a lack of trimming. Thus, the weak [PSI+] variant showed much more trimming than the strong [PSI+] variant.
The extent of trimming of the prion seeds in the weak and strong [PSI+] variants during the 3 h of Hsp104 induction was quantified in Table 1. For each time point, the number of cells with numerous foci and the number of cells with none or almost no foci (<5 foci) were counted both before stress and after stress. The percentage of trimmed cells was calculated as the difference between the percentage of cells with few foci before stress and the percentage of cells with few foci after stress. Consistent with our prediction, there was much more trimming in the weak [PSI+] variant than the strong [PSI+] variant. For example, 50% of the weak [PSI+] variant cells, but only 10% of the strong [PSI+] variant cells, were trimmed after 3 h in galactose medium. As expected, the percent of cured [psi−] cells as determined from the plating assay was equivalent to the percent of cells with few or no foci after stress. Therefore, not only is curing much faster in the weak than in the strong [PSI+] variant, but also trimming of the prion seeds is much faster in the weak than in the strong [PSI+] variant.
Table 1.
Trimming of the foci seeds by Hsp104 in strong and weak [PSI+] variants
The L2888 and SY80 [PSI+] variants were grown overnight in raffinose medium and at T = 0 were transferred to galactose medium to induce overexpression of Hsp104 from the GAL1 promoter. At the indicated times, cells were fixed either before stress or after stress, and Z-stacks confocal microscopy was used to count foci. More than 300 cells were counted for each time point. The % [psi−] cells were determined from the red/white colony assay. The percent trimmed cells were calculated as the difference between cells with <5 foci before stress and <5 foci after stress.
| [PSI+] variant | Time | <5 foci before stress | <5 foci after stress | Trimmed | [psi−] |
|---|---|---|---|---|---|
| h | % | % | % | % | |
| Weak | 0 | 0 | 0 | 0 | 0 |
| Strong | 0 | 0 | 0 | 0 | 0 |
| Weak | 1 | 9 | 2 | 7 | 2 |
| Strong | 1 | 0 | 0 | 0 | 0 |
| Weak | 2 | 30 | 10 | 20 | 12 |
| Strong | 2 | 0 | 0 | 0 | 0 |
| Weak | 3 | 76 | 23 | 53 | 24 |
| Strong | 3 | 12 | 0 | 12 | 0 |
Role of the C-terminal domain of Hsp104 in the curing of [PSI+] by Hsp104 overexpression
We next examined how overexpression of different Hsp104 constructs affected the trimming and curing of the [PSI+] variants. It was previously shown that the N-terminal domain of Sc-Hsp104 is essential for curing by overexpression (21), but it has not yet been determined whether the C-terminal domain of Sc-Hsp104 is important for this curing. Specifically, we were interested in whether the acidic C-terminal extension of Hsp104, which is highly conserved among the members of the fungal Hsp104 family, has an essential role in curing by overexpression. Therefore, we made several C-terminal deletions of Sc-Hsp104, the shortest was 7 amino acids long and the longest was 52 amino acids long; the latter causes removal of the entire acidic C-terminal extension (27).
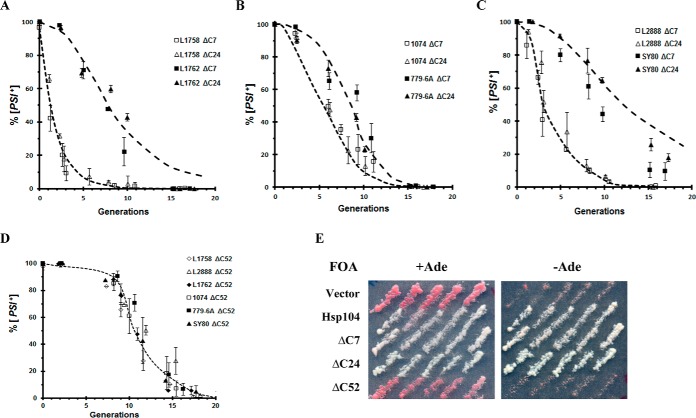
In Fig. 2, A–C, the rates of curing of the different weak and strong [PSI+] variants by overexpression of either the Sc-Hsp104ΔC7 or Sc-Hsp104ΔC24 fragments were compared with curing by overexpression of full-length Sc-Hsp104 (dashed lines). The kinetics of curing by these two truncated fragments was not significantly different from that by the full-length protein. Therefore, the C-terminal 24 amino acids of Sc-Hsp104 are not required for curing by Hsp104 overexpression nor do they affect the rate of curing by Hsp104 overexpression. As expected, overexpression of these C-terminal truncated Hsp104 fragments caused trimming of the fluorescent foci concomitant with the curing of [PSI+] (data not shown). These results are consistent with the results of Glover and co-workers (27) showing that the C-terminal 22 amino acids of Sc-Hsp104 are not required for Hsp104 to confer thermotolerance in yeast.
Figure 2.
Curing of different [PSI+] variants was measured upon overexpression of different Hsp104 fragments. A–C, curing of different [PSI+] variants was measured in yeast overexpressing Sc-Hsp104ΔC7 or Sc-Hsp104ΔC24 from the GAL1 promoter. The dashed lines are the curing curves from Fig. 1A obtained by overexpressing full-length Sc-Hsp104. D, Sc-Hsp104ΔC52 was overexpressed in different [PSI+] variants. Curing was measured at the indicated times using the red/white colony assay. E, 5-fluoroorotic acid (5-FOA) shuffle experiment using 1408 yeast in which the pJ312 plasmid, which encodes Sc-Hsp104 under the control of the S. cerevisiae HSP104 promoter on a URA3-based centromeric plasmid, was shuffled with empty vector or plasmids expressing full-length Sc-Hsp104 or the indicated C-terminal truncations of Sc-Hsp104. The full-length and fragments of Sc-Hsp104 were under the control of the Sc-HSP104 promoter. Only constructs that support [PSI+] propagation grow on plates without adenine.
Deletion of the entire C-terminal acidic extension yielded an Hsp104ΔC52 fragment that, when overexpressed in the presence of endogenous levels of full-length Hsp104, cured all of the different [PSI+] variants, but with very different kinetics from that of overexpression of full-length Sc-Hsp104. As shown in Fig. 2D, there was a long lag prior to the curing by Hsp104ΔC52, indicating that when this fragment is overexpressed, it is acting as a dominant-negative mutant similar to the Hsp104(KT) mutant (37). Because a dominant-negative Hsp104 mutant cannot sever the prion seeds, the kinetics show a lag prior to curing because it takes a number of generations to dilute out the seeds by cell division (37). Consistent with this fragment being a dominant-negative mutant, it does not propagate [PSI+] when expressed from the HSP104 promoter, whereas the Sc-Hsp104ΔC7 and Sc-Hsp104Δ24 fragments propagate [PSI+]. This was determined with the use of a shuffle assay (38, 39) in which only yeast having Hsp104 fragments that propagate [PSI+] are viable on 5-fluoroorotic acid plates without adenine (Fig. 2E). The fact that this fragment is acting as a dominant-negative mutant indicates that it must be oligomerizing, which is unexpected based on the in vitro finding from Glover and co-workers (27) that showed oligomer assembly required the C-terminal 38 amino acids of Sc-Hsp104. Oligomerization may occur in vivo as opposed to in vitro because either the fragment is expressed at higher concentrations or the endogenous full-length Sc-Hsp104 is also present in vivo. In either case, the highly acidic C-terminal region of Sc-Hsp104 is essential for the curing of [PSI+] by overexpression of Sc-Hsp104.
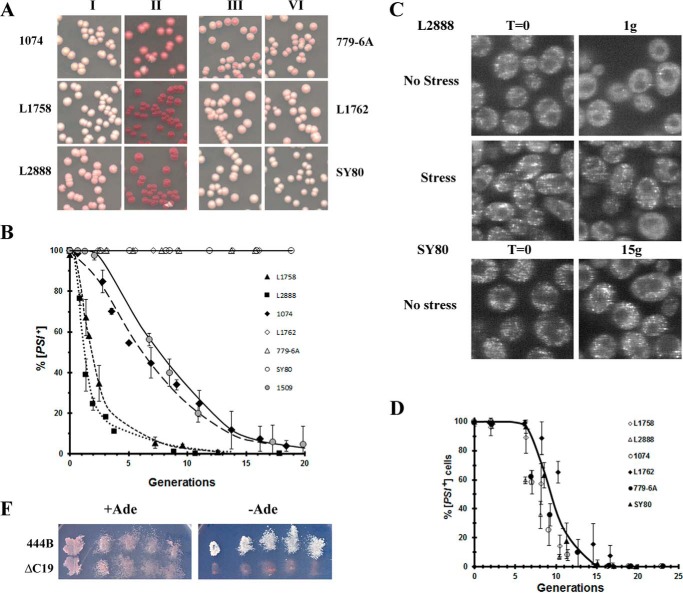
The next question is whether other Hsp104 constructs also require the C-terminal acidic extension to cure by overexpression, in particular the 444B chimera, which was previously shown to propagate [PSI+] (3, 24). The 444B chimera differs from Sc-Hsp104 in that it has the NBD2 and C-terminal domains from ClpB, the Escherichia coli paralog of Hsp104 (3, 24). Importantly, the C-terminal domain of ClpB is 51 amino acids shorter than the C-terminal domain of Sc-Hsp104, and it does not contain an acidic extension. Following overnight overexpression of 444B from the GAL1 promoter, the curing of the different [PSI+] variants was assayed by plating the cells on ½ YPD medium. The red/white colony assay (Fig. 3A) showed curing of the weaker [PSI+] variants (L1758[PSI+], L2888[PSI+], and 1074[PSI+]DM), which established that the C-terminal acidic extension is not necessary for curing by overexpression. However, overexpression of 444B did not cure the stronger [PSI+] variants (L1762[PSI+], SY80[PSI+], and 779-6A[PSI+]DM) regardless of the yeast background. Even after 15 generations in galactose, there was no significant curing of these strong variants (Fig. 3B).
Figure 3.
Overexpression of 444B cures weak [PSI+] variants but not strong [PSI+] variants. A, effect of overexpression of 444B in different [PSI+] variants. Yeast in raffinose medium were grown overnight in galactose medium to induce 444B expression. Yeast were plated on ½ YPD medium before (columns I and III) and after induction of 444B (columns II and IV). Note [psi−] yeast in the 779-6A and the 74D-694 backgrounds give different red hues on ½ YPD plates. B, curing of different [PSI+] variants was measured in yeast overexpressing 444B. 444B was expressed from the GAL1 promoter in different [PSI+] variants and plated at the indicated times. Curing of [PSI+] by overexpression of 444B was also done in the 1509 [PSI+] DM yeast. The 1509 yeast was derived by integrating 444B into the HSP104 chromosomal locus of the 1074 yeast. C, overexpression of 444B causes loss of detectable foci in weak but not strong [PSI+] variants. Fluorescence images of GFP-labeled Sup35 are of weak L2888 [PSI+] variant overexpressing 444B for one generation and strong SY80 [PSI+] variant overexpressing 444B for 15 generations. The L2888 [PSI+] yeast were imaged both before stress and after stress. D, rate of curing of different [PSI+] variants was determined upon overexpression of 444BΔC19. Curing was measured at the indicated times using the red/white colony assay. E, 5-fluoroorotic acid (5-FOA) shuffle experiment using 1408 yeast in which the pJ312 plasmid, which encodes Sc-Hsp104 under the control of the S. cerevisiae HSP104 promoter on a URA3-based centromeric plasmid., was shuffled with plasmids expressing either 444B or 444BΔC19. The 444B and 444BΔC19 were expressed using the Sc-HSP104 promoter. Only constructs that support [PSI+] propagation grow on plates without adenine.
Based on our model that trimming is required for curing of [PSI+] by Hsp104 overexpression, 444B would be expected to trim the foci of the weak, but there should be no detectable trimming of the strong [PSI+] variants. This is confirmed by the fluorescent images shown in Fig. 3C. There is no visible trimming of the fluorescent foci of the strong SY80[PSI+] variant cells after 15 generations in galactose medium, whereas 70% of the weak L2888[PSI+] variant cells showed diffuse GFP fluorescence after one generation in galactose medium, at a time when less than 20% of these cells were cured. Furthermore, consistent with trimming, in most cells the fluorescent foci reappeared after stress.
To further examine the effect of the C-terminal domain on the curing of [PSI+] by Hsp104 overexpression, we measured the time course of curing of the different weak [PSI+] variants by overexpression of 444B. Interestingly, this overexpression cured all of the weak [PSI+] variants with a time course similar to that of overexpression of Sc-Hsp104 (compare Fig. 3B with 1A). Therefore, the C-terminal domain of Sc-Hsp104 is not necessary for curing [PSI+] by Hsp104 overexpression, although its loss does reduce the trimming activity of Hsp104 so the strong variants are not cured.
Interestingly, comparison of the ClpB and Hsp104 C-terminal domains by two-protein BLAST analysis showed that these domains were 71% homologous and 51% identical. However, there was no homology between the C-terminal 52 amino acids of Sc-Hsp104 and the C-terminal 19 amino acids of ClpB. This observation that the Sc-Hsp104ΔC52 fragment acted as a dominant-negative Hsp104 mutant implied that the 444BΔC19 would also act as a dominant-negative Hsp104 mutant and indeed this was the case. When the truncated chimera was overexpressed from the GAL1 promoter, it cured all of the different [PSI+] variants with a long lag prior to curing (Fig. 3D), indicating that the 444BΔC19 fragment acts as a dominant-negative mutant, similar to Hsp104-KT. Consistent with this fragment being a dominant-negative mutant, the shuffle assay (Fig. 3E) shows that expression of 444BΔC19 from the HSP104 promoter does not propagate [PSI+] unlike the full-length chimera (3, 24). These results show that among members of the ClpB/Hsp104 protein family, the function of the C-terminal domain is conserved, although not the primary amino acid sequence.
Curing of [PSI+] by Sp-Hsp104 and Ca-Hsp104 overexpression
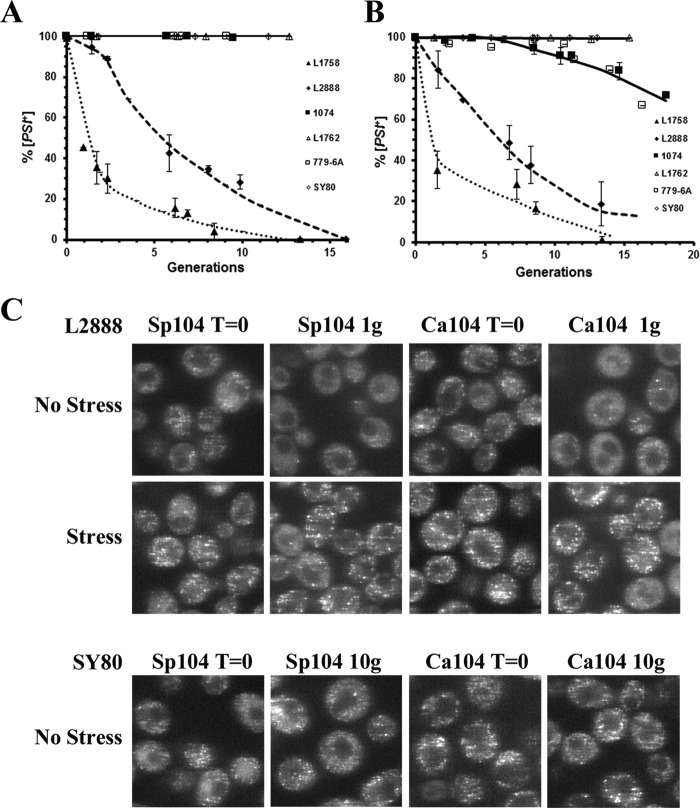
Members of the fungal Hsp104 family show a high conservation of primary amino acid sequence. However, overexpression Sp-Hsp104 did not cure [PSI+] in two studies using a single [PSI+] variant (29, 40). Because overexpression of 444B cured weak but not strong [PSI+] variants, this raised the possibility that Sp-Hsp104 might behave similarly. Therefore, we overexpressed Sp-Hsp104 in many different [PSI+] variants to determine whether curing occurred. Similar to the results obtained when 444B was overexpressed, overexpression of Sp-Hsp104 from the GAL1 promoter cured the weaker but not the stronger [PSI+] variants (Fig. 4A). The rates of curing of the weak L1758 and L2888[PSI+] variants by overexpression of Sp-Hsp104 was slower than that obtained with 444B, and unlike 444B, it did not cure the 1074 [PSI+]DM variant.
Figure 4.
Overexpression of Sp-Hsp104 or Ca-Hsp104 cures weak [PSI+] variants but not strong [PSI+] variants. A, curing of different [PSI+] variants by overexpression of Sp-Hsp104 was measured as a function of generation time. B, curing of different [PSI+] variants by overexpression of Ca-Hsp104 was measured as a function of generation time. C, overexpression of either Sp-Hsp104 or Ca-Hsp104 causes loss of detectable foci in the weak L2888 [PSI+] variant but not the strong [PSI+] SY80 variant. Fluorescence imaging of GFP-labeled Sup35 of weak L2888 [PSI+] variant overexpressing either Sp-Hsp104 or Ca-Hsp104 after overnight incubation in raffinose (T = 0) or for one generation in galactose medium is shown. The latter cells were stressed for 1 h in 5 mm guanidine. Top panel, fluorescence imaging of GFP-labeled Sup35 of strong SY80 [PSI+] variant overexpressing either Sp-Hsp104 or Ca-Hsp104 after overnight incubation in raffinose (T = 0) or for 10 generations in galactose medium is shown.
In contrast to Sp-Hsp104, overexpression of Hsp104 from C. albicans has been reported to cure [PSI+] (28). However, it is not clear whether overexpression of Ca-Hsp104 would cure the strong [PSI+] variants that we are using in this study. Therefore, Ca-Hsp104 was overexpressed from the GAL1 promoter, and curing was measured using the same [PSI+] variants used throughout this study. As shown in Fig. 4B, the curing of different [PSI+] variants by overexpression of Ca-Hsp104 was very similar to overexpression of Sp-Hsp104; it cured the weaker [PSI+] variants, but not the stronger [PSI+] variants and cured the weak variants at a slower rate than the rate obtained with 444B. Therefore, Sp-Hsp104 and Ca-Hsp104 are like 444B in that they do not cure the strong [PSI+] variants but do cure the weak variants, although somewhat more poorly than 444B.
Because overexpression of Sp-Hsp104 and Ca-Hsp104 cured the weak, but not the strong [PSI+] variants, our model for curing by overexpression predicts that these fungal Hsp104 homologs should trim the prion seeds of the weak but not the strong [PSI+] variants. This was confirmed by confocal imaging of the fluorescent foci in yeast overexpressing either Sp-Hsp104 or Ca-Hsp104. As shown in Fig. 4C, there was a marked loss of detectable foci in the weak L2888[PSI+] yeast expressing either Sp-Hsp104 or Ca-Hsp104 for one generation, a time period when the red/white colony assay showed that the yeast were still more than 90% [PSI+]. Furthermore, the foci became prominent again in the majority of the cells after stress, showing that the seeds were indeed trimmed by these Hsp104 homologs. In contrast, when the fungal Hsp104 homologs were overexpressed in the strong SY80[PSI+] yeast, fluorescent foci were visible even after 10 generations (Fig. 4C), which shows no detectable trimming. Therefore, overexpression of Sp-Hsp104 and Ca-Hsp104 can trim the prion seeds and cure the weak [PSI+] variants like overexpression of Sc-Hsp104. However, there is neither visiblet trimming of the prion seeds nor curing the strong [PSI+] variants by overexpression of these fungal Hsp104 homologs unlike overexpression of Sc-Hsp104.
Effect of decrease trimming activity on growth of strong [PSI+] variants
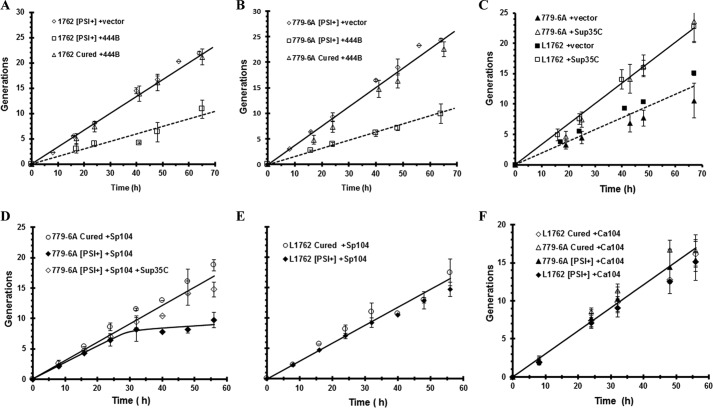
A reduction in the trimming activity of Hsp104 leads to a decrease in the soluble pool of Sup35, which may impact the growth of yeast if the soluble pool becomes limiting because Sup35 is an essential protein. In fact, this was the case when 444B was overexpressed in the strong [PSI+] variants. The growth rate was reduced by ∼50% in the L1762[PSI+] and 779-6A[PSI+]DM variants overexpressing 444B compared with [psi−] yeast overexpressing 444B or the [PSI+] vector control (Fig. 5, A and B). The slow growth was due to a reduction in the free Sup35 pool because the growth defect was rescued by expression of the soluble C-terminal domain of Sup35, the domain responsible for the translation termination activity of Sup35 (Fig. 5C). Evidently, overexpression of 444B, even in yeast expressing endogenous levels of Sc-Hsp104, causes the soluble pool of Sup35 to become a limiting factor for growth.
Figure 5.
Growth of strong [PSI+] variants was measured in yeast overexpressing 444B, Sp-Hsp104, or Ca-Hsp104. A, overexpression of 444B causes a slow growth phenotype in 1762 [PSI+] variant. B, overexpression of 444B causes a slow growth phenotype in 779-6A [PSI+]DM variant. Yeast growth in galactose medium as a function of time is shown for [PSI+] yeast expressing 444B (closed squares), [PSI+] yeast with empty vector (open diamonds), and cured yeast expressing 444B (open triangles). C, overexpression of 444B causes a slow growth phenotype in strong [PSI+] variants due to sequestration of Sup35. 444B was overexpressed in either the 779-6A [PSI+]DM variant or the L1762 [PSI+] variant. Growth was measured in [PSI+] cells expressing 444B containing an empty vector plasmid (solid symbols) or a plasmid expressing the C-terminal fragment of Sup35 (open symbols). D, Sp-Hsp104 was overexpressed in yeast with the 779-6A [PSI+]DM variant (solid diamonds), in cured yeast (open circles), or in yeast expressing the Sup35 C-terminal domain of Sup35 (open diamonds). E, Sp-Hsp104 was overexpressed in yeast with the 1762 [PSI+] variant (solid symbols) or in cured yeast (open symbols). F, Ca-Hsp104 was overexpressed in yeast with the 779-6A [PSI+]DM variant (solid triangles) or in cured yeast (open triangles) and in yeast with the 1762 [PSI+] variant (solid diamonds) or in cured yeast (open diamonds). Growth was monitored by optical density, and cells were constantly diluted to maintain them in early log phase. The different [PSI+] variants were cured in guanidine.
In general, overexpression of these fungal Hsp104 homologs had less effect on the growth of the strong [PSI+] variants than 444B even though they had reduced trimming activity in comparison with Sc-Hsp104. After overnight induction of Sp-Hsp104 expression, the 779-6A[PSI+]DM variant showed a reduction in growth rate, which was rescued by expressing the C-terminal Sup35 fragment (Fig. 5D). However, there was no significant growth defect from overexpressing Sp-Hsp104 in the L1762[PSI+] variant (Fig. 5E). Similarly, no significant growth defect was observed in yeast overexpressing Ca-Hsp104 in either the L1762[PSI+] or the 779-6A[PSI+]DM variant (Fig. 5F). Therefore, 444B causes a greater reduction in the free Sup35 pool than these fungal Hsp104 homologs. This could be due to a difference in the extent of trimming and/or interactions of Hsp104 with other chaperones altering the free Sup35 pool.
Trimming of the prion seeds by low levels of Sc-Hsp104
Our results show that curing of [PSI+] by Hsp104 overexpression is dependent on both the [PSI+] variant and the source of the fungal Hsp104 homolog. Specifically, Sc-Hsp104 has greater trimming activity than Sp-Hsp104 and Ca-Hsp104, and therefore, unlike these Hsp104 homologs, its overexpression can cure the strong [PSI+] variants. The question then arises as to whether a lower level of Sc-Hsp104 would act like overexpression of Sp-Hsp104 and Ca-Hsp104 in that it would neither trim nor cure the strong [PSI+] variants. To achieve low levels of Hsp104 overexpression, we used the TET-ON promoter, which induces about 3-fold overexpression of Sc-Hsp104, compared with the GAL1 promoter, which induces 10–20-fold overexpression (14). This relatively low level of Hsp104 overexpression was previously shown to trim the prion seeds and cures the relatively weak 1074 [PSI+] variant (14), as well as the weak L2888[PSI+] variant (Fig. 6, A and B).
Figure 6.
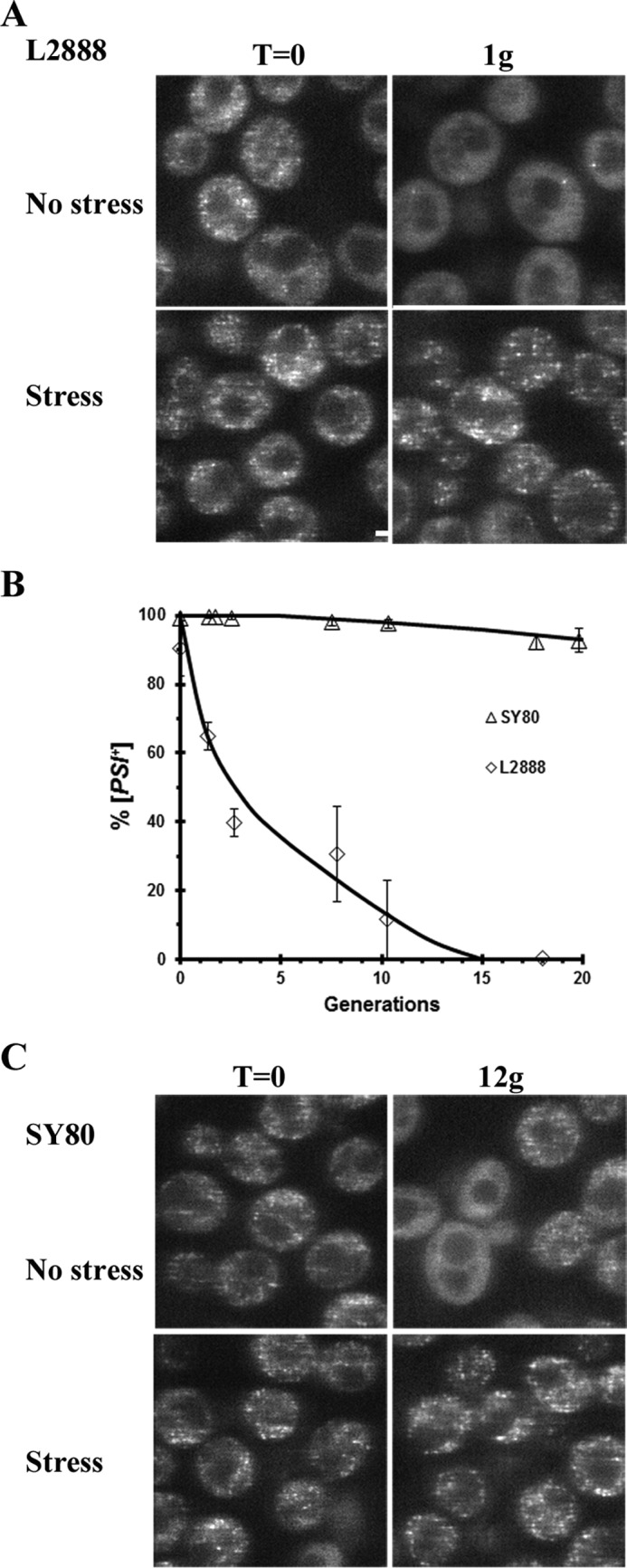
Low levels of Sc-Hsp104 cures and trims the weak L2888 [PSI+] variant but only trims the strong SY80 [PSI+] variant. A, GFP-labeled Sup35 was imaged in the weak L2888 [PSI+] overexpressing Sc-Hsp104 from the TET-ON promoter after 1 h of Sc-Hsp104 induction by 10 μg/ml doxycycline. B, curing of the weak L2888 [PSI+] variant and strong SY80 [PSI+] variant was measured as a function of generation time when Sc-Hsp104 is overexpressed from the TET-ON promoter. C, GFP-labeled Sup35 was imaged in the strong SY80 [PSI+] variant overexpressing Sc-Hsp104 from the TET-ON promoter. The SY80 [PSI+] cells were imaged 12 generations after induction of Sc-Hsp104 overexpression. Imaging was done in both non-stressed and stressed cells.
Unlike the weak L2888 [PSI+] variants, the strong SY80[PSI+] variant does not show significant curing by overexpression of low levels of Sc-Hsp104. Even overexpressing low levels of Hsp104 for 12 generations, the yeast were still 95% [PSI+] based on the plating assay (Fig. 6B). Given these results, we expected little or no trimming of the foci, but surprisingly, roughly half of the cells either had fine foci or no foci, whereas in the other half of the population, the foci were clearly evident. This heterogeneity in yeast was detected after four generations of Hsp104 induction and persisted even after 10 generations of Hsp104 overexpression. Consistent with the lack of curing, prominent foci were visible once the cells were stressed (Fig. 6A). It is not clear why only approximately half of the cells showed trimming, but one possibility is that the level of Hsp104 overexpression differs in different cells, perhaps due to the variation in the number of centromeric plasmids expressing Hsp104 per cell (41). Interestingly, the fact that there was no curing even though there was trimming suggests that trimming is a reversible reaction. If trimming were an irreversible reaction, all of the cells would have cured over time because the slow trimming of the seeds would constantly decrease the size of the seeds. Therefore, in addition to Sup35 monomers being trimmed from the prion seeds, there must also be rebinding of the soluble Sup35 in the cytosol to the seeds. This, in turn, means that the trimming does not go to completion, resulting in the lack of curing of the [PSI+] prion.
Discussion
To extend our understanding of the mechanism of [PSI+] curing by Hsp104 overexpression, we examined the ability of different Hsp104 homologs at varying levels of overexpression to cure a wide range of [PSI+] variants. In our previous study (14), we found no significant difference in the fraction of cured mother and daughter cells when [PSI+] was partially cured by Hsp104 overexpression. These results do not fit with a model in which curing is caused by asymmetric segregation, as was recently proposed by Tuite and co-workers (42), based on their measurement of the number of prion seeds in mother and daughter cells. They found a subpopulation of cells had an unequal distribution of seeds between mother and daughter cells. However, this will not lead to a marked increase in the fraction of cured daughter cells because even a cell that is left with only one seed will remain [PSI+]. Instead, we observed that curing of [PSI+] by Hsp104 overexpression was dependent on the trimming of the prion seeds by Hsp104, which decreased the size of the seeds without changing their number. Specifically, we found that Hsp104 mutants that did not trim the foci, such as the Hsp104(T160M) mutant, did not cure [PSI+] when overexpressed (14). Consistent with these results, in vitro studies of Shorter and co-workers (43) showed that low concentrations of Hsp104 caused fragmentation of the amyloid Sup35 fibers, which in turn generated new prion seeds. In contrast, high concentrations of Hsp104 caused dissolution of the fibers into noninfectious aggregates and soluble protein (15, 43). Interestingly, unlike full-length Hsp104, a high concentration of an N-terminal Hsp104 deletion mutant, a mutant that does not cure [PSI+] when overexpressed (21), caused only fragmentation and not dissolution of the Sup35 fibers. These results are consistent with fragmentation and dissolution being the in vitro analogs of severing and trimming, respectively.
We first examined the curing of [PSI+] by Hsp104 overexpression in different [PSI+] variants. Overexpression of Sc-Hsp104 cured weak [PSI+] variants faster than strong [PSI+] variants, as was shown previously (31). [PSI+] variants differ in their biophysical properties such as thermal stability, indicating that the variants have different conformations (32). This has been confirmed by structural studies showing that the [PSI+] variants differ in the location of the turns between the β-strands in their amyloid core (33, 34). Because strong [PSI+] variants cure more slowly than weak [PSI+] variants, this suggests that the conformational differences between the weak and strong variants not only affect the severing but also the trimming activity of Hsp104; an increased rate of trimming of the weak variants would be expected based on their faster curing by Hsp104 overexpression. This was in fact the case. Consistent with the difference in the rates of curing between the weak and strong [PSI+] variants by Hsp104 overexpression, Hsp104 trimmed the fluorescent foci of the weak [PSI+] variants faster than foci of the strong [PSI+] variants. 2 h after induction of Hsp104 overexpression by galactose, a time when the yeast were still mostly [PSI+], about one-fifth of the yeast had trimmed foci in the weak [PSI+] variant, while it took 8 h before a comparable percentage of trimming occurred in the strong [PSI+] variant. The observation that there is faster trimming of the foci in the weak than in the strong [PSI+] variant may be due to variant-dependent binding of Hsp104 to the ends of the fibers. In fact, Lindquist and co-workers (44) found at least two classes of binding sites for Hsp104 on amyloid fibers composed of the NM-domains of Sup35. One of the binding sites is dependent on the strength of the [PSI+] variant with weak variants containing a greater fraction of such sites. Another possibility is soluble Sup35 binds with greater affinity to fibers in the strong than in the weak conformation.
A second line of evidence that Hsp104 overexpression trims the seeds of the weak [PSI+] variants faster than the strong [PSI+] variants comes from data showing that the fluorescent foci of the weak [PSI+] variants are trimmed at lower levels of Hsp104 overexpression than the foci of the strong [PSI+] variant. Specifically, the weak [PSI+] variant showed trimming of all of its prion seeds, followed by [PSI+] curing when Sc-Hsp104 was overexpressed only ∼3-fold from the TET-ON promoter (14). In marked contrast, at these low levels of Sc-Hsp104 overexpression, the fluorescent foci showed a varied appearance in the strong [PSI+] variant. In about half of the population, the foci remained prominent as if no trimming occurred, whereas in the rest of the population the foci were barely detectable above the background fluorescence. As discussed under “Results,” this heterogeneity is probably due to a variable number (1–3) of centromeric plasmids per cell (41). Interestingly, even cells with barely detectable foci failed to cure over a long time, which suggests that trimming is a reversible reaction. Not only is Sup35 dissociating from the foci, but there must also be rebinding of Sup35 from the soluble pool. Otherwise, the strong [PSI+] variants would slowly cure over time as the seeds were completely trimmed by Hsp104. The reversibility of trimming is also shown by the observation that the trimmed foci become prominent when the cells are stressed. Again, this must be due to Sup35 monomers rebinding to as well as dissociating from the prion seeds. Therefore, when trimming was not detected when 444B, Sp-Hsp104, and Ca-Hsp104 were overexpressed in the strong [PSI+] variants, there could still be dissociation of monomers from the fiber ends, but the rebinding occurs at a comparable rate as the rate of dissociation.
A third line of evidence that, like curing, the trimming activity of Hsp104 is dependent on the [PSI+] variant comes from overexpression of different Hsp104 constructs. Our data show that the 444B chimera retains its trimming activity even though it lacks a C-terminal acidic extension, a highly conserved region of the Hsp104 fungal homologs that is important for oligomer assembly (27). Overexpression of 444B trimmed the foci of the weak [PSI+] variants, and this trimming was followed by [PSI+] curing with a similar time course as occurred when full-length Sc-Hsp104 was overexpressed. Therefore, the C-terminal acidic extension is not necessary for the trimming or curing of the weak [PSI+] variants. However, the strong [PSI+] variant showed no net trimming of the foci and no curing when 444B was overexpressed. Similarly, although overexpression of the fungal Hsp104 homologs from S. pombe and C. albicans both trimmed the foci of the weak [PSI+] variants and cured these variants, there was no net trimming of the foci or curing of the strong [PSI+] variants. Importantly, prior to this study, the curing of [PSI+] by the Hsp104 homologs was considered an all or none process (28–30), i.e. overexpression of Sc-Hsp104 and Ca-Hsp104 was considered to cure all of the [PSI+] variants, whereas overexpression of Sp-Hsp104 did not cure any of the [PSI+] variants. However, we have now established that Ca-Hsp104 and Sp-Hsp104 cure the weak but not the strong [PSI+] variants. There is the question of whether the GAL1 promoter is expressing sufficient levels of these fungal Hsp104 homologs and 444B. Importantly, curing is not the only phenotype that we observed; both 444B and Sp-Hsp104 produced a slow growth phenotype when overexpressed in strong [PSI+] variants, which shows sufficient expression levels to produce a phenotype. However, we did not observe a phenotype with Ca-Hsp104 overexpression in strong [PSI+] variants, but it seems highly unlikely that it is not expressed from the GAL1 promoter at comparable levels to the other Hsp104 paralogs because these fungal Hsp104 proteins all show high homology.
Although the curing of [PSI+] by Hsp104 overexpression is dependent on the trimming activity of Hsp104, other factors play a role in curing by overexpression. For example, it has been shown that the curing of [PSI+] by Hsp104 overexpression is also dependent on the Hsp90 chaperone and the Sti1 cochaperone (18, 19), but the function of these proteins in the curing process has yet to be determined. It is interesting that the [PSI+] prion is the only yeast prion that is cured by Hsp104 overexpression. Because Sup35 is an essential protein, the ability of Hsp104 to trim the [PSI+] prion seeds may have evolved to maintain a sufficient pool of soluble Sup35, i.e. as different [PSI+] variants are propagated, the steady-state number of seeds and the free pool of Sup35 is maintained in the cell by the severing and trimming activities of Hsp104. In fact, when trimming was markedly reduced, as occurred when 444B was overexpressed in the strong [PSI+] variants, a slow growth defect was obtained due to a lack of soluble Sup35 protein. The fact that a low level of Hsp104 overexpression is sufficient to cure the weak [PSI+] variants raises the possibility that when very weak [PSI+] variants spontaneously arise, they are cured by the endogenous level of Hsp104. In contrast, very strong [PSI+] variants have been shown to be suicidal due to the lack of a free Sup35 pool (45). Therefore, there may only be a narrow window of [PSI+] variants between weak and strong variants that are mitotically stable in yeast, which, in turn, may explain the extremely low incidence of [PSI+] in wild yeast isolates (46).
Experimental procedures
Yeast strains
The yeast strains were either derivatives of 779-6A (MATa, kar1-1, SUQ5, ade2-1, his3Δ202, leu2Δ1, trp1Δ63, ura3-52) or the 74D-694 (MATa ade1-14, trp1-289 his3-Δ200 ura3-52 leu2-3,112). Yeast expressing the GFP-labeled version of Sup35 (NGMC) instead of the wild-type Sup protein (NMC) was integrated into the genomic locus of these two strains. The following yeast expressed NGMC instead of NMC: 1074 yeast was derived from the 779-6A background strain (13); L2888 and SY80 were derived from the 74D-696 strains (47, 48). The 1509 yeast strain was derived from the 1074 by integrating 444B into the chromosomal HSP104 locus. Yeast strain 1408, derived from 779-6A, contains a deletion of the HSP104 gene (39). This strain carries the pJ312 plasmid, which encodes Sc-Hsp104 under the control of the S. cerevisiae HSP104 promoter on a URA3-based centromeric plasmid.
Prions
All yeast strains were [PIN+]. The different [PSI+] variants in the 74D-694 yeast background were as follows: L1758, L2888, L1762, and SY80 (47–49). The [PSI+]DM prion present in the 779-6A background was maintained in the 1074 and 1509 yeast (50).
Plasmids
A pRS315 centromeric plasmid was used to express the full-length Sc-Hsp104 and Sc-Hsp104 fragments from the GAL1 promoter. The GAL1 promoter was used to overexpress 444B, SpHsp104, and Ca Hsp104 using the following vectors: pMR180 for 444B (24); a pRS415 vector for SpHsp104 (29); and a pRS314 vector for Ca-Hsp104. Plasmid pMR117 and pMR170 was used to express Sc-Hsp104 and Sp-Hsp104 from the S. cerevisiae HSP104 promoter, respectively (29). The TET-ON promoter was used to express relatively low levels of Sc-Hsp104 (14). The C terminus of Sup35 was expressed from a centromeric plasmid (pRPM02) under the control of the TET-OFF promoter (45). For the shuffle experiments, we used plasmid pMR117 encoding Sc-HSP104 (39), pMR122 encoding 444B (39), or the indicated engineered fragments. The latter proteins were expressed from the Sc-HSP104 promoter on a centromeric plasmid. All truncations were made using the QuikChange Lightning Site-directed Mutagenesis kit (Agilent) and then sequenced (Macrogen).
Measuring the rate of [PSI+] curing
Our strains use the white/red and adenine-independent/dependent phenotypes to monitor [PSI+]. The nonsense mutation in either the ade2-1 or ade1-14 allows for growth in the absence of adenine and confers white/pink colony color on media containing a limiting concentration of adenine. The [psi−] cells are adenine auxotrophs and appear red on limiting adenine. Curing by overexpression of Hsp104 was done by growing the yeast overnight in raffinose medium prior to addition of galactose medium as described previously (14) Yeast growth was monitored by the absorbance at 600 nm and the plated on ½ YPD. Colonies that had any white were scored as [PSI+], whereas completely red colonies were scored as [psi−]. Induction of Sc-Hsp104 expression from the TET-ON promoter was by addition of 10 μg/ml doxycycline (Sigma). All experiments were repeated a minimum of three times, and the data were then plotted as the average and standard deviation for each time point.
Imaging of GFP-labeled Sup35 in yeast
Cells were imaged on a Zeiss live confocal microscope using a piezoelectric stage to obtain Z-stacks with a ×100 objective in 8-well 25-mm2 chambered coverslips (Lab-Tek, Rochester, NY). Z-stack confocal images were acquired (10 slices at 0.6 μm intervals) of all samples using the same confocal settings. Cells were stressed by addition of 5 mm guanidine for 1.5 h (14). When indicated in the figure legends, yeast were fixed in 4% paraformaldehyde (Sigma). Super-resolution images were obtained with the Delta Vision OMX microscope (GE Healthcare) using a ×60, 1.42 NA objective.
Author contributions
X. Z., R. R., R. S., J. A., S. S., and K. C. performed the experiments and analyzed the data. E. E. provided critical advice regarding the design of the project and the revision of the manuscript. L. G. supervised the project and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Susan Liebman (University of Nevada) for all the different [PSI+] variants in the 74D-694 yeast background; Dr. Daniel Masison (NIDDK, National Institutes of Health) for making yeast strains; Dr. Reed Wickner (NIDDK, National Institutes of Health) for the Sup35-C-terminal fragment expression vector; Dr. Michael Reidy (NIDDK, National Institutes of Health) for the 444B expression vector; and Dr. Mick Tuite (University of Kent, Canterbury, Kent, UK) for the Ca-Hsp104 clone. We also thank Dr. Xufeng Wu and the NHLBI microscopy core for assistance.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- NBD
- nucleotide-binding domain.
References
- 1. Doyle S. M., and Wickner S. (2009) Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34, 40–48 [DOI] [PubMed] [Google Scholar]
- 2. Wickner R. B., Shewmaker F. P., Bateman D. A., Edskes H. K., Gorkovskiy A., Dayani Y., and Bezsonov E. E. (2015) Yeast prions: structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 79, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tipton K. A., Verges K. J., and Weissman J. S. (2008) In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 32, 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferreira P. C., Ness F., Edwards S. R., Cox B. S., and Tuite M. F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369 [DOI] [PubMed] [Google Scholar]
- 5. Eaglestone S. S., Ruddock L. W., Cox B. S., and Tuite M. F. (2000) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 240–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox B., Ness F., and Tuite M. (2003) Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165, 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., and Liebman S. W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 268, 880–884 [DOI] [PubMed] [Google Scholar]
- 8. Patino M. M., Liu J. J., Glover J. R., and Lindquist S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622–626 [DOI] [PubMed] [Google Scholar]
- 9. Paushkin S. V., Kushnirov V. V., Smirnov V. N., and Ter-Avanesyan M. D. (1996) Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 10. Greene L. E., Park Y. N., Masison D. C., and Eisenberg E. (2009) Application of GFP-labeling to study prions in yeast. Protein Pept. Lett. 16, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Y. X., Masison D. C., Eisenberg E., and Greene L. E. (2006) Application of photobleaching for measuring diffusion of prion proteins in cytosol of yeast cells. Methods 39, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satpute-Krishnan P., and Serio T. R. (2005) Prion protein remodelling confers an immediate phenotypic switch. Nature 437, 262–265 [DOI] [PubMed] [Google Scholar]
- 13. Park Y. N., Morales D., Rubinson E. H., Masison D., Eisenberg E., and Greene L. E. (2012) Differences in the curing of [PSI+] prion by various methods of Hsp104 inactivation. PLoS ONE 7, e37692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park Y. N., Zhao X., Yim Y. I., Todor H., Ellerbrock R., Reidy M., Eisenberg E., Masison D. C., and Greene L. E. (2014) Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot. Cell 13, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shorter J., and Lindquist S. (2006) Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23, 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chernoff Y. O., Newnam G. P., Kumar J., Allen K., and Zink A. D. (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19, 8103–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Newnam G. P., Wegrzyn R. D., Lindquist S. L., and Chernoff Y. O. (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19, 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reidy M., and Masison D. C. (2010) Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] Prions by Hsp104. Mol. Cell. Biol. 30, 3542–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moosavi B., Wongwigkarn J., and Tuite M. F. (2010) Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 27, 167–179 [DOI] [PubMed] [Google Scholar]
- 20. Kiktev D. A., Patterson J. C., Müller S., Bariar B., Pan T., and Chernoff Y. O. (2012) Regulation of chaperone effects on a yeast prion by cochaperone sgt2. Mol. Cell. Biol. 32, 4960–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hung G.-C., and Masison D. C. (2006) N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., and Tsai F. T. (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 [DOI] [PubMed] [Google Scholar]
- 23. Sielaff B., and Tsai F. T. (2010) The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 402, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miot M., Reidy M., Doyle S. M., Hoskins J. R., Johnston D. M., Genest O., Vitery M. C., Masison D. C., and Wickner S. (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. U.S.A. 108, 6915–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cashikar A. G., Schirmer E. C., Hattendorf D. A., Glover J. R., Ramakrishnan M. S., Ware D. M., and Lindquist S. L. (2002) Defining a pathway of communication from the C-terminal peptide-binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760 [DOI] [PubMed] [Google Scholar]
- 26. Tkach J. M., and Glover J. R. (2004) Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J. Biol. Chem. 279, 35692–35701 [DOI] [PubMed] [Google Scholar]
- 27. Mackay R. G., Helsen C. W., Tkach J. M., and Glover J. R. (2008) The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry 47, 1918–1927 [DOI] [PubMed] [Google Scholar]
- 28. Zenthon J. F., Ness F., Cox B., and Tuite M. F. (2006) The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot. Cell 5, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reidy M., Sharma R., and Masison D. C. (2013) Schizosaccharomyces pombe disaggregation machinery chaperones support Saccharomyces cerevisiae growth and prion propagation. Eukaryot. Cell 12, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Helsen C. W., and Glover J. R. (2012) Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J. Biol. Chem. 287, 542–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., and Liebman S. W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanaka M., Chien P., Naber N., Cooke R., and Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 33. Gorkovskiy A., Thurber K. R., Tycko R., and Wickner R. B. (2014) Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. U.S.A. 111, E4615–E4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong S. H., and King C. Y. (2015) Amino acid proximities in two Sup35 prion strains revealed by chemical cross-linking. J. Biol. Chem. 290, 25062–25071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toyama B. H., Kelly M. J., Gross J. D., and Weissman J. S. (2007) The structural basis of yeast prion strain variants. Nature 449, 233–237 [DOI] [PubMed] [Google Scholar]
- 36. Tanaka M., Collins S. R., Toyama B. H., and Weissman J. S. (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 37. Ness F., Ferreira P., Cox B. S., and Tuite M. F. (2002) Guanidine hydrochloride inhibits the generation of prion “Seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22, 5593–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boeke J. D., Trueheart J., Natsoulis G., and Fink G. R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 39. Reidy M., Miot M., and Masison D. C. (2012) Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics 192, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sénéchal P., Arseneault G., Leroux A., Lindquist S., and Rokeach L. A. (2009) The Schizosaccharomyces pombe Hsp104 disaggregase is unable to propagate the [PSI] prion. PLoS ONE 4, e6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jordan B. E., Mount R. C., and Hadfield C. (1996) Determination of plasmid copy number in yeast. Methods Mol. Biol. 53, 193–203 [DOI] [PubMed] [Google Scholar]
- 42. Ness F., Cox B. S., Wongwigkarn J., Naeimi W. R., and Tuite M. F. (2017) Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartition of [PSI+] propagons. Mol. Microbiol. 104, 125–143 [DOI] [PubMed] [Google Scholar]
- 43. Sweeny E. A., Jackrel M. E., Go M. S., Sochor M. A., Razzo B. M., DeSantis M. E., Gupta K., and Shorter J. (2015) The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57, 836–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frederick K. K., Debelouchina G. T., Kayatekin C., Dorminy T., Jacavone A. C., Griffin R. G., and Lindquist S. (2014) Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol. 21, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McGlinchey R. P., Kryndushkin D., and Wickner R. B. (2011) Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. U.S.A. 108, 5337–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Halfmann R., Jarosz D. F., Jones S. K., Chang A., Lancaster A. K., and Lindquist S. (2012) Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mathur V., Hong J. Y., and Liebman S. W. (2009) Ssa1 overexpression and [PIN+] variants cure [PSI+] by dilution of aggregates. J. Mol. Biol. 390, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Satpute-Krishnan P., Langseth S. X., and Serio T. R. (2007) Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 5, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma J., and Liebman S. W. (2012) [PSI(+)] prion variant establishment in yeast. Mol. Microbiol. 86, 866–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wickner R. B., Kryndushkin D., Shewmaker F., McGlinchey R., and Edskes H. K. (2012) Study of amyloids using yeast. Methods Mol. Biol. 849, 321–346 [DOI] [PMC free article] [PubMed] [Google Scholar]