Abstract
Coordinated regulation of innate immune responses is necessary in all metazoans. In Drosophila the Imd pathway detects Gram-negative bacterial infections through recognition of diaminopimelic acid (DAP)-type peptidoglycan and activation of the NF-κB precursor Relish, which drives robust antimicrobial peptide gene expression. Imd is a receptor-proximal adaptor protein homologous to mammalian RIP1 that is regulated by proteolytic cleavage and Lys-63-polyubiquitination. However, the precise events and molecular mechanisms that control the post-translational modification of Imd remain unclear. Here, we demonstrate that Imd is rapidly Lys-63-polyubiquitinated at lysine residues 137 and 153 by the sequential action of two E2 enzymes, Ubc5 and Ubc13-Uev1a, in conjunction with the E3 ligase Diap2. Lys-63-ubiquitination activates the TGFβ-activated kinase (Tak1), which feeds back to phosphorylate Imd, triggering the removal of Lys-63 chains and the addition of Lys-48 polyubiquitin. This ubiquitin-editing process results in the proteasomal degradation of Imd, which we propose functions to restore homeostasis to the Drosophila immune response.
Keywords: antimicrobial peptide (AMP), Drosophila, mass spectrometry (MS), NF-κB (NF-κB), phosphorylation, ubiquitylation (ubiquitination), Imd, Tak1, ubiquitin editing
Introduction
The Imd pathway is largely responsible for the robust antimicrobial peptide (AMP)2 induction observed after septic bacterial infection in Drosophila and thus is critically important for defense against invading pathogens (1, 2). This pathway is triggered by diaminopimelic acid (DAP)-type peptidoglycan (PGN) from bacterial cell walls. Immune-responsive cells recognize PGN through two peptidoglycan recognition proteins (PGRPs): the cell surface receptor PGRP-LC and the cytosolic receptor PGRP-LE (3). PGN recognition by these receptors leads to the cleavage of Imd, a key adaptor protein in this pathway, by the caspase 8-like protease Dredd (4). Once cleaved, Imd associates with the E3 ligase Diap2 and is rapidly ubiquitinated. This modification leads to the activation of the Drosophila homologs of the Tak1 and IKK (5) and ultimately to the activation of the NF-κB precursor Relish and induction of AMP genes expression.
Ubiquitination is a critical regulator of innate immune signaling, especially NF-κB pathways in mammals and insects. The number and topology of ubiquitin conjugations determines the fate of substrate proteins. For example, Lys-48-polyubiquitination targets proteins to proteasome for degradation (6), whereas Lys-63-polyubiquitin chains often function as scaffolds in signaling pathways, recruiting and activating downstream factors (7, 8). Ubiquitination requires the sequential action of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). Whereas E3s are critical for substrate recognition, E2s play central role in determining chain topology (9, 10). In the Drosophila immune response, Lys-63-polyubiquitination of Imd plays a crucial role relaying signals to downstream kinases. In particular, we previously demonstrated that Imd polyubiquitination requires the E3 ligase Diap2 as well as the E2 ubiquitin-conjugating enzymes Ubc5 (Effete) and the Ubc13 (Bendless)-Uev1a complex (5). Effete, also known as UbcD1, is a Drosophila member of the yeast Ubc4/5 family along with the human E2s in the Ube2D (UbcH5) group (11–13). Bendless (UbcD3) is the Drosophila homolog of mammalian Ubc13/Ube2N, which dimerizes with ubiquitin enzyme variants (Uevs) to generate Lys-63 chains (12–14). However, the molecular mechanisms by which these two E2s function together in Imd polyubiquitination remain unclear.
The MAP3 kinase Tak1, complexed with the Drosophila Tab2 homolog (15), is known to function downstream of ubiquitinated Imd (5). Tab2 contains a conserved Lys-63-polyubiquitin binding domain (16, 17), suggesting activation of Tak1/Tab2 complex by association with Lys-63-polyubiquitinated Imd. Tak1 is required for activation of Drosophila IKK complex, which is essential for activation of NF-κB precursor Relish (18–20), the key transcription factor leading to induction of AMP genes.
In addition to ubiquitination, phosphorylation is another common type of post-translational modification observed in signaling pathways. Signal transduction often relies on cascades of kinase activation and phosphorylation. Previous research suggests that Imd is also phosphorylated upon immune stimulation (5). However, it is still unknown what kinases are responsible for Imd phosphorylation or what functional relevance this modification may have for immune signaling and defense.
Before this work it was demonstrated that Imd is polyubiquitinated and phosphorylated, yet no connection has been made between these types of post-translational modifications. Here we confirm that Imd is rapidly cleaved and Lys-63-polyubiquitinated upon immune stimulation and further demonstrate that this is followed by removal, or deubiquitination, of the Lys-63 chains and the addition of Lys-48-polyubiquitin. This ubiquitin editing strongly correlates with Imd phosphorylation and requires the Lys-63-activated MAP3K Tak1, creating a feedback loop that culminates in the proteasomal destruction of Imd.
Results
Imd is Lys-63- and Lys-48-polyubiquitinated as well as phosphorylated upon immune stimulation
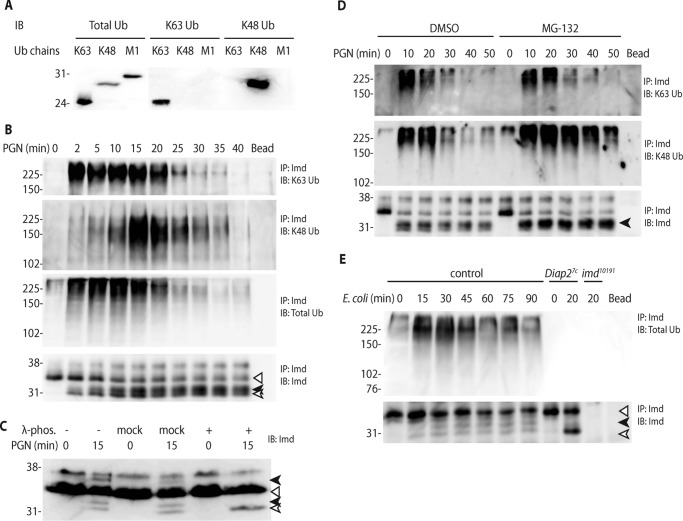
Previously, we have shown that PGN stimulation leads to the caspase-dependent cleavage and Lys-63-polyubiquitnation of the adaptor protein Imd (5). Our earlier work suggests that Imd was Lys-63-polyubiquitinated but not Lys-48-conjugated. On the other hand, another report suggested that Imd is modified by both types of polyubiquitin chains (21). To examine the post-translational modifications of Imd more closely, a new assay was developed whereby a single Imd immunoprecipitation could be examined for both Lys-63 and Lys-48 chains with the use of chain-specific antibodies (22) (Fig. 1A). Taking advantage of the immune-inducible Drosophila S2* cell line (23, 24), we stimulated these cells with PGN across a time course from 0 to 40 min. These assays showed rapid (within 2 min) Lys-63-polyubiquitination followed by intensive Lys-48 polyubiquitination peaking at 15 min, both of which progressively decreased and disappeared at later time points (Fig. 1B). The antibody detecting total ubiquitin showed a pattern that combined the Lys-63 and Lys-48 kinetics but slightly overrepresenting of Lys-63 modification, consistent with hypersensitive detection of synthetic Lys-63 Ub chains by this antibody (Fig. 1A). In addition to full-length (Fig. 1, open triangle) and cleaved Imd (open arrow), other inducible forms of Imd were observed (black arrows). λ-Phosphatase treatment combined with higher resolution SDS-PAGE demonstrated that these particular forms of Imd are phosphorylated (Fig. 1C). In these analyses, a fast migrating phospho-form is always observed (Fig. 1B and lower black arrow in Fig. 1C), which presumably represents phosphorylation of cleaved Imd. Often, a high molecular weight phospho-form is also observed, presumably due to phosphorylation of full-length Imd (upper black arrow in Fig. 1C).
Figure 1.
Imd is Lys-63- and Lys-48-polyubiquitinated as well as phosphorylated upon immune stimulation. A, specificity of ubiquitin chain antibodies. 100 ng of synthetic Lys-63, Lys-48, and Met-1 tetra-ubiquitin chains were each immunoblotted (IB) with antibody specific to total ubiquitin, Lys-63 ubiquitin, and Lys-48 ubiquitin. B, two types of Imd ubiquitination induced by PGN stimulation. S2* cells were stimulated with PGN for the indicated times. Imd was immunoprecipitated (IP) from whole cell lysate, and immunoblotted with antibody specific to Lys-63, Lys-48, or total ubiquitin as well as to Imd. C, Imd phosphorylation induced by PGN stimulation. Lysates from S2* cells stimulated with PGN were treated with λ-phosphatase as indicated before analysis by immunoblot. D, proteasomal degradation of Imd triggered by Lys-48 modification. S2* cells were treated with vehicle control (DMSO) or the proteasome inhibitor MG-132 before PGN stimulation for indicated times. Imd modifications were assayed as above. Dipt, diptericin. E, Imd modifications in adult flies. Control (w1118), Diap27c, and imd10191 adult flies were challenged with live E. coli for the indicated times. Endogenous Imd was assayed as above. All blots are representative of at least three independent experiments. In all cases, the open triangle marks the unmodified full-length Imd, the open arrow marks cleaved Imd, and the black arrow marks phosphorylated Imd.
Lys-48-polyubiquitination traffics conjugated proteins to the proteasome and/or autophagolysosome for degradation. To determine whether Lys-48 conjugation is involved in the turnover of Imd through the proteasome pathway, S2* cells were treated with MG-132, a proteasome inhibitor, before stimulation with PGN (Fig. 1D). This treatment led to persistent Imd Lys-48-polyubiquitination but little change in the Lys-63 kinetics. Interestingly, the phosphorylated Imd also accumulated to higher levels in the MG-132-treated cells (bottom panel, Fig. 1D). These results indicate that Imd is degraded by proteasome after conjugation of Lys-48-polyubiquitin chains and suggest that phosphorylation might be linked to these ubiquitination/degradation events. On the other hand, blocking of autophagy by Atg1 RNAi did not cause any Imd accumulation (data not shown), suggesting that autophagy is not part of the degradation machinery for Imd.
To further analyze these patterns in vivo, adult animals were infected with Escherichia coli, and whole animal lysates were assayed by Imd immunoprecipitation and immunoblotting for post-translational modifications. Diap2 deficient (Diap27c) and Imd null (imd10191) flies were included for comparison, relative to a control strain. Similar to S2* cells, Imd exhibited clear and transient infection-induced cleavage, ubiquitination, and phosphorylation in adult flies, whereas Diap2-deficient flies showed increased accumulation of cleaved Imd with a complete absence of ubiquitination or phosphorylation (Fig. 1E). The Lys-48- and Lys-63-specific antibodies did not have the sensitivity required for use in these whole animal assays (data not shown). Together, these cell-based and whole animal data show that Imd is multiply modified after immune activation with cleavage, two types of ubiquitination, phosphorylation, and ultimately proteasome-mediated degradation.
Lys-137 and Lys-153 are the sites for Imd ubiquitin conjugation
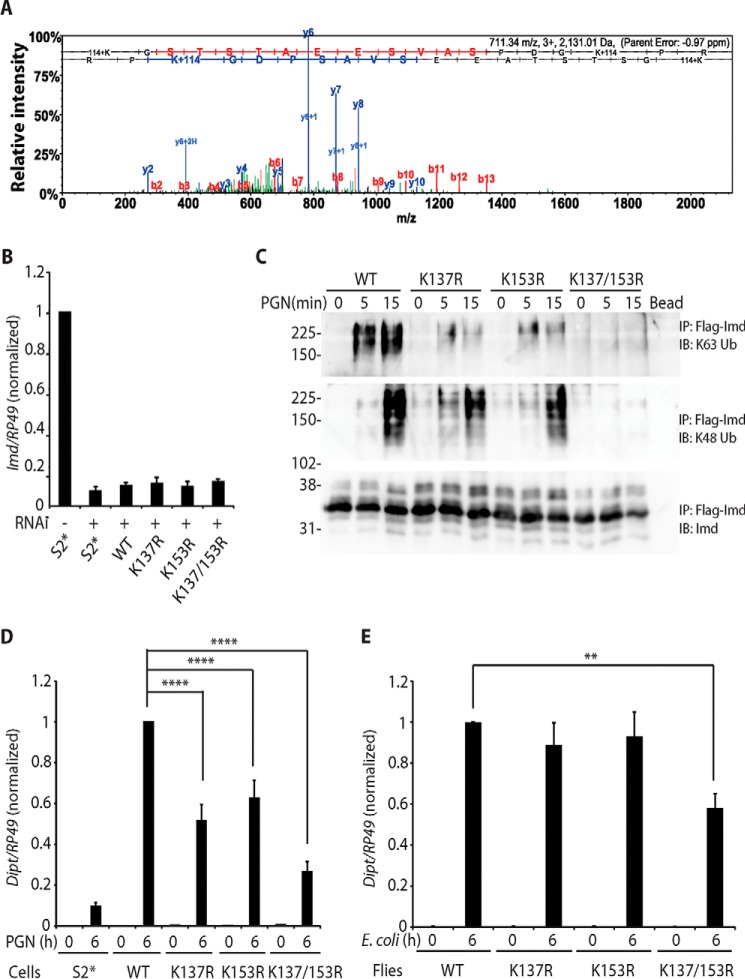
Next we sought to identify the ubiquitin conjugation sites on endogenous Imd with immunopurification and mass spectrometry. Two rounds of similar assays showed that lysine residues 137 and 153 were conjugated with ubiquitin, as indicated by diglycine tags after trypsin digestion and tandem MS (Fig. 2A, K+114 in the spectrum).
Figure 2.
Lys-137 and Lys-153 are the sites for Imd ubiquitin conjugation. A, Imd ubiquitination at Lys-137/Lys-153. Endogenous polyubiquitinated Imd was immunopurified and analyzed by tandem mass spectrometry. Ubiquitination sites were identified by their diglycine tags (K+114). A representative spectrum from two biologically independent mass spectrometric analyses is shown, with multiple diglycine-tagged peptides in each. B, Imd 3′-UTR targeting RNAi is ∼90% efficient. Parental S2* cell and stable cell line expressing Imd wild type (WT), K137R, K153R, or K137R/K153R were treated with dsRNA targeting Imd 3′-UTR. Endogenous Imd expression level was monitored by qRT-PCR with primers targeting Imd 5′-UTR. Data are shown as the mean ± S.D. from 3 independent experiments. C and D, inhibition of polyubiquitination and signaling by mutation of Lys-137 and/or Lys-153 in S2* cells. Stable cell lines expressing FLAG-Imd WT, K137R, K153R, or K137R/K153R were stimulated with PGN for indicated times. Endogenous Imd was knocked down by dsRNA targeting Imd 3′-UTR. Conjugation of FLAG-Imd was assayed by FLAG-immunoprecipitated (IP) and immunoblotting (IB) (C). The blot is representative of three independent experiments. Diptericin expression was monitored by qRT-PCR in D. Data are shown as the mean ± S.D. from three independent experiments. ****, p < 0.0001 (one-way ANOVA). E, inhibition of signaling by mutation of Lys-137 and Lys-153 in flies. An imd10191 (null) strain carrying transgenic WT, K137R, K153R, or K137R/K153R imd were stimulated with E. coli by septic infection at indicated. Diptericin expression was monitored by qRT-PCR. **, p < 0.01 (one-way ANOVA). Data are shown as the mean ± S.D. from three independent experiments.
To verify the result from mass spectrometry analysis, stable cell lines expressing Imd WT, K137R, K153R, or K137R/K153R were generated and transfected with dsRNA targeting the 3′-UTR of Imd to knock down the endogenous protein (RNAi efficiency is shown in Fig. 2B). PGN-stimulated Imd Lys-63- and Lys-48-polyubiquitination were completely absent when both lysine residues were mutated to arginine, whereas only Lys-63-polyubiquitination showed a considerable decrease when either Lys-137 or Lys-153 were singly mutated to arginine (Fig. 2C). Moreover, the signaling pathway activity, as measured by induction of the antimicrobial peptide gene Diptericin, showed ∼40–50% reduction caused by either lysine mutation and 70% reduction by the double lysine mutation (Fig. 2D). By comparison, the control parental cell line showed 10% residual Diptericin induction in these knockdown conditions, indicative of the small amount of imd transcript that remained after RNAi treatment (Fig. 2B). These results demonstrate that ubiquitination at Lys-137 and Lys-153 is critical for Imd signaling in S2* cells.
To further explore the importance of Lys-137/Lys-153 in vivo, imd10191 (null) flies expressing wild-type, K137R, K153R, or K137R/K153R imd were generated by PhiC31-mediated transgenesis. E. coli-triggered induction of Diptericin was significantly inhibited when both lysine residues are substituted with arginine (Fig. 2E), further highlighting the role of these two residues in Imd signaling in vivo.
Imd ubiquitination requires Diap2 and multiple E2s
Our previously published work showed that the E3 ligase Diap2 is required for Imd Lys-63-polyubiquitination (5). Now, using antibodies specific to Lys-63 and Lys-48 ubiquitin, we demonstrate that Imd Lys-63-polyubiquitination is prevented by knocking down Diap2, which leads to accumulation of cleaved Imd and a failure to generate Lys-48-conjugated Imd (Fig. 3A). Coincident with this lack of ubiquitination, Diptericin induction is also blocked with Diap2 knockdown (Fig. 3, B and C, for RNAi efficiency). The accumulation of cleaved Imd in this assay as well as in adult Diap2 mutant animals (Fig. 1E), strongly argues that Imd cleavage occurs before Lys-63-polyubiquitination.
Figure 3.
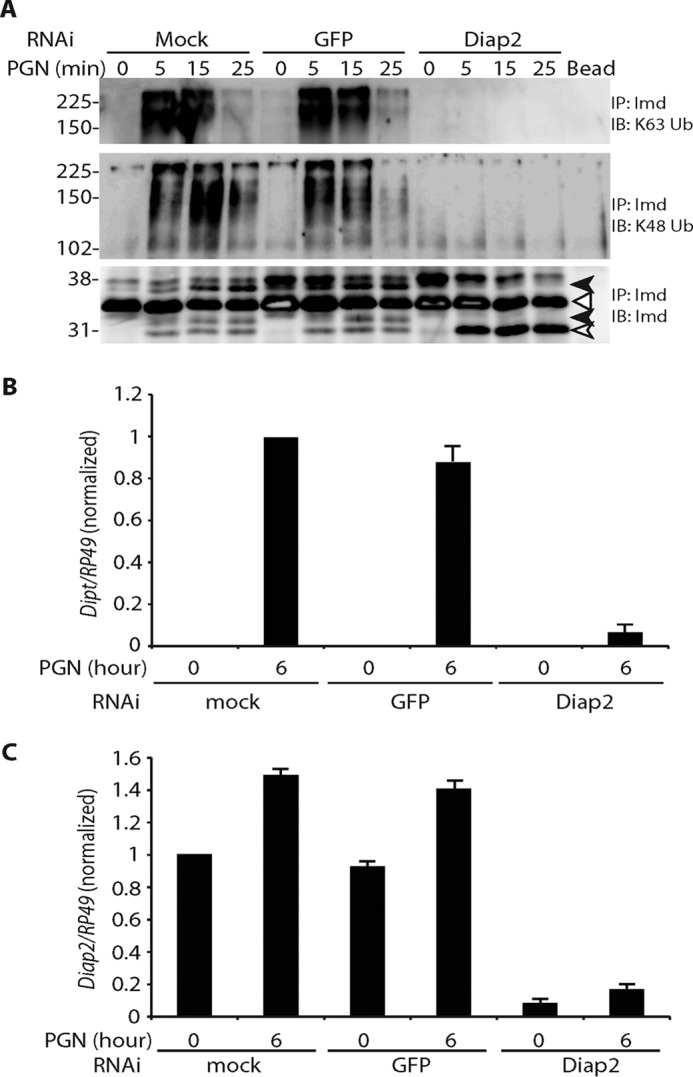
Imd ubiquitination requires Diap2. A, requirement of Diap2 in Imd polyubiquitination. S2* cells were treated with dsRNA targeting GFP or Diap2 followed by PGN stimulation for the indicated times. Imd modifications were assayed as above. The open triangle marks unmodified full-length Imd, the open arrow marks cleaved Imd, and the black arrow marks phosphorylated Imd. Diap2 knockdown leads to the accumulation of cleaved, but unmodified, Imd. IP, immunoprecipitated; IB, immunoblotting. B and C, efficiency and effect of Diap2 RNAi. S2* cells were treated with dsRNA targeting GFP or Diap2. Expression level of Diptericin (Dipt, B) or Diap2 (C) before and after 6 h of immune stimulation was monitored by qRT-PCR. Data are shown as the mean ± S.D. from three independent experiments.
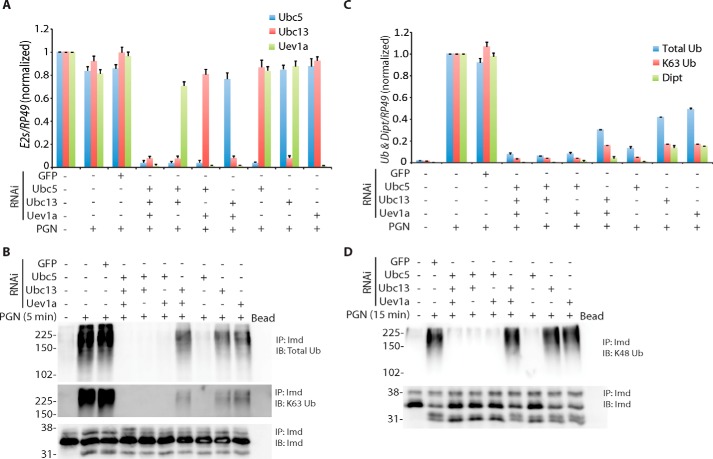
To examine the detailed mechanism of Imd ubiquitination, we sought to more thoroughly characterize the role of two different E2 enzymes previously implicated: Ubc5 and Ubc13-Uev1a. RNAi targeting each of these E2s was optimized such that knockdown was at least 90% efficient, as measured by qRT-PCR (Fig. 4A). Each E2 was knocked down individually or in all possible combinations. Strikingly, total and Lys-63-polyubiquitination was completely abolished by Ubc5 depletion alone, whereas a low level (20–50%) of ubiquitination signal (both total and Lys-63) remained after knockdown of Ubc13 and/or Uev1a (Fig. 4B and intensity quantification in 4C). The induction of Diptericin showed a similar pattern (Fig. 4C). Knockdown of Ubc5 prevented Diptericin induction, whereas cells depleted of Ubc13 and/or Uev1a still induced a low level (5–15%). Similarly, Lys-48-polyubiquitination was also blocked in the absence of Ubc5 (Fig. 4D). On the other hand, the level of Lys-48-polyubiquitination observed with Ubc13 and/or Uev1a RNAi was mostly unchanged (Fig. 4D), suggesting that neither of these two E2s is involved in Imd Lys-48-polyubiquitination. Together, these data indicate that Imd Lys-63-polyubiquitination requires two E2s, which play distinct roles in the process, as discussed in more detail below.
Figure 4.
Two-step ubiquitination of Imd. A, efficiency and specificity of E2 RNAi. S2* cell were transfected with dsRNA targeting GFP, Ubc5, Ubc13, or Uev1A in all possible combinations as indicated. In all cases 30 μg of combined dsRNA was transfected. Expression levels of targeted E2 mRNAs was monitored by qRT-PCR. RNAi was highly specific. Data are shown as the mean ± S.D. from three independent experiments. B and C, differentiated roles of E2s in Imd polyubiquitination. S2* cells were treated with dsRNA targeting GFP or E2s in all combinations as indicated followed by a 5-min PGN stimulation. B, Imd modifications were assayed as above. IP, immunoprecipitated; IB, immunoblotting. C, total and Lys-63 ubiquitin intensities from immunoblotting were quantified and plotted along with Diptericin expression measured by qRT-PCR from the same experiments. Data are shown as the mean ± S.D. from three independent experiments. D, Imd Lys-48-polyubiquitination catalyzed by E2s. Same RNAi assay was done as above with PGN stimulation for 15 min. All blots are representatives of three independent experiments.
Tak1 is required for Imd phosphorylation and ubiquitin editing
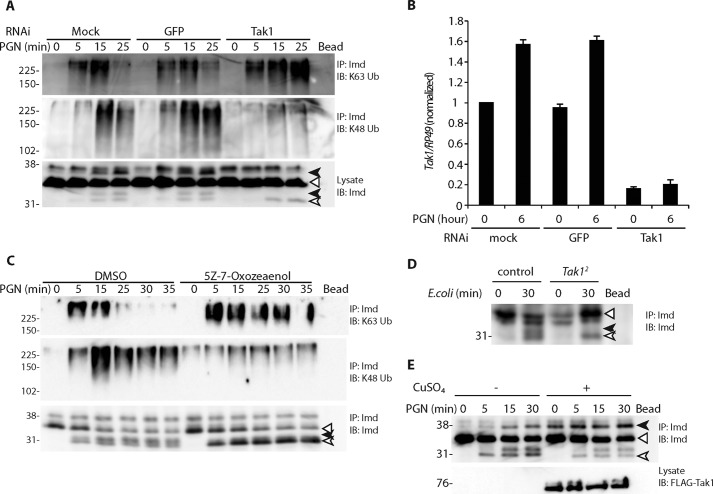
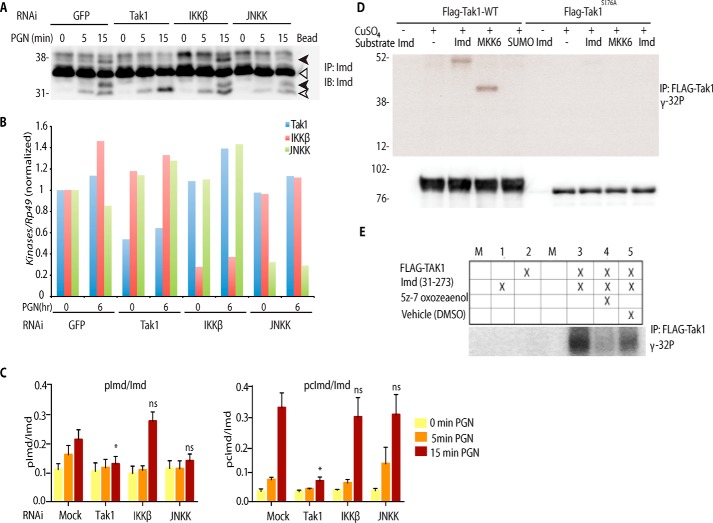
In addition to ubiquitination, Imd is phosphorylated upon immune stimulation, and the timing of this modification tracks with Lys-48-ubiquitination (Fig. 1B). Tak1, a Lys-63-polyubiquitin-activated MAP3K that functions downstream of Imd Lys-63-polyubiquitination, is implicated in Imd phosphorylation (25). To determine if Tak1 is required for Imd phosphorylation, S2* cells were treated with dsRNA targeting GFP or Tak1 followed by PGN stimulation (Fig. 5A, RNAi efficiency shown in Fig. 5B). Imd phosphorylation and Lys-48-polyubiquitination were lost in the Tak1 knockdown cells, whereas the Imd Lys-63-polyubiquitination persisted into later time points. Similarly, inhibition of Tak1 by (5Z)-7-oxozeaenol showed persistent accumulation of Imd Lys-63-polyubiquitination, reduced Imd phosphorylation, loss of Lys-48-polyubiquitination, and accumulation of cleaved unphosphorylated Imd (Fig. 5C). These results demonstrate that Tak1 is required for Imd phosphorylation, Lys-63-deubiquitination, and the addition of Lys-48-polyubiquitination.
Figure 5.
Tak1 is required for Imd phosphorylation and ubiquitin editing. A–C, requirement of Tak1 for Imd phosphorylation and ubiquitin editing. S2* cells were treated with dsRNA (A and B) or the Tak1 inhibitor (5Z)-7-oxozeaenol (C), followed by PGN stimulation for indicated times. Modifications of endogenous Imd were assayed as previously described. Tak1 RNAi efficiency was monitored by qRT-PCR (B). Data are shown as mean ± S.D. from three independent experiments. IP, immunoprecipitated; IB, immunoblotting. D, requirement of Tak1 in Imd phosphorylation in vivo. Control (yw;wt) and yw;Tak12 adult flies were challenged with live E. coli by septic infection for indicated times. Endogenous Imd was assayed as above. E, Tak1 expression is sufficient to drive Imd phosphorylation. Stable cell lines expressing FLAG-Tak1 from the copper-inducible metallothionein promoter were treated with CuSO4 (500 μm) for 6 h followed by PGN stimulation, as indicated. Phosphorylation of endogenous Imd was assayed by immunoprecipitated and immunoblotting. Tak1 protein level was monitored by lysate immunoblotted against FLAG. All blots are representative of at least three independent experiments. In all cases, the open triangle marks unmodified full-length Imd, the open arrow marks cleaved Imd, and the black arrows mark phosphorylated Imd.
To further confirm the functional relevance of Tak1 in Imd modification in vivo, control or Tak1-deficient flies (Tak12) were challenged with E. coli and analyzed for Imd phosphorylation (Fig. 5D). The Tak12 strain showed a lack of Imd phosphorylation, indicating that Tak1 is required for Imd phosphorylation in vivo.
To determine if ectopic expression of Tak1 is sufficient to drive Imd phosphorylation, a stable cell line expressing FLAG-Tak1 from the Cu2+-inducible metallothionein promoter was treated with CuSO4 for 6 h followed by PGN stimulation. As shown in Fig. 5E, Tak1 overexpression leads to robust phosphorylation of full-length Imd before PGN stimulation.
Tak1 directly phosphorylates Imd
To determine whether Tak1 is solely responsible for Imd phosphorylation, S2* cells were treated with dsRNA targeting two downstream kinases in the Imd-signaling pathway, IKKβ (encoded by ird5) or JNKK (encoded by hep). Unlike Tak1, depletion of neither IKKβ nor JNKK significantly affected the PGN-induced phosphorylation of Imd (Fig. 6, A and B and multiple blots quantified in C), indicating these kinases are not involved in phosphorylating Imd. However phosphorylation of full-length Imd was slightly (but not significantly) reduced in cells when JNKK was knocked, leaving open the possibility that JNKK contributes partially to phosphorylation of full-length, but not cleaved, Imd. To address whether Tak1 directly phosphorylates Imd, in vitro immunoprecipitation kinase assays were performed. FLAG-tagged Tak1 WT or kinase dead Tak1S176A was immunoprecipitated with anti-FLAG-agarose and used in a kinase reaction with recombinant SUMO-tagged Imd (or SUMO or MKK6 as controls) as substrates along with radiolabeled ATP (Fig. 6C). WT Tak1 but not Tak1S176A phosphorylated Imd and MKK6 but not SUMO. Furthermore, the addition of (5Z)-7-Oxozeaenol, a selective inhibitor of Tak1, to the in vitro kinase reaction inhibited Imd phosphorylation (Fig. 6D). Together these experiments support the conclusion that Tak1 directly phosphorylates Imd.
Figure 6.
Tak1 directly phosphorylates Imd. A and B, Imd phosphorylation is dependent on Tak1 rather than IKKβ or JNKK. S2* cell were transfected with dsRNA targeting GFP, Tak1, IKKβ (Ird5), or JNKK (Hep) followed by PGN stimulation, as indicated. Endogenous Imd phosphorylation was assayed as above. IP, immunoprecipitated; IB, immunoblotting. B, RNAi efficiency, from the assay displayed in above, was monitored by qRT-PCR. The open triangle marks unmodified full-length Imd, the open arrow marks cleaved Imd, and the black arrow marks phosphorylated Imd. C, immunoblot band intensity ratio from five independent replicate Imd phosphorylation assays, similar to the blot shown above, were quantified. Data are shown as the mean ± S.E., and statistical significance was determined by one-way ANOVA followed by Dunnett's post-test, comparing mock RNAi to each kinase knockdown. *, p < 0.05. ns, not significant. D, Tak1 phosphorylates Imd in vitro. Stable cell lines expressing FLAG-Tak1 WT or S176A (kinase dead) from the Cu2+ inducible metallothionein promoter were treated with CuSO4 (500 μm) for 6 h. FLAG-Tak1 was immunoprecipitated and incubated with [γ-32P]ATP and substrates as indicated. IP samples were also immunoblotted against FLAG-Tak1 as the internal loading control. E, in vitro phosphorylation of Imd is inhibited by the addition o the Tak1 inhibitor, (5Z)-7-oxozeanol, to the reaction. Kinase assays are representative of three independent experiments; M, mock.
Discussion
In previous work we demonstrated that Imd is cleaved, Lys-63-polyubiquitinated, and phosphorylated upon immune stimulation (5). Although this earlier study did not find Lys-48-polyubiquitin chains, others have reported evidence of both Lys-63- and Lys-48-Imd modifications (21). However, the overall dynamics of, and interconnections between these Imd post-translational modifications remained unclear. Here we show that PGN stimulation of S2* cells leads to five different Imd modifications: proteolytic cleavage, Lys-63-polyubiquitination, phosphorylation, Lys-63-deubiquitination, and Lys-48-polyubiquitination, which leads to degradation of Imd through proteasome. These immune-triggered signaling events are robust and incredibly rapid, with Imd cleavage and Lys-63-polyubiquitination occurring as early as 2 min after PGN stimulation. Although Lys-63 modification peaks early and then steadily declines, Lys-48-conjugation appears later, along with phosphorylation, and declines in a proteasome-dependent manner. These kinetics argue that Imd is sequentially conjugated with Lys-63 then Lys-48 ubiquitin, so-called ubiquitin editing, as has been reported for IRAK1 and RIP1 in mammalian innate immune signaling pathways (22).
In addition to ubiquitination, two slow-migrating species of Imd were detected and shown to be phosphorylated forms. Judging by their size, these two phospho-forms appear to be derived from either full-length Imd (upper) or cleaved Imd (lower) (Fig. 1C). Interestingly, we also observe persistence of phosphorylated Imd in the proteasome-inhibited samples (Fig. 1D), suggesting that Imd is both Lys-48-polyubiquitinated and phosphorylated before entering proteasome. Tak1 is required for these phosphorylation events as well as for ubiquitin editing, demonstrating a key role for this MAP3K in a negative feedback loop.
Conjugation of ubiquitin usually occurs on lysine side chains of target proteins, and mass spectrometry of immunopurified endogenously expressed Imd identified Lys-137 and Lys-153 as the sites of ubiquitin linkage. Note, our mass spectrometry analysis includes 50% coverage of Imd, and a cluster of four lysine residues at the very C terminus were not observed. Substitution of Lys-137 and Lys-153 residue with arginine prevented signal-induced ubiquitination of Imd in S2* cells and reduced expression of the AMP gene Diptericin in both cells and flies. These results demonstrate that both lysine residues are required for Lys-63-polyubiquitination and downstream signaling events. In S2* cells, mutation of single lysine residue led to a partial reduction of Lys-63-ubiquitination and a partial reduction of AMP gene induction. Surprisingly, a single lysine mutation did not correspondingly reduce Imd Lys-48-polyubiquitination, whereas the double lysine mutation completely blocked it. These results suggest that even the reduced signal, mediated by a single Lys-63-chain, is sufficient to trigger a robust feedback response with Lys-48-chain formation. On the other hand, a complete block in Lys-63-chains prevents Tak1 activation, which in turn fails to promote the ubiquitin editing of Imd. These findings are consistent with results observed with knockdown of Ubc5, Ubc13, and Uev1a. Ubc5 depletion prevented all Lys-63 ubiquitination and signaling (as measured by Diptericin induction), and subsequent Lys-48 modification was absent, whereas the Ubc13 and/or Uev1a knockdown showed residual Lys-63 chains and greatly reduced Diptericin expression but robust Lys-48-ubiquitination. These results also suggest that Lys-48-polyubiquitination may occur on lysine residues beyond Lys-137 and Lys-153, although more detailed mass spectrometric analyses is required to map these sites more thoroughly.
On the other hand, only double lysine mutation leads to significant reduction of AMP expression in adult flies, and this reduction is not as robust as in cultured cells. This pattern suggests that activation of the NF-κB protein Relish and its transcriptional targets do not solely rely on ubiquitination of Imd Lys-137 /153. Other possible ubiquitination targets include the upstream caspase Dredd, which has been shown to be critical for signaling (26), or the E3 ligase Diap2. This redundancy may represent multiple parallel mechanisms that contribute to the NF-κB activation. Furthermore, tissue-specific immune differences may contribute to the discrepancy in Diptericin induction between macrophage-derived S2* cells and whole flies, in which the fat body rather than hemocytes is the major organ for inducible expression of AMPs.
Previous work has suggested that Diap2 is the E3 ligase for Imd ubiquitination (5). With the advantage of ubiquitin linkage-specific antibodies, data presented here show that Diap2 is required for Imd Lys-63-polyubiquitination and signaling, as measured by induction level of Diptericin. Moreover, the accumulation of cleaved but non-ubiquitinated Imd, in the Diap2 depleted cells and flies, provides further evidence that ubiquitination is downstream of Imd cleavage and highlights the role of Diap2 as the critical E3 in the Lys-63-modification of Imd. In addition, Imd is no longer Lys-48-modified when Diap2 is removed, suggesting that either Diap2 is also involved in Lys-48-conjugation or the failure of Lys-63-polyubiquitination leads to the loss of Lys-48-polyubiquitination. Given the role of Tak1 in this ubiquitin editing event, we favor the latter hypothesis and speculate another E3 is likely involved in the Lys-48 conjugation.
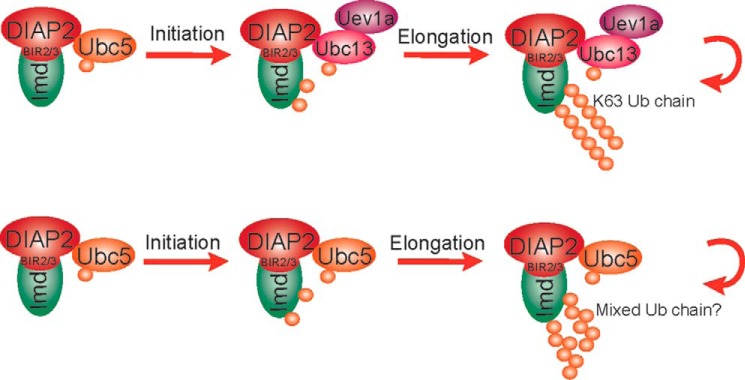
In addition to E3 ligases, E2 ubiquitin-conjugating enzymes are the other key factors in the ubiquitin conjugation reaction. We previously showed that Ubc5 and Ubc13-Uev1a are all involved in Imd ubiquitination (5). However, the mechanism by which these E2s collaborated was unclear. Results from in vitro reconstituted ubiquitination assays suggested a two-step reaction model for ubiquitin conjugation with different E2s (27–30). In particular, it was shown that some E2s, such as Ubc5, are effective at the initial ubiquitination of substrates but are ineffective at generating long chains, whereas other E2s, like Ubc13/Uev1a, are efficient at generating long ubiquitin chains but fail to conjugate substrate proteins. Thus, these two types of E2s can work together to generate long ubiquitin chains conjugated to target proteins. We propose that Imd undergoes ubiquitin chain initiation and elongation catalyzed by two separate E2s (Fig. 7). Once cleaved, Imd interacts with Diap2 through its baculovirus inhibitor repeat (BIR) repeats and is first modified by Ubc5-mediated substrate ubiquitination on lysines 137 and 153. Subsequently, the E2 complex of Ubc13-Uev1a pairs with an E3 (possibly Diap2 although another unidentified E3 is not excluded) and switches the reaction to the chain elongation mode, during which additional ubiquitin molecules are attached to the substrate-linked ubiquitin in a Lys-63-specific manner (31–35). In the absence of Ubc13/Uev1a, Ubc5 alone is still able to elongate the polyubiquitin chains but less efficiently and with unknown linkages.
Figure 7.
Model of two-step reaction in Imd Lys-63-polyubiquitination. Imd is Lys-63-polyubiquitination by E2s. Imd is mono-ubiquitinated by Ubc5 and subsequently elongated with Lys-63 linkage by Ubc13-Uev1a. Alternatively, Ubc5 alone is able to catalyze both mono-ubiquitination and chain elongation with unknown linkage.
Induction of Diptericin expression generally tracks with the Lys-63-polyubiquitination signal (but not the total Ub signal) observed in various E2 knockdown cells (Fig. 4C). The one exception is the samples in which Ubc13 and Uev1a are both knocked down and Ubc5 is still available. These samples display a similar Lys-63 intensity as the single Ubc13 or Uev1a RNAi lanes, but Diptericin induction is lower, close to background levels. Ubc5 alone is known to conjugate ubiquitin without linkage specificity, a random polyubiquitin chain that consist of all seven types of lysine linkages (29). Because the Lys-63 antibody recognizes Lys-63-linked diubiquitin (22), it is possible that Ubc5-mediated random ubiquitin chain elongation generates some Lys-63 diubiquitin linkages that are detected by this antibody but are unable support signaling due to their altered topology and limited amounts of Lys-63-linkages. More detailed biochemical characterization of these Ubc5-catalyzed chains is required to confirm this hypothesis.
Lys-48 modification of Imd shows a subtle difference relative to the Lys-63 chains. Again, knockdown of Ubc5 causes complete blocking of Lys-48 conjugation, but depletion of Ubc13/Uev1a has no effect. The failure of Lys-48 modification in the Ubc5-depleted cells may have two possible underlying causes. First, the complete lack of Lys-63 chains will fail to activate Tak1 and the subsequent ubiquitin editing feedback loop, whereas the Ubc13 and/or Uev1a RNAi display some residual Lys-63 activity and thus can trigger Tak1 and the feedback response. Alternatively, Ubc5 might be directly required for Imd Lys-48-polyubiquitination as shown in the degradation of proteins in multiple Drosophila pathways including eye development, maintenance of germ-line stem cells and apoptosis (36). These are not mutually exclusive possibilities.
Phosphorylation of Imd appears to be a major regulator of these ubiquitin-editing events. Knockdown of Tak1 prevents Imd phosphorylation in S2* cells and in adult flies. Moreover, immune-purified Tak1 can directly phosphorylate recombinant Imd in vitro, whereas neither JNKK nor IKK is required for phosphorylation of cleaved Imd, strongly arguing that Tak1 directly modifies Imd. RNAi depletion or drug inhibition of Tak1 prevents Lys-63-deubiquitination and the subsequent Lys-48-polyubiquitination/proteasome-mediated degradation, leading to accumulation of cleaved but unphosphorylated Imd, presumably an intermediate during chain editing. From these results, we infer that Tak1-mediated phosphorylation of Imd is required for ubiquitin editing. Future studies will reveal the underlying mechanisms by which phosphorylation triggers Lys-63-deubiquitination and Lys-48 chain conjugation. Nonetheless, these results are consistent with earlier reports of Imd regulation by Lys-63-deubiquitination and degradation (21).
Considering the results presented here together with earlier studies, we propose the following model of Imd signal activation and subsequent down-regulation (Fig. 8). One of the earliest events after PGN stimulation is the rapid cleavage of Imd by the caspase-8 homolog DREDD at Asp-30 (5). Cleaved Imd then interacts with BIR repeats of the E3 ligase Diap2 and is Lys-63-polyubiquitinated through the sequential action of Ubc5, for substrate conjugation, and Ubc13-Uev1a, for catalyzing long Lys-63 chains. These Lys-63-polyubiquitin chains are then likely to activate the Tak1/Tab2 kinase complex through the conserved Lys-63-binding motif in Tab2, which in turns signals through IKK complex to activate the NF-κB precursor Relish (20). Relish is central for the robust induction of AMP gene transcription. Meanwhile, Tak1 also mediates a retrograde signal that phosphorylates Imd and triggers ubiquitin editing and leads to the degradation of Imd through proteasome. This regulatory interaction between Tak1 and Imd represents a novel homeostatic loop whereby the Drosophila immune response is rapidly activated but also quickly shutdown. Future studies are necessary to determine function of this feedback loop relative to other feedback mechanisms reported for the Imd pathway (21, 37–45).
Figure 8.
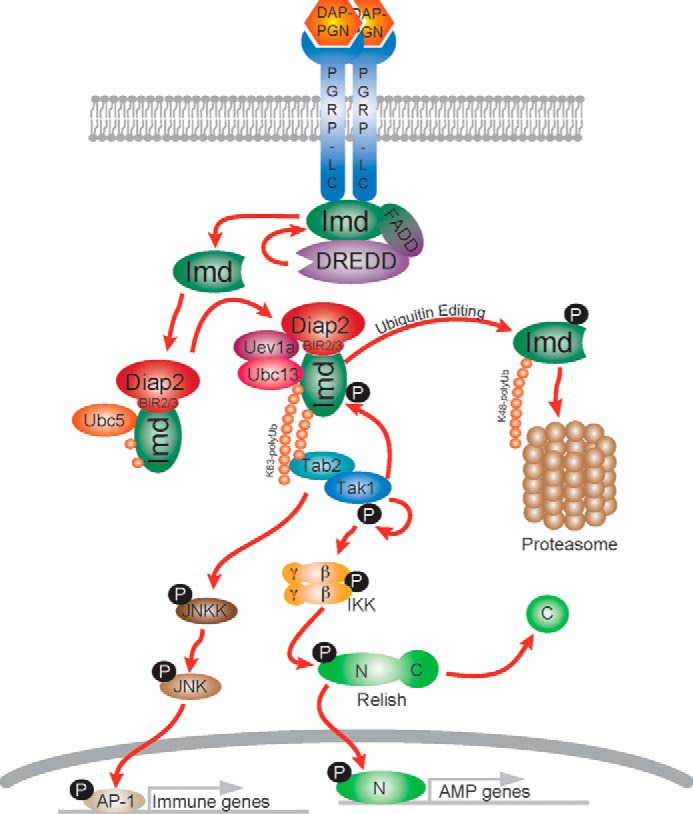
Model of ubiquitin editing in the Imd-signaling pathway. Upon immune stimulation, Imd undergoes rapid proteolytic cleavage and Lys-63-polyubiquitination in a two-step manner. Although ubiquitin-activated Tak1 signals to downstream kinases and NF-κB protein Relish, it also phosphorylates Imd and triggers a feedback loop to promote ubiquitin editing, leading to proteasome-mediated degradation of Imd.
Experimental procedures
Fly stocks
The following fly strains were used in this work: w1118, Diap27c (46), imd10191 (47), yw;wt and yw;Tak12 (18).
Antibodies
Anti-Lys-63 (Apu3) and anti-Lys-48 (Apu2) ubiquitin antibodies were purchased from EMD Millipore (Billerica, MA). Total ubiquitin antibody (P4D1) was purchased from Santa Cruz Biotechnology (Dallas, TX). Anti-FLAG M2 Affinity Gel and anti-FLAG antibody were purchased from Sigma. Polyclonal Imd antibody was prepared by immunizing rabbits with peptide (31AAPVDDNEPDN41) according to the affinity-purified package offered by New England Peptide (Gardner, MA).
Peptidoglycan preparation
A 5-liter culture of E. coli strain 1106 was collected and resuspended with 4% SDS solution before boiling at 100 °C for 30 min. Traces of SDS were removed by repeated centrifugation and resuspending with sterilized water. Then the lysate was sequentially treated with DNase, α-amylase, and Pronase. These enzymes were removed by repeated centrifugation and resuspending with sterilized water. At last the lysate was treated with sonification and lyophilization and prepared as 1 mg/ml stock.
Immunoprecipitation and immunoblotting protein assay
Cells or flies were lysed in standard lysis buffer containing 10% glycerol, 1% Triton X-100, 20 mm Tris, 150 mm NaCl, 24 mm β-glycerol phosphate, 2 mm EDTA, 1 mm 1,4 dithiothreitol (DTT), 1 mm sodium orthovanadate, 5 mm N-ethylmaleimide, 1× protease inhibitor mixture. Total protein lysates was immunoprecipitated overnight at 4 °C followed by immunoblotting at room temperature.
Proteasome inhibition assay
S2* cells were treated with 20-hydroxyecdysone (1 μm) for 24 h before treatment of 10 μm MG-132 (Sigma) for 6 h followed by PGN stimulation and protein assay.
λ-Phosphatase assay
S2* cells were stimulated with E. coli PGN for the indicated times before lysis. The whole cell lysates (100 ng for each condition) were treated without or with λ-phosphatase for 30 min at 30 °C followed by immunoblotting with anti-Imd antibody.
Protein purification for mass spectrometry
S2* cells were stimulated with PGN for 0 and 15 min and lysed with the buffer described above. 20 mg of total protein lysate was immunoprecipitated overnight at 4 °C with anti-Imd antibody. Samples were washed twice with 5 ml of lysis buffer and eluted with 200 μl of elution buffer (1 m citric acid, pH 2.2). Elutes were separated by SDS-PAGE and stained by Coomassie Blue. The corresponding bands were cut out for in-gel trypsin digestion and analyzed by tandem mass spectrometry.
In-gel digestion (MS)
Gel slices were cut into 1 × 1-mm pieces and placed in 1.5-ml Eppendorf tubes with 1 ml of water for 1 h. The water was removed, and 50 μl of 250 mm ammonium bicarbonate was added. For reduction, 5 μl of a 45 mm solution of DTT was added, and the samples were incubated at 50 C for 30 min. The samples were cooled to room temperature and then for alkylation, 5 μl of a 100 mm iodoacetamide solution was added and allowed to react for 30 min. The gel slices were washed 2× with 1-ml water aliquots. The water was removed, and 1 ml of 50:50 (50 mm ammonium bicarbonate:acetonitrile) was placed in each tube, and samples were incubated at room temperature for 1 h. The solution was then removed, and 200 μl of acetonitrile was added to each tube at which point the gel slices turned opaque white. The acetonitrile was removed, and gel slices were further dried in a SpeedVac concentrator. Gel slices were rehydrated in 50 μl of 2 ng/μl trypsin (Sigma) in 0.01% ProteaseMAX Surfactant (Promega), 50 mm ammonium bicarbonate. An additional aliquot of 50 mm ammonium bicarbonate was added to fully submerge the gel slices. Samples were incubated at 37 °C for 21 h. The supernatant of each sample was then removed and placed in a separate 1.5-ml Eppendorf tube. Gel slices were further dehydrated with 100 μl of 80:20 (acetonitrile:1% formic acid). The extract was combined with the supernatants of each sample. The samples were then dried down in a SpeedVac concentrator (Savant Instruments, Inc.).
LC/MS/MS on Orbitrap Velos
Tryptic peptides were dissolved in 20 μl of 0.1% TFA, and a 3.5-μl aliquot was directly injected onto a custom packed trap column (2 cm × 100 μm C18). Peptides were then eluted and sprayed from a custom-packed emitter (75 μm × 25 cm C18) with a linear gradient from 100% solvent A (0.1% formic acid in 5% acetonitrile) to 35% solvent B (0.1% formic acid in acetonitrile) in 90 min at a flow rate of 300 nl/min on a Proxeon Easy nanoLC system directly coupled to a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Data-dependent acquisitions were set up according to an experiment where full MS scans from 350 to 2000 m/z were acquired in the Orbitrap at a resolution of 60000 followed by 10 MS/MS scans acquired in the LTQ ion trap instrument.
Data analysis (MS)
The raw data files were processed with Proteome Discoverer v. 1.3 (Thermo Scientific) into peak lists and then searched against the Drosophila Index of the UniProt database using the Mascot Search engine version 2.3 (Matrixsciences, Ltd.). Parent mass tolerances were set to 10 ppm, and fragment mass tolerances were set to 0.5 Da. The variable modifications of acetyl (protein N-term), Gly-Gly on lysine, pyroglutamic for N-term glutamine, carbamidomethylation of cysteine, and oxidation of methionine were used. Mascot search results were loaded into the Scaffold software (Proteome Software, Inc.) to compare sample results.
RNAi
DsRNA was produced using T7 RiboMAX Express Large Scale RNA Production System (Promega). S2* cells were split to 0.5 × 106 cells/ml and incubated for ∼24 h at 27 °C. 2 μg/ml dsRNA was delivered by calcium phosphate transfection, and cells were allowed to recover for 18 h at 27 °C. 1 μm 20-hydroxyecdysone was then added to the cells for 24 h. Finally, cells were treated with peptidoglycan (10 μg/ml) before harvesting for experiments.
Imd lysine mutants assay
Stable cell line expressing FLAG-Imd WT, K137R, K153R, or K137R/K153R were generated and then transfected with dsRNA targeting Imd 3′-UTR to knock down the endogenous Imd. Cells were allowed to recover for 18 h at 27 °C before treatment of 20-hydroxyecdysone (1 μm). Finally, cells were stimulated with peptidoglycan (10 μg/ml) and harvested for the experiments.
E2 RNAi
Each sample was transfected with 10 μg of dsRNA per target gene, and the total dsRNA used in each sample was equalized to 30 μg by adding GFP dsRNA. Cells were allowed to recover for 18 h at 27 °C before treatment of 20-hydroxyecdysone (1 μm). Finally cells were stimulated with peptidoglycan (10 μg/ml) and harvested for the experiments.
Transgenic Drosophila
PhiC31 integrase-mediated transgenic rescue construct included the Imd coding region and about 3 kb of the surrounding gene region. This fragment was PCR-amplified and cloned into pattB vector. Lysine residues 137 and 153 were substituted with arginine individually or in combination, generating four donor plasmids: pattB-WT, -K137R, -K153R, and -K137R/K153R. To simplify the screening process, each plasmid contained a silent mutation between lysine 137 and 153, creating a SacII restriction digestion site. Confirmed transgenic flies were crossed with a double balancer and the null allele imd10191 and homozygous flies were collected for experiments.
RNA analysis
Total RNA was extracted with TRIzol (Invitrogen), treated with DNase I, and purified by phenol/chloroform. Transcripts from the gene of interest were monitored by qRT-PCR (Bio-Rad). The gene expression level in each sample was normalized to RP49. Data from 3 biological independent experiments were collected, normalized with control samples (set as 1), and plotted as the mean ± S.D.
Tak1 inhibitor assay
S2* cells were treated with 20-hydroxyecdysone (1 μm) for 24 h before treatment of 2 μm (5Z)-7-oxozeaenol (Sigma) for 3 h followed by PGN stimulation and protein assay.
Kinase assay
Recombinant proteins His6-SUMO-Imd(31–273), His6-SUMO and His6-MKK6-K28A were purified with nickel magnetic beads (EMD Millipore). Stable cell lines expressing FLAG-tagged Tak1 WT or S176A (kinase dead) from the Cu2+-inducible metallothionein promoter were treated with CuSO4 (500 μm) for 6 h before lysing with lysis buffer as described above. 100–400 ng of total protein lysate from each cell line was immunoprecipitated with anti-FLAG antibody for 2 h at 4 °C. Kinases were then washed twice with 500 μl of lysis buffer and twice with 500 μl of kinase buffer (200 mm HEPES, 200 mm β-glycerol phosphate, 100 mm MgCl2, 500 mm NaCl, 10 mm DTT, 1 mm sodium orthovanadate, 1 μm okadaic acid). Finally, kinases were incubated in kinase buffer containing 200 μm cold ATP, 5 μCi of [γ-32P]ATP, and substrate (100 ng each) at 30 °C for 30 min. Samples were separated by SDS-PAGE, fixed with 10% methanol, 10% acidic acid, dried, and autoradiographed. For inhibitor studies, in vitro kinase assay were performed as described above, with the addition of (5Ζ)-7-Οxozeaenol (or DMSO vehicle) to the reaction immediate before the addition of recombinant Imd substrate.
Author contributions
L. C. and N. S. conceived the work and wrote the manuscript. L. C. performed the majority of the experiments. S. M. and F. R. performed the other experiments. A. N. contributed to the manuscript preparation. N. P. provided an important initial observation. J. L. and S. S. performed the mass spectrometry analysis. N. S. supervised all aspects of the project.
Acknowledgments
We thank David Schneider for providing the imd10191 fly line and Pascal Meier for the Diap27c fly line. Tak12 stock obtained from the Bloomington Drosophila Stock Center (National Institutes of Health Grant P40OD018537) was used in this study. We also thank Pascal Meier and Meike Broemer for providing the recombinant His6-SUMO-Imd(31–273) construct.
This work was supported, in whole or in part, by National Institutes of Health Grant AI060025 (to N. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AMP
- antimicrobial peptide
- PGN
- peptidoglycan
- PGRP
- peptidoglycan recognition protein
- IKK
- IκB kinase
- dsRNA
- double-stranded RNA
- ANOVA
- analysis of variance
- JNKK
- JNK kinase
- qRT
- quantitative real-time
- Ub
- ubiquitin
- Uev
- ubiquitin enzyme variant.
References
- 1. Ferrandon D., Imler J. L., Hetru C., and Hoffmann J. A. (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874 [DOI] [PubMed] [Google Scholar]
- 2. Lemaitre B., and Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 3. Kaneko T., Yano T., Aggarwal K., Lim J. H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W. E., Oh B. H., Kurata S., and Silverman N. (2006) PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723 [DOI] [PubMed] [Google Scholar]
- 4. Kim C. H., Paik D., Rus F., and Silverman N. (2014) The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J. Biol. Chem. 289, 20092–20101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paquette N., Broemer M., Aggarwal K., Chen L., Husson M., Ertürk-Hasdemir D., Reichhart J. M., Meier P., and Silverman N. (2010) Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-κB signaling. Mol. Cell 37, 172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glickman M. H., and Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 7. Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., and Chen Z. J. (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548 [DOI] [PubMed] [Google Scholar]
- 8. Ea C. K., Deng L., Xia Z. P., Pineda G., and Chen Z. J. (2006) Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 9. David Y., Ziv T., Admon A., and Navon A. (2010) The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285, 8595–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., and Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Treier M., Seufert W., and Jentsch S. (1992) Drosophila UbcD1 encodes a highly conserved ubiquitin-conjugating enzyme involved in selective protein degradation. EMBO J. 11, 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones D., Crowe E., Stevens T. A., and Candido E. P. (2002) Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michelle C., Vourc'h P., Mignon L., and Andres C. R. (2009) What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 68, 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., and Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 15. Zhuang Z. H., Sun L., Kong L., Hu J. H., Yu M. C., Reinach P., Zang J. W., and Ge B. X. (2006) Drosophila TAB2 is required for the immune activation of JNK and NF-κB. Cell. Signal. 18, 964–970 [DOI] [PubMed] [Google Scholar]
- 16. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., and Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 17. Zhou R., Silverman N., Hong M., Liao D. S., Chung Y., Chen Z. J., and Maniatis T. (2005) The role of ubiquitination in Drosophila innate immunity. J. Biol. Chem. 280, 34048–34055 [DOI] [PubMed] [Google Scholar]
- 18. Vidal S., Khush R. S., Leulier F., Tzou P., Nakamura M., and Lemaitre B. (2001) Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB-dependent innate immune responses. Genes Dev. 15, 1900–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silverman N., Zhou R., Erlich R. L., Hunter M., Bernstein E., Schneider D., and Maniatis T. (2003) Immune activation of NF-κB and JNK requires Drosophila TAK1. J. Biol. Chem. 278, 48928–48934 [DOI] [PubMed] [Google Scholar]
- 20. Ertürk-Hasdemir D., Broemer M., Leulier F., Lane W. S., Paquette N., Hwang D., Kim C. H., Stöven S., Meier P., and Silverman N. (2009) Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thevenon D., Engel E., Avet-Rochex A., Gottar M., Bergeret E., Tricoire H., Benaud C., Baudier J., Taillebourg E., and Fauvarque M. O. (2009) The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe 6, 309–320 [DOI] [PubMed] [Google Scholar]
- 22. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., et al. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 23. Schneider I. (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353–365 [PubMed] [Google Scholar]
- 24. Samakovlis C., Asling B., Boman H. G., Gateff E., and Hultmark D. (1992) In vitro induction of cecropin genes–an immune response in a Drosophila blood cell line. Biochem. Biophys. Res. Commun. 188, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 25. Paquette N., Conlon J., Sweet C., Rus F., Wilson L., Pereira A., Rosadini C. V., Goutagny N., Weber A. N., Lane W. S., Shaffer S. A., Maniatis S., Fitzgerald K. A., Stuart L., and Silverman N. (2012) Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 12710–12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meinander A., Runchel C., Tenev T., Chen L., Kim C. H., Ribeiro P. S., Broemer M., Leulier F., Zvelebil M., Silverman N., and Meier P. (2012) Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 31, 2770–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christensen D. E., Brzovic P. S., and Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 28. Rodrigo-Brenni M. C., and Morgan D. O. (2007) Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 29. Windheim M., Peggie M., and Cohen P. (2008) Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem. J. 409, 723–729 [DOI] [PubMed] [Google Scholar]
- 30. Williamson A., Wickliffe K. E., Mellone B. G., Song L., Karpen G. H., and Rape M. (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 106, 18213–18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wooff J., Pastushok L., Hanna M., Fu Y., and Xiao W. (2004) The TRAF6 RING finger domain mediates physical interaction with Ubc13. FEBS Lett. 566, 229–233 [DOI] [PubMed] [Google Scholar]
- 32. Andersen P. L., Zhou H., Pastushok L., Moraes T., McKenna S., Ziola B., Ellison M. J., Dixit V. M., and Xiao W. (2005) Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamothe B., Besse A., Campos A. D., Webster W. K., Wu H., and Darnay B. G. (2007) Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J. Biol. Chem. 282, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petroski M. D., Zhou X., Dong G., Daniel-Issakani S., Payan D. G., and Huang J. (2007) Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. J. Biol. Chem. 282, 29936–29945 [DOI] [PubMed] [Google Scholar]
- 35. Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., and Chen Z. J. (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergmann A. (2010) The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ. 17, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aggarwal K., Rus F., Vriesema-Magnuson C., Ertürk-Hasdemir D., Paquette N., and Silverman N. (2008) Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 4, e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Basbous N., Coste F., Leone P., Vincentelli R., Royet J., Kellenberger C., and Roussel A. (2011) The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to down-regulate the Imd pathway. EMBO Rep. 12, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guntermann S., Primrose D. A., and Foley E. (2009) Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev. Comp. Immunol. 33, 127–134 [DOI] [PubMed] [Google Scholar]
- 40. Kaneko T., Goldman W. E., Mellroth P., Steiner H., Fukase K., Kusumoto S., Harley W., Fox A., Golenbock D., and Silverman N. (2004) Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649 [DOI] [PubMed] [Google Scholar]
- 41. Khush R. S., Cornwell W. D., Uram J. N., and Lemaitre B. (2002) A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr. Biol. 12, 1728–1737 [DOI] [PubMed] [Google Scholar]
- 42. Kim M., Lee J. H., Lee S. Y., Kim E., and Chung J. (2006) Caspar, a suppressor of antibacterial immunity in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 16358–16363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kleino A., Myllymäki H., Kallio J., Vanha-aho L. M., Oksanen K., Ulvila J., Hultmark D., Valanne S., and Rämet M. (2008) Pirk is a negative regulator of the Drosophila imd pathway. J. Immunol. 180, 5413–5422 [DOI] [PubMed] [Google Scholar]
- 44. Lhocine N., Ribeiro P. S., Buchon N., Wepf A., Wilson R., Tenev T., Lemaitre B., Gstaiger M., Meier P., and Leulier F. (2008) PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158 [DOI] [PubMed] [Google Scholar]
- 45. Werner T., Borge-Renberg K., Mellroth P., Steiner H., and Hultmark D. (2003) Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J. Biol. Chem. 278, 26319–26322 [DOI] [PubMed] [Google Scholar]
- 46. Leulier F., Lhocine N., Lemaitre B., and Meier P. (2006) The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol. Cell. Biol. 26, 7821–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pham L. N., Dionne M. S., Shirasu-Hiza M., and Schneider D. S. (2007) A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 3, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]