Abstract
Trypanosoma cruzi, the etiological agent of Chagas disease, is a protozoan parasite with a complex life cycle involving a triatomine insect and mammals. Throughout its life cycle, the T. cruzi parasite faces several alternating events of cell division and cell differentiation in which exponential and stationary growth phases play key biological roles. It is well accepted that arrest of the cell division in the epimastigote stage, both in the midgut of the triatomine insect and in vitro, is required for metacyclogenesis, and it has been previously shown that the parasites change the expression profile of several proteins when entering this quiescent stage. However, little is known about the metabolic changes that epimastigotes undergo before they develop into the metacyclic trypomastigote stage. We applied targeted metabolomics to measure the metabolic intermediates in the most relevant pathways for energy metabolism and oxidative imbalance in exponentially growing and stationary growth-arrested epimastigote parasites. We show for the first time that T. cruzi epimastigotes transitioning from the exponential to the stationary phase exhibit a finely tuned adaptive metabolic mechanism that enables switching from glucose to amino acid consumption, which is more abundant in the stationary phase. This metabolic plasticity appears to be crucial for survival of the T. cruzi parasite in the myriad different environmental conditions to which it is exposed during its life cycle.
Keywords: cell growth, cell metabolism, energy metabolism, metabolomics, Trypanosoma cruzi, Chagas disease, epimastigotes, oxidative imbalance
Introduction
Cell growth, both in natural environments or in in vitro culture, usually presents in two phases: the exponential phase where cells divide at a roughly constant rate, and the stationary phase where cells slow down or stop the cell cycle and division. During the exponential phase, cells go through the well described canonical cell cycle of G1 → S → G2 → M. In the stationary phase, cells leave the regular cell cycle to enter a resting state known as quiescence, which is highly relevant because it corresponds to the physiological state in which most cells from both unicellular and multicellular organisms spend most of their life (1).
Trypanosoma cruzi, the etiological agent of Chagas disease, is a protozoan parasite with a complex life cycle involving a triatomine insect and mammals. Throughout its life cycle, the T. cruzi parasite faces several alternated events of cell division and cell differentiation. Briefly, insects are infected during a blood meal from a mammal having non-dividing forms of the parasite, denominated trypomastigotes, present in their blood. Once in the midgut of the triatomine insect, the trypomastigotes differentiate to replicative non-infective epimastigotes, which proliferate and colonize the digestive tube. After the blood meal, nutrients are consumed by the intestinal epithelium, the parasite, and the intestinal microbiota population; therefore, the replicative epimastigotes eventually face nutrient starvation. Under this situation of metabolic stress, cell division is arrested, and the parasites adhere to the intestinal epithelium along the midgut. In the terminal portion of the digestive tube, epimastigotes start the process to differentiate to infective, non-dividing metacyclic trypomastigotes (2). Interestingly, this differentiation process, called metacyclogenesis, is fueled by amino acids present in the urine at the distal portion of the triatomine's intestine such as proline (Pro), aspartate (Asp), and glutamate (Glu), among others (3). In a new blood meal on a mammalian host the infected triatomine, which usually defecates near the biting point, will contaminate the skin or mucosa with metacyclic trypomastigotes expelled along with their feces, allowing the parasite to internalize and establish infection in a new host (2).
It is well accepted that arrest of the cell division in the epimastigote stage, both in vivo and in vitro, is required for metacyclogenesis, and it has been previously shown that the parasites change the expression profile of several proteins when entering this quiescent stage (4). In addition, a previous work comparing exponential and stationary phases of T. cruzi epimastigotes (EPE5 and SPE, respectively) has assessed the physiological changes occurring at each stage and how some genes are regulated during the epimastigote growth to understand their possible participation in the metacyclogenesis (4–6). However, little is known about metabolic changes that occur along with this physiological transition, and there are no reported studies supporting a broader view of the adaptive changes that epimastigotes undergo before they develop into metacyclic trypomastigote stage.
Early studies have focused on specific enzymes involved in energy metabolism as well as a specific subset of metabolites (7). However, a systematic analysis of the metabolic changes associated with energy metabolism that occurs during exponential and stationary phases of growth in T. cruzi epimastigotes has not been performed. We hypothesized that T. cruzi epimastigotes finely orchestrate a metabolic switch, especially related to the energy metabolism, when exposed to nutritional starvation. Here, we show for the first time T. cruzi epimastigotes transitioning from exponential to stationary phase (Fig. 1) present a distinct metabolic profile regarding their energy metabolism and redox imbalance, including amino acids, carbohydrates, and the tricarboxylic acids (TCA) cycle intermediates.
Figure 1.
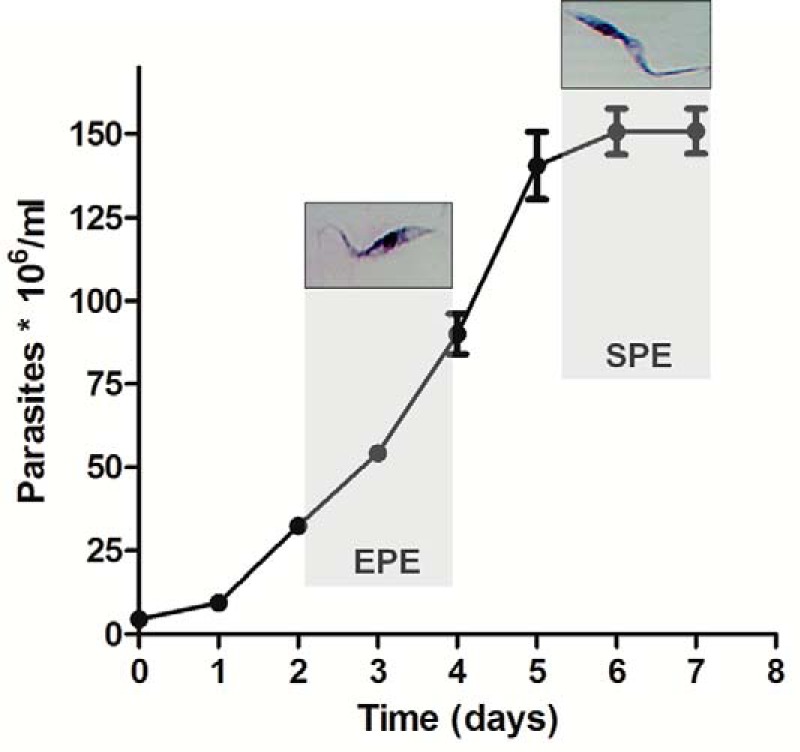
Representative growth curve of T. cruzi epimastigotes in culture indicating the EPE and SPE growth phases where samples were collected for targeted metabolomic analyses. A representative image of light microscopy is shown for each phase.
Results
Comprehensive targeted metabolomics
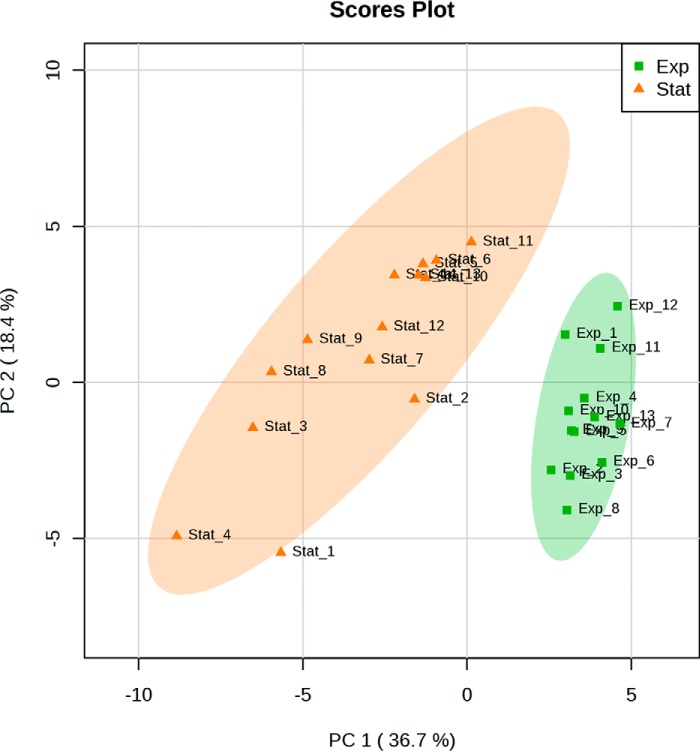
To assess the main metabolic changes between exponentially growing and stationary arrested epimastigote T. cruzi parasites, we focused on a set of 47 metabolic intermediates from the most relevant pathways for energy metabolism and oxidative imbalance. Selected metabolites (supplemental Table S1) were analyzed in positive and negative modes using two different chromatography separations. Our initial hypothesis was that EPE and SPE constitute two different populations in terms of energy metabolome composition due to their adaptation to different nutritional conditions. To assess this hypothesis, we performed a principal component analysis (PCA) where results are displayed as score plots, and each point represents a sample that when clustered together indicates similar metabolite composition based on the selected metabolites for the targeted analysis (Fig. 2). PCA analysis revealed a clear separation between the two growth phases of T. cruzi epimastigotes. This result strongly supports the proposed hypothesis that T. cruzi EPE and SPE present different energy and oxidative imbalance-related metabolic states. Interestingly, our results also revealed a greater metabolic heterogeneity among stationary phase cells than exponential phase cells.
Figure 2.
PCA model score plot of LC-MS/MS data from analyzed T. cruzi epimastigotes samples comparing exponential (n = 13, squares) and stationary (n = 14, triangles) phases outlines a separation between the two phases.
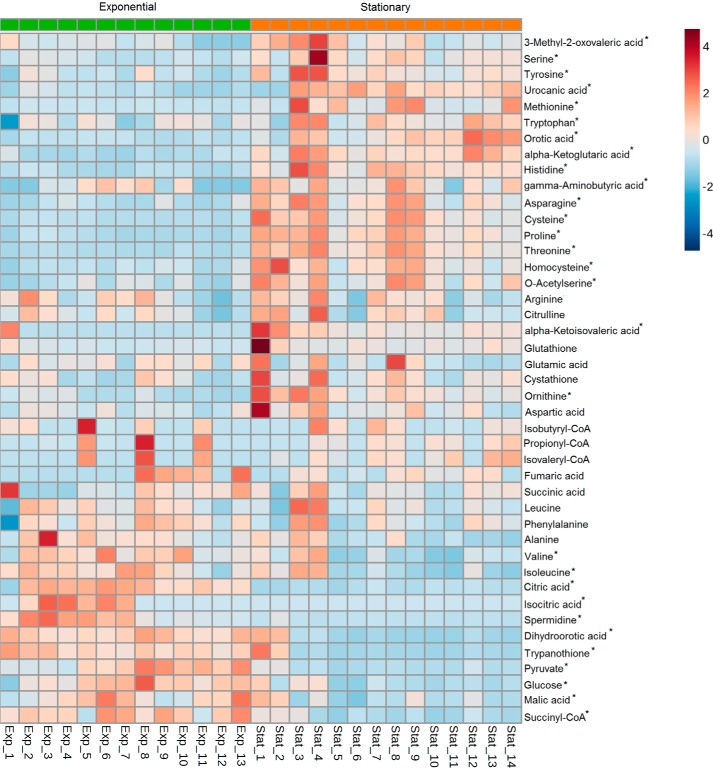
To further identify the underlying metabolites responsible for the segregation between both T. cruzi EPE and SPE, a heat map analysis was performed. In contrast to the scores plots, a heat map displays the actual data values that allow visualization of changing patterns in metabolite levels across samples and across experimental conditions. The heat map analysis showed that amino acids and their metabolic intermediates were increased in the stationary phase when compared with exponential phase, whereas glycolysis and TCA intermediates were reduced (Fig. 3 and supplemental Fig. 1). Consequently, we pursued a more detailed analysis for each metabolic pathway to assess specific changes that occur when parasites are exposed to nutritional starvation.
Figure 3.
Heat map of metabolites detected in T. cruzi epimastigotes from selected metabolic pathways. Each line represents a metabolite detected by LC/MS-MS analysis, and values are represented as the metabolite/internal standard area ratio of MS signals detected by MRM normalized to the cell numbers. Columns represent each technical replicate from seven independent experiments in exponential phase (green labeled on the top-left side) and stationary phase (orange labeled on the top-right side). *, metabolites that presented a significant difference between exponential and stationary phases according to the Benjamini and Hochberg test (supplemental Table S2). The colors vary from deep blue (very low data value) to dark brown (extremely high data value).
Energy metabolism: Glycolysis and TCA
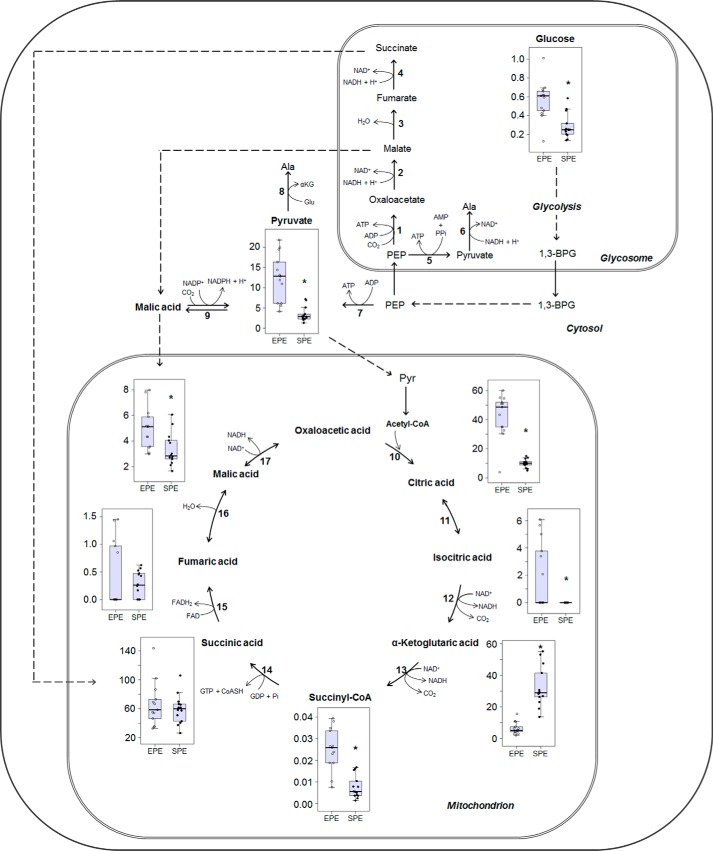
To study in more detail the metabolic switch from a glucose-based to an amino acid-based metabolism in T. cruzi epimastigotes, we measured the relative levels of the first and last metabolites of glycolysis, i.e. glucose and pyruvate (Pyr), and all the intermediates of the TCA cycle, except oxaloacetate that could not be resolved by any of the two chromatographic methods used for this study (Fig. 4). As shown in the heat map (Fig. 3), both glycolysis and most of the TCA-related metabolite levels decreased during the transition of parasites from EPE to SPE. The relatively higher levels of glucose during the exponential phase as compared with the stationary phase could be explained by its uptake from the extracellular medium by the exponentially replicating cells due to its presence in high concentrations in the fresh medium (2 mg/ml). Notably, the intracellular concentration of glucose was still detected in SPE instead of being completely consumed as we would expect because extracellular glucose has been depleted at that stage (8). This finding may indicate that in SPE either amino acids are serving for de novo synthesis of glucose to maintain a certain intracellular level or its consumption is arrested.
Figure 4.
Glycolysis and TCA cycle intermediate levels compared in exponential and stationary phases of T. cruzi epimastigotes. Values correspond to the metabolite/internal standard area ratio of MS signals detected by MRM normalized to the cell numbers. * indicates significant differences between EPE and SPE according to the Benjamini and Hochberg test (supplemental Table S2). Numbers indicate the corresponding enzymes. 1, Phosphoenolpyruvate carboxykinase; 2, malate dehydrogenase (glycosomal); 3, fumarate hydratase (glycosomal); 4, NAD-linked fumarate reductase; 5, pyruvate-phosphate dikinase; 6, alanine dehydrogenase; 7, pyruvate kinase; 8, alanine aminotransferase; 9, NADP-linked malic enzyme; 10, citrate synthase; 11, aconitase; 12, isocitrate dehydrogenase; 13, α-ketoglutarate dehydrogenase; 14, succinyl CoA-synthetase; 15, succinate dehydrogenase; 16, fumarase; 17, malate dehydrogenase. Dashed arrows indicate several enzymatic reactions.
Alongside a decrease in the glucose content, the relative abundance of Pyr as well as the metabolic intermediates canonically linked to the TCA cycle, such as isocitrate, citrate, succinyl-CoA, and malate, were significantly reduced in SPE when compared with EPE (Fig. 4 and supplemental Table S2). By contrast, α-ketoglutarate (α-KG) was the only derivative from the amino acid oxidation that appeared to exhibit around six times higher abundance in the SPE. The increased levels of α-KG are compatible with an increase of amino acids catabolism in SPE (Fig. 5). T. cruzi epimastigotes depend on different catabolic routes leading to α-KG generation for energy production, such as the Pro (9, 10) or His (11) oxidation pathways, both rendering Glu that can be converted into α-KG through a reductive deamination or by transamination of the –NH2 to Pyr forming Ala (12). It worth mentioning that several metabolites are present simultaneously in different subcellular compartments such as malate, which is present in the glycosome, cytoplasm, and mitochondrion (Fig. 4). Because we measured the total level of a metabolite, the detected changes in their levels are the result of all metabolic contributions independently of their localization.
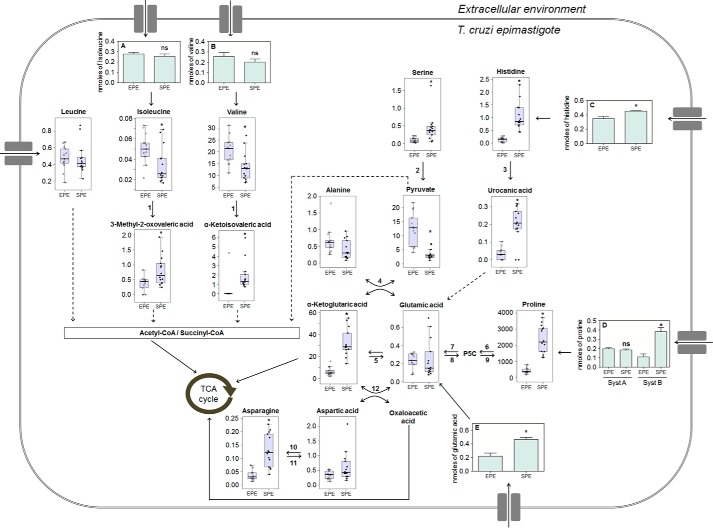
Figure 5.
Metabolic network of selected amino acids and their intermediates in EPE and SPE phases of T. cruzi epimastigotes. Transport of amino acids are represented by light green bars, and values correspond to nanomoles of Ile (A), Val (B), His (C), Pro (D), and Glu (E) incorporated by 2.0 × 107 parasites in EPE and SPE. Values of metabolite levels correspond to the metabolite/internal standard area ratio of MS signals detected by MRM normalized to the cell numbers. * indicates metabolites with a significant difference between EPE and SPE according to the Benjamini and Hochberg test (supplemental Table S2). Differences in the transport of amino acids were found as statistically significant by using unpaired Student's t test (proline System B p < 0.001, histidine p < 0.041, glutamate p < 0.01). N.S. indicates that no significant variation was observed in the transport of the corresponding amino acid. Dashed arrows represent more than one enzymatic reaction. Numbers indicate the corresponding enzymes: 1, tyrosine aminotransferase; 2, serine/threonine dehydratase; 3, histidine ammonia-lyase; 4, alanine aminotransferase; 5, glutamate dehydrogenase; 6, proline dehydrogenase; 7, pyrroline-5-carboxylate dehydrogenase; 8, pyrroline-5-carboxylate synthetase; 9, pyrroline-5-carboxylate reductase; 10, asparagine synthetase; 11, asparaginase; 12, aspartate aminotransferase.
Glutamate metabolism
Glu and α-KG constitute the main metabolic bridge between amino acid metabolism and the TCA cycle. Interestingly, our results showed that the levels of Glu remained nearly constant in both growth phases. As shown in Fig. 5, it is worth stressing that Glu is a branching point for several metabolic pathways. Epimastigotes are able to acquire Glu through a variety of processes, which include its uptake from the extracellular medium, biosynthesis by reductive amination of α-KG catalyzed by glutamate dehydrogenase, which is NADH- and NADPH-dependent, transamination reactions, or by oxidizing other amino acids such as Pro, His, and glutamine (Gln). Notably, both Glu uptake and its precursors such as α-KG, Pro, His, Asp and its derivative asparagine were detected at higher levels in the SPE as compared with EPE (Fig. 5). In addition, the uptake of Pro and His was also increased in SPE. Because their main metabolic fate is Glu, our results suggest that the metabolism in SPE shifts toward Glu production and further oxidation in the TCA cycle through α-KG. However, it is also possible that the concomitant increase of Pro, His, and Glu uptake could lead to an accumulation of these precursors. In both cases, our findings are in agreement with the hypothesis that Glu or its 2-oxoacid derivative is critically involved in the parasite's survival in this phase.
Histidine metabolism
T. cruzi is the only trypanosomatid equipped with putative genes encoding the four enzymatic steps connecting His to Glu as follows: His ammonia-lyase, urocanate hydratase, imidazolone propionase, and formiminoglutamase. Recently, it was shown that this parasite can fully oxidize His, producing CO2 and triggering an increase of the mitochondrial membrane potential and O2 consumption (11). In addition, His is an essential amino acid in T. cruzi; therefore, its intracellular levels depend on both its uptake and degradation rates. Our results show that the intracellular levels of His and urocanate, the first metabolic intermediate in the His metabolism, were significantly increased in SPE as compared with EPE. Therefore, we measured His transport-specific activity in both EPE and SPE populations, and it was slightly increased in SPE when compared with EPE (Fig. 5 and supplemental Table S2). The small increase in His uptake could contribute to the increased intracellular levels of His and urocanate in SPE.
Proline metabolism
Our results showed that the intracellular levels of Pro were significantly increased in SPE as compared with EPE (Fig. 5 and supplemental Table S2). Pro levels were shown to be critical in several biological processes in T. cruzi epimastigotes, such as differentiation, resistance to oxidative imbalance, as well as response to nutritional and thermal stress (10, 13–16). Differently from His, epimastigotes were able to synthesize Pro. Therefore, the balance of Pro levels is more complex than His and is determined by the interplay among its degradation, biosynthesis from Glu, and uptake rates through two active transporters. Similar to most organisms, Pro degradation to Glu occurs through two enzymatic steps where Pro is first converted into Δ1-pyrroline-5-carboxylate (P5C) by a Pro dehydrogenase with the production of FADH2 followed by conversion into Glu by P5C dehydrogenase (P5CDH) with the production of NADH (Fig. 5 and supplemental Table S2). Both enzymatic steps by themselves are able to energize the mitochondria, promoting the production of ATP without requiring the deamination of Glu and further full oxidation through the TCA cycle and oxidative phosphorylation to obtain energy. Differently from other organisms, this pathway is not branched because there is no functional ornithine aminotransferase in T. cruzi epimastigotes able to interconvert P5C and ornithine (12, 17).
The lack of an ornithine aminotransferase interconverting ornithine and P5C indicates that all of the oxidized Pro have to be converted into P5C/γGS and Glu and that all the biosynthesized Pro have to be produced from Glu. Thus, the relative gene expression levels and specific enzymatic activities of P5CR (Pro biosynthesis) and P5CDH (Pro degradation) both in EPE and SPE could indicate the predominant direction of Pro metabolism (biosynthesis or degradation) in each phase (Fig. 6). Interestingly, both protein and enzymatic activity levels of P5CR were higher in the EPE than in the SPE, whereas the opposite was observed for P5CDH (Fig. 6, B and C). This suggests an up-regulation of the synthesis of Pro during the exponential phase, which could drive the metabolism to a pre-adaptive accumulation of this amino acid. It is worth mentioning that Pro can be consumed in SPE to support metacyclogenesis and can be used by metacyclic trypomastigotes to energize the invasion of the mammalian hosts (18). In fact, from all the selected metabolites analyzed in this study, Pro presented the highest levels, measured as metabolite/internal standard area ratio of MS signals detected by MRM normalized to the cell numbers, in both T. cruzi EPE and SPE (Fig. 5). Because we detected differences between Pro synthesis and degradation in EPE and SPE, we evaluated the contribution of its uptake in both growth phases. Previously, we described and characterized two Pro transport systems, designated A and B (19). Both systems had different kinetic and thermodynamic characteristics, showing substrate saturations at 0.75 mm (system A) and 3 mm (systems A + B). Thus, transport activities for both systems were measured at substrate-saturating concentrations. Interestingly, system A did not show variations in Pro uptake, but system B increased ∼3 times its activity (Fig. 5). The increase in the Pro uptake may be compensating for the simultaneous up-regulation of Pro consumption and down-regulation of Pro synthesis, avoiding a futile cycle, and could be responsible for Pro accumulation in SPE.
Figure 6.
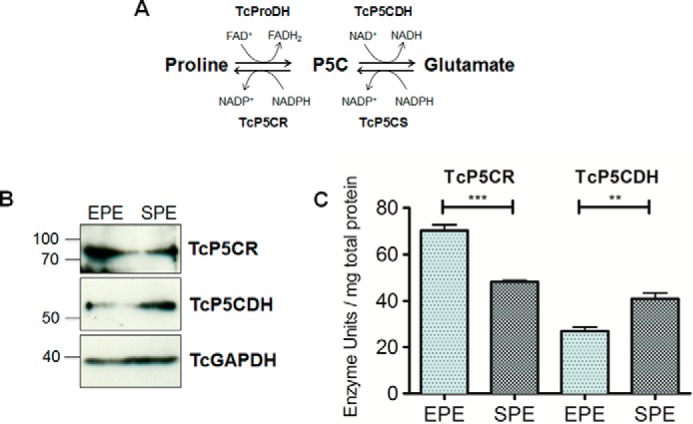
Proline metabolism is finely regulated in T. cruzi epimastigotes transitioning from exponential to stationary phase of growth. A, summary of the main intermediates involved in the proline (Pro) glutamate (Glu) interconversion pathway of T. cruzi. Proline oxidation to P5C is catalyzed by a FAD-dependent proline dehydrogenase (TcProDH). Subsequently, P5C is spontaneously converted into γ-glutamate semialdehyde and then oxidized to glutamic acid by a NAD(P)+-dependent Δ1-pyrroline-5-carboxylate dehydrogenase (TcP5CDH). Glutamic acid can be reduced into P5C in two enzymatic steps catalyzed by a bi-functional P5C-synthetase hydrolyzing ATP and oxidizing NADPH. P5C produced from glutamic acid can be reduced into proline by a P5C-reductase with oxidation of NADPH. B, analysis of expression of two proline metabolism enzymes (TcP5CDH and TcP5CR) by Western blotting. Protein extracts were prepared from parasites (1 × 107 parasites per lane) in EPE or SPE phase of growth. Proteins were blotted and probed with corresponding polyclonal antibodies produced in mouse against TcP5CS (83 kDa), TcP5CDH (63 kDa), and glyceraldehyde-3-phosphate dehydrogenase, used as loading control (TcGAPDH-39 kDa). C, comparison of enzymatic activities in parasites from EPE and SPE. Cell-free protein homogenates were prepared as described previously and used in the enzymatic test. Enzymatic rates were determined spectrophotometrically by following the changes in absorbance (λ340 nm) of NADPH oxidation (decrease) or NAD+ reduction (increase) in the TcP5CR or TcP5CDH assays, respectively. One enzyme unit represents 1 μmol of NADP+ or NADH produced per min, accordingly. Bar represents means ± S.D. from four biological replicates. Differences were found as statistically significant by using unpaired Student's t test. Significance values are depicted by ***, p < 0.001, or **, p < 0.05, by comparing exponential over stationary values.
Branched chain amino acid metabolism
Leucine (Leu), isoleucine (Ile), and valine (Val) are branched-chain amino acids (BCAA), and the only ones involved in the energy production in T. cruzi that are not directly connected to Glu (20). These amino acids are oxidized to acetyl- or methylmalonyl-CoA and further oxidized through the TCA cycle. Similar to most eukaryotic cells, BCAA are essential for T. cruzi. Therefore, their intracellular levels also depend on their uptake and metabolism. As described previously, all three BCAA are taken up by the same transporter (21), and their specific transport activity did not change in SPE when compared with EPE (Fig. 5). The fact that the intracellular levels for Ile and Val are slightly reduced, while Leu remained constant in SPE compared with EPE, indicates that Ile and Val catabolism is more active than Leu during the stationary phase (Fig. 5).
Thiol-containing amino acids
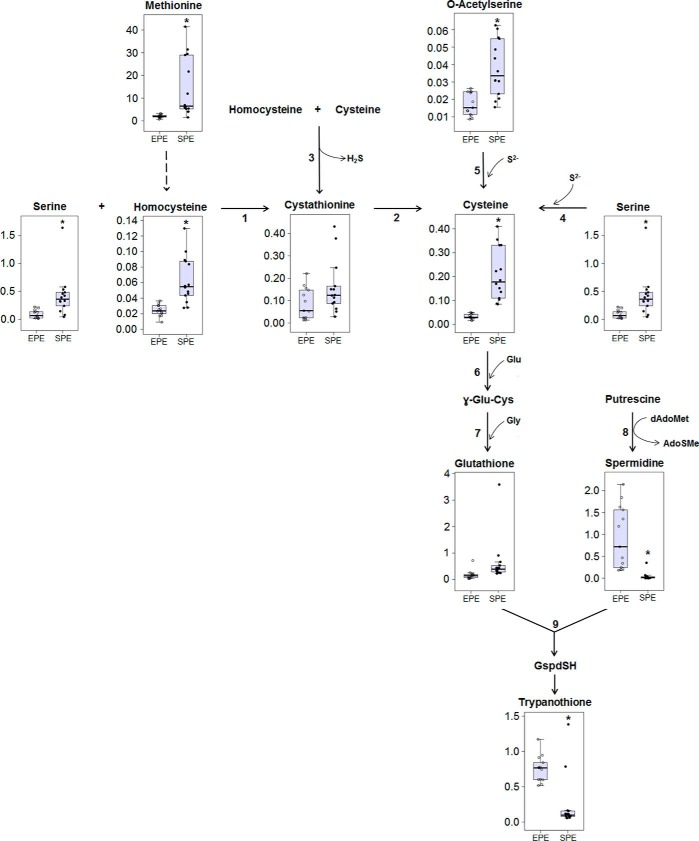
An important group of metabolites related to energy metabolism and defense against oxidative stress are polyamines and the thiol-containing amino acids. To assess potential variations of these thiol-containing metabolites between EPE and SPE, we measured the levels of intermediates involved in the different routes for cysteine (Cys) biosynthesis as well as potential variations in the levels of glutathione, spermidine, and trypanothione (TSH). The latter represents the main antioxidant agent in the redox metabolism in trypanosomatids. As shown in Fig. 7, the relative abundance of the intermediates belonging to the reverse trans-sulfuration pathway (RTP) (serine, homocysteine, and cystathionine) as well as those related to de novo synthesis of cysteine (serine and O-acetylserine) were increased in SPE as compared with EPE. These results strengthened previous findings showing that unlike most living organisms T. cruzi displays an unusual redundancy in cysteine production by the co-existence of de novo synthesis and the RTP. As schematized in Fig. 7, precursors such as serine, acetyl-CoA, and O-acetylserine are required for de novo synthesis of cysteine. This is a two-step pathway catalyzed by serine acetyltransferase and cysteine synthase, and both enzymes have been found to be functional in T. cruzi (22, 23). Interestingly, cystathionine β-synthase (CBS), which catalyzes the first step of the RTP, is distinguished from its mammalian counterpart by its broader substrate specificity. In addition to catalyzing the condensation of serine with homocysteine to produce cystathionine, the T. cruzi CBS can also act as a serine sulfhydrylase and a cysteine synthase. Given that cystathionine γ-lyase is also functional in T. cruzi, cystathionine is expected to be also cleaved to produce cysteine, the end product of the RTP.
Figure 7.
Thiol-containing metabolite levels and cellular redox state in EPE and SPE. Schematic representation of cysteine and trypanothione biosynthetic pathways. The reactions catalyzed by 1 (cystathionine-β-synthase) and 2 (cystathionine-γ-lyase) correspond to the canonical RTP. The metabolic step 3 is also catalyzed by CBS to produce cystathionine and H2S via condensation of homocysteine and cysteine, the latter being the preferred substrate. Reaction 4 illustrates CBS contribution to the de novo synthesis of cysteine by its serine sulfhydrylase activity. Reaction 5 depicts involvement of cysteine synthase and CBS in the de novo synthesis of cysteine, 6, glutamylcysteine synthetase; 7, glutathione synthetase; 8, spermidine synthase; 9, trypanothione synthetase. Dashed arrows represent more than one enzymatic reaction. Values correspond to the metabolite/internal standard area ratio of MS signals detected by MRM normalized to the cell numbers. *, indicates significant differences between EPE and SPE according to the Benjamini and Hochberg test (supplemental Table S2).
In contrast, synthesis of TSH is known to be essential for neutralizing free radicals, thereby contributing to the redox homeostasis in all of the developmental stages of T. cruzi. However, our results unexpectedly showed that the availability of Glu and the higher abundance of cysteine in SPE were not reflected in an increase in the relative abundance of TSH in SPE (Fig. 7). It is possible that the synthesis of TSH is limited by the significantly lower abundance of spermidine in SPE, which is essential to accomplish the final steps in TSH biosynthesis.
T. cruzi is the only eukaryotic organism unable to synthesize polyamines de novo because it lacks ornithine and arginine decarboxylases, the enzymes catalyzing the first step in polyamines biosynthesis (24, 25). Interestingly, despite there being no evidence of the urea cycle in the parasite's genome, ornithine and citrulline were detected in both growth phases, and ornithine levels were around four times higher in SPE compared with EPE, whereas arginine and citrulline levels were similar between both phases (supplemental Fig. 1). Spermidine is involved in cell cycle control in trypanosomatids (26). As mentioned above, our results showed that the intracellular levels of spermidine were decreased in SPE compared with EPE (Fig. 7). This finding is in agreement with the fact that in SPE the cellular replication process, one of the most polyamines demanding cellular processes, is arrested.
Discussion
In this work, we applied a targeted metabolomic approach to assess the metabolic changes that occur in T. cruzi epimastigote parasites transitioning from the exponential to the stationary phase of growth. Herein, we established a correlation between the relative abundance of assessed metabolites with a metabolic process resulting from the nutritional conditions that parasites have to face during exponential and stationary growth. We carefully normalized the data, and the presented results have been obtained from independent biological replicates that also included new media preparation with components from different batches. Therefore, the patterns observed among metabolites represent a specific response to nutritional starvation under the experimental conditions used for this study.
The first systematic analysis of the different phases of a growth curve was reviewed by J. Monod in his classical work “The Growth of Bacterial Cultures” (27). In that seminal work, the author established that bacterial growth constitutes “a method for study of bacterial physiology and biochemistry” ruling out the argument that growth phases are referred to as just a laboratory phenomenon. General aspects of growth curves analysis described by Monod (27) are valid not only for prokaryotic organisms but also applies to most unicellular organisms, including unicellular fungi and protozoa.
Cells of exponential and stationary phases of growth correspond to different physiological states (1); therefore, it is expected that these differences will be reflected at the metabolic level. In the specific case of T. cruzi, the occurrence of these two well-defined phases of parasite growth in the gut environment of its natural triatomine host was recently documented by analyzing the development of the parasite in the different portions of the insect gut (28). Dias Fde et al. (28) showed that parasite densities remain constant over time in the anterior and posterior midgut, whereas parasites in the hindgut exhibited an exponential growth profile within the first 5 days of post-infection. It should be stressed that parasites reported as being in the stationary phase of growth were epimastigotes because metacyclic trypomastigotes started to appear at day 7 post-infection and did not constitute more than 7% of the total cells at that time. These findings by Dias Fde et al. (28) are consistent with a previous review of the stationary phase phenomena suggesting that a potential metabolic switch in parasites during the stationary phase may represent a pre-adaptive stage for metacyclogenesis, a critical differentiation step inside its invertebrate host (4). As a first approach to assess in more detail these adaptive metabolic changes, we performed a targeted metabolomic analysis of 47 metabolites to address the hypothesis that EPE and SPE cells constitute two different populations in terms of metabolite composition. From the selected metabolites, spermine, putrescine, acetyl-CoA, and β-methylcrotonyl-CoA were not detected in EPE nor SPE under our experimental conditions. The data presented in this work support our hypothesis (Fig. 2). Interestingly, PCA analysis revealed greater variation among SPE samples than EPE samples. Among the factors that can contribute to differences in the metabolite profile is the fact that the stationary phase is defined by the absence of net growth of the cell population, which is composed of a heterogeneous population of replicating, quiescent, and dying cells. Replicating and quiescent cells can be undergoing a combination of complex stress-resistant strategies, from the production of anti-oxidant defenses to autophagy, contributing to the variations observed at the metabolic level among SPE samples.
We initially hypothesized that T. cruzi epimastigotes finely orchestrate a metabolic switch, especially related to energy metabolism, when exposed to nutritional starvation. Glycolysis and the TCA cycle are considered the major metabolic pathways for production of ATP and reducing equivalents in most mitochondrion-bearing organisms. Exponentially growing T. cruzi epimastigotes are not an exception because all the TCA cycle intermediates were detected in EPE, and enzymes involved in the TCA cycle are predicted to be functional (7). Moreover, epimastigotes can use either glucose or amino acids as a source of energy for growth, although glucose is preferred during the exponential phase, and amino acids are preferred during the stationary phase (7, 29, 30). Consistent with these previous findings, our results showed the expected variations for free glucose and Pyr, by comparing EPE and SPE first and last metabolites of the glycolytic pathway. As mentioned above, glucose was still detected in SPE despite extracellular glucose having been depleted at that stage (8), which may indicate that in SPE either amino acids are serving for de novo synthesis of glucose to maintain a certain intracellular level or glucose consumption is arrested. T. cruzi has the two essential enzymes for gluconeogenesis: a reversible phosphoenolpyruvate carboxykinase, which indicates that gluconeogenesis from several amino acids may occur, and a fructose-1,6-bisphosphatase (31, 32). However, the parasite lacks a reserve polysaccharide. A detailed metabolic analysis of this pathway has not been performed, and our results suggest a potentially critical role of intracellular levels of glucose in this parasite, which certainly warrants further studies.
Remarkably, succinate and fumarate did not show significant differences between EPE and SPE (Fig. 4). Succinate can be converted to fumarate by succinate dehydrogenase, and fumarate reductase converts fumarate into succinate. Succinate dehydrogenase is the only TCA enzyme that is a structural component of the respiratory chain. In addition, this enzyme is part of the fully functional complex II in T. cruzi, which functions as the first respiratory chain complex because complex I is not functional in this organism (12). Moreover, a fumarate reductase activity was previously described in T. cruzi mitochondrial fractions that can reversibly reduce fumarate into succinate (29). This enzyme is present in several bacteria, protozoa, yeasts, and other eukaryotes that are submitted to low O2 tension in some or all of the natural environments in which they live. Fumarate reductase contributes to the TCA to proceed in a reverse mode under anaerobic conditions, so NAD+ can be regenerated from NADH-stimulating glycolysis and ATP production by regulation of the levels of substrate phosphorylation (7, 29). Because of these overlapped enzymatic activities of fumarate reductase and succinate dehydrogenase, where the main difference is the cofactor used as donor or acceptor of electrons, some controversy was generated regarding whether both enzymes are present in the parasite or both reactions are catalyzed by the same enzyme. A cytoplasmic dihydroorotate dehydrogenase with a unique fumarate reductase activity was described in T. cruzi, resolving this point (17). Interestingly, despite the total levels of fumarate and succinate not changing in EPE and SPE, a significant reduction in dihydroorotate and an increase in orotate were detected in SPE as compared with EPE (Fig. 4 and supplemental Fig. 1). An increased level of orotate, a precursor for pyrimidine biosynthesis, was surprising because RNA and DNA biosynthesis are expected to be reduced in SPE. These results support a previous hypothesis that fumarate/succinate balance may regulate the redox homeostasis in this parasite and that dihydroorotate dehydrogenase is a potential drug target due to its unique activity (17). One potential explanation for a tight regulation of succinate/fumarate levels could be that variations of their levels could unbalance the electron flow through the respiratory chain, modifying the mitochondrial inner membrane potential. In addition, it could affect the ATP levels in the following two ways: 1) through complex V of the respiratory chain, which depending on the polarization state of the mitochondria can operate in the ATP synthesis mode using the H+ gradient as driving energy or in the H+ pump mode re-establishing ΔΨ by ATP hydrolysis, or 2) through the availability of NAD+ to keep glycolysis working. In summary, the fact that fumarate and succinate levels remained constant in EPE and SPE, despite other TCA and pyrimidine intermediates levels changing when comparing EPE and SPE, supports the hypothesis that their homeostasis is critical for the cell survival.
As mentioned above, a metabolic switch from glucose to amino acid consumption occurs when EPE progresses to SPE and Glu is an intermediate of oxidation when Pro or His are used as energy sources. Our results showed a significant increase in His and its first metabolic product, urocanate (Fig. 5). Because there are no alternative routes for His degradation besides Glu synthesis, and neither biochemical evidence nor putative genes encoding enzymes for His biosynthesis were found in T. cruzi, the observed changes can only be attributed to an increase in His uptake. In fact, transport experiments showed increased transport of His in SPE when compared with EPE (Fig. 5). This is the first time that a systematic analysis of the metabolic changes induced by nutritional stress is assessed in T. cruzi epimastigotes during their exponential and stationary phases of growth. Our results are in agreement with recent findings regarding the metabolic environment in the gut of the insect host where a reduced availability of oxygen implies anaerobic fermentation of glucose and amino acid carbon sources. Interestingly, His is the predominant free amino acid in the triatomine feces (33), which correlates with our results showing that His uptake and its metabolism is increased in SPE as a metabolic adaptation under nutritional starvation.
In contrast, interpretation of the high levels of Pro detected in our analysis is more complex because (i) putative genes for Pro biosynthesis from Glu are present in the T. cruzi genome, and Pro biosynthesis was recently demonstrated in this parasite;6 and (ii) Pro uptake occurs through two different transporters (19). Therefore, changes in the free levels of Pro in SPE as compared with EPE are the result of an interplay among consumption, biosynthesis, and uptake through two active transporters. Our results showed that despite Pro consumption increases in SPE, intracellular levels of Pro are also increased in these cells (Fig. 6); therefore, an increase in Pro biosynthesis and/or uptake should be considered. In fact, both protein levels and enzymatic activity of the enzymes required to convert Glu into Pro were reduced, which is consistent with directing Pro to oxidation, whereas both Pro transport systems notably increased their activity. This suggests a predominant synthesis of Pro during the exponential phase, driving the metabolism to a pre-adaptive accumulation of this amino acid that can be consumed in SPE to support metacyclogenesis, and it can be used by metacyclic trypomastigotes to energize the invasion of the mammalian hosts (18). Altogether, our results support the hypothesis that when parasites progress from EPE to SPE, cells finely orchestrate a metabolic switch when exposed to nutritional starvation as indicated by the reduction of glucose levels and an increased transport of Pro and His.
Besides Pro and His, BCAA can be also oxidized through the TCA cycle through their conversion into acetyl-CoA. It is worth mentioning that acetyl-CoA was not detected either in EPE or SPE. All three BCAA are taken up by the same transporter, and because the first steps of their degradation pathways are also catalyzed by the same enzymes, it would be expected that their levels would follow a similar pattern in EPE and SPE. Surprisingly, the intracellular levels of Ile and Val were reduced in SPE as compared with EPE, whereas Leu remained constant (Fig. 5). These results strongly suggest a differential metabolic regulation of Leu degradation compared with Ile and Val. The fact that both Val and Ile levels are reduced and their derivatives α-keto-isovalerate and 3-methyl-2-oxovalerate are increased in SPE indicates an increased consumption of these amino acids with production of the corresponding ketoacids that are the products of a transamination reaction that, differently from other organisms, in T. cruzi could be catalyzed by a tyrosine or aspartate aminotransferase.7 The biological relevance of a preferential use of Ile and Val over Leu remains to be addressed and may unveil novel potential drug targets.
As mentioned before, polyamines and the thiol-containing amino acids are an important group of metabolites related to energy metabolism and the defense against oxidative stress. For the first time, our findings presented herein provide experimental evidence that a reduced synthesis of polyamines has a negative impact on TSH levels (Fig. 7). The significantly increased levels of cysteine detected in SPE would compensate in some way for the strong need of reducing power for detoxification of reactive oxygen and nitrogen species that are more actively produced in SPE. It is noteworthy to mention that T. cruzi CBS is able to catalyze different reactions leading to H2S production that could play signaling or regulatory functions on varied proteins by a covalent modification of their essential thiols, including those related to the RTP and de novo synthesis of cysteine. It is important to keep in mind that both processes take place in the cytosol. The findings reported here show for the first time the co-existence of intermediates related with de novo synthesis of cysteine such as O-acetylserine in parallel with those belonging to the RTP such as cystathionine. This observation alongside previous results showing the functionality of the enzymes involved in de novo synthesis as well as the RTP (23) provide strong evidence for the operability of both routes in T. cruzi. In addition to the described metabolic uniqueness, our results showed that these processes are differentially used in both phases of epimastigote growth. Interestingly, recently it was suggested that epimastigote proliferation requires an oxidative environment to happen, whereas differentiation occurs in a more reductive setting (34).
Interestingly, ornithine levels were around four times higher in SPE compared with EPE, whereas arginine and citrulline levels were similar between both phases (supplemental Fig. 1). Recently, a novel arginine/ornithine transporter expressed in intracellular compartments was reported in T. cruzi (35). Because there is no evidence of the urea cycle in the parasite's genome, the novel transporter is considered to be physiologically involved in arginine homeostasis throughout the T. cruzi life cycle. Ornithine and citrulline are likely being uptaken from the liver infusion tryptose media; however, the increase ornithine levels detected are a real response to starvation as mentioned above. Different biological roles of polyamines beyond metabolism have been proposed such as contribution to osmotic stress protection by functioning as “osmolytes” and different mechanisms for stress protection, including acidic stress (36). Nevertheless, the potential biological role of polyamines beyond metabolism in T. cruzi remains largely unknown with new functions waiting to be unveiled.
In summary, our results provided strong evidence that T. cruzi epimastigotes exhibit a finely tuned adaptive metabolic mechanism that allows the switch from highly reduced and energy-rich metabolites, such as glucose, to more oxidized energy-poorer nutrients such as amino acids, which are the most abundant nutrients in the stationary phase of T. cruzi epimastigote growth. This metabolic plasticity appears to be crucial for the T. cruzi parasite's survival throughout the myriad of different environmental conditions that parasites go through during their life cycle, and likely there is a reason for certain metabolites to be more or less abundant at certain stages. A deep knowledge of their metabolic capabilities and their changes during the life cycle would likely reveal key metabolic checkpoints as novel targets for future therapeutic intervention and epidemiological control of Chagas disease.
Experimental procedures
Chemicals
All chemicals and compounds were of analytical grade unless stated otherwise. Internal standards l-[13C6,15N2]lysine, l-[15N2]phenylalanine, l-[15N2]glutamic acid, and [1,4-13C2] succinic acid were purchased from Cambridge Isotope Laboratories (Tewksbury, MA). 2-D1-Methionine was purchased from CDN Isotopes (Pointe-Claire, Quebec, Canada). Mass spectroscopy grade acetonitrile, methanol, ammonium formate, and formic acid (99%) were purchased from Fisher Scientific (Hampton, NH). Mass spectroscopy grade water was prepared with a Millipore Milli-Q Plus system equipped with an LC-Pak® cartridge (EMD Millipore, Billerica, MA). All metabolites used as analytical standards listed on supplemental Table S1 were purchased from Sigma unless stated otherwise. Culture media and fetal calf serum (FCS) were purchased from Invitrogen.
Experimental design and sample collection
T. cruzi CL strain clone 14 epimastigotes (37) were maintained by sub-culturing every 48 h in liver infusion tryptose medium supplemented with 10% FCS at 28 °C. Exponential phase epimastigotes were obtained from a 24-h culture starting at 2.5 × 107 parasites/ml (Fig. 1). Stationary phase epimastigotes were obtained from an exponential culture at 5.0 × 106 parasites/ml and maintained for 4 days without media change. Parasites were harvested by centrifugation at 4,000 rpm for 10 min and washed twice with cold PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4) to remove metabolites that were in excess in the medium that can mask smaller changes in the internal metabolome (38). An aliquot was separated for cell counting with a Neubauer chamber, and the remainder of the sample was immediately frozen by immersion in ethanol/dry ice bath. Cell pellets were stored at −80 °C until metabolite extraction was performed. Seven independent biological replicates were obtained for each growth phase of epimastigotes. Two or three technical replicates for each biological replicate were processed for metabolite extraction and analyzed by LC-MS/MS.
Sample preparation for targeted metabolomics
Metabolites were extracted from 13 samples of T. cruzi epimastigote collected during the exponential phase and 14 samples of T. cruzi epimastigote collected during the stationary phase. During metabolite extractions, samples were kept on ice. Metabolite extraction protocol was adapted from Ref. 39. Briefly, extraction was initiated by adding 400 μl (sample up to 5 × 107 parasites) or 800 μl (sample ≥5 × 107 parasites) of extraction mixture containing methanol/water (1:1, v/v) and the necessary amount of internal standards to obtain the final concentration indicated after metabolite extraction (l-[13C6,15N2]lysine (2 μm), l-[15N2]phenylalanine (1 μm), l-[15N2]glutamic acid (5 μm), [1,4-13C2]succinic acid (5 μm) and 2-D1-methionine (5 μm)). The cell pellet was mixed for 5 min with a vortex at 4 °C. Then, three cycles of sonication on ice for 2 min and incubation on dry ice for 5 min were performed. Finally, samples were centrifuged at 12,000 rpm for 20 min at room temperature to recover the supernatant and dried in a SpeedVac set at room temperature. Samples were stored at −80 °C until analysis by LC-MS/MS when samples were resuspended in water and filtered using a MultiScreen Ultracel-10 filter plate (EMD Millipore, Billerica, MA). Injections of 10 μl were performed for both standards and samples. Standards mix was freshly prepared from stocks and diluted in water.
Data acquisition and analysis
To perform targeted metabolomics on a broad spectrum of metabolites of central metabolic pathways, such as glycolysis, tricarboxylic acids cycle, amino acids, and low molecular weight thiols, different chromatographic separations were implemented. Two previously described methods using ultra-high performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) were used for analysis of the metabolites described on supplemental Table S1 (40, 41). Briefly, separations and analyses were performed using a Waters ACQUITY H-class UPLC (Waters) liquid chromatography system in tandem with a XEVO TQ-MS mass spectrometer (Waters) equipped with an electrospray ionization (ESI) source operating both in the positive and negative ion modes. The LC system was equipped with a quaternary pump, and the autosampler was set at 10 °C. The standards and samples were separated using the following two gradient elution chromatographic separations.
Method A
A waters ACQUITY UPLC HSS T3 column (1.8 μm, 2.1 × 100 mm) with an in-line filter was maintained at 45 °C. Mobile phase A consisted of 100% water and 0.2% formic acid (v/v); mobile phase B consisted of 100% methanol and 0.1% formic acid (v/v). The gradient elution was as follows: 0–1 min isocratic 98% A, 1–4 min linear from 2 to 70% B, 4–5 min linear from 70 to 90% B, 5–6 min isocratic 90% B, 6–7 min linear from 90 to 2% B. The 98% mobile phase A was maintained for 3 min for column equilibration. The flow rate was set at 0.3 ml/min (40).
Method B
A Waters ACQUITY UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) with an in-line filter was maintained at 40 °C. Mobile phase A consisted of 25 mm ammonium formate in 98% water and 2% acetonitrile, pH 8.2; mobile phase B consisted of 5 mm ammonium formate in 98% acetonitrile and 2% water. The gradient elution was as follows: 0–0.2 min isocratic 100% A; 0.2–1.08 min linear from 0 to 30% B; 1.08–6.63 min linear from 30 to 55% B; 6.63–7.20 min linear from 55 to 100% B; 7.20–9.46 min isocratic 100% B; 9.46–9.75 min linear from 0 to 100% A; and 9.75–11.45 min isocratic 100% A for column equilibration. The flow rate was set at 0.31 ml/min (41).
For the MS analysis using LC method A, the capillary voltage was set at 3.0 kV for positive ion mode and 3.7 kV for negative ion mode. For method B, the capillary voltage was set at 3.75 kV for positive ion mode. The source and desolvation gas temperatures of the mass spectrometer were set at 150 and 500 °C, respectively. The desolvation gas (N2) was set at 800 liters/h. Relative quantitative determination was performed in ESI negative and positive ion mode using MRM mode. The ion transitions, cone voltage, and collision energy used for ESI-MS/MS analysis were determined using MassLynx V4.1 Intellistart software (supplemental Table S1). Data were acquired using MassLynx V4.1 software and processed using TargetLynxTM application manager (Waters). Relative quantitative determination (response) was performed by determining the metabolite/internal standard area ratio of MS signals detected by MRM. Responses were further normalized to cell number.
Statistical analysis
The PCA, heat map, and t test for metabolomics data were performed using MetaboAnalyst 3.0 (42, 43). One-way analysis of variance followed by the Tukey post-test was used for statistical analysis. The Benjamini and Hochberg procedure was used to analyze differences between groups, and a false discovery rate of 0.05 was used to identify statistically significant differences. After performing the multivariate analysis of the samples using MetaboAnalyst 3.0, one technical replicate outlier from the exponential phase was removed from the subsequent analysis (44).
Amino acids uptake in EPE and SPE
Parasites were cultured as described above and harvested by centrifugation at 4,000 rpm for 10 min, washed twice with cold PBS, counted, and adjusted to a final concentration of 20 × 107 cells/ml and distributed in 100-μl aliquots. Transport assays for each tested amino acid was initiated by the addition of 100 μl of 3 mm of the corresponding unlabeled amino acid in PBS traced with the corresponding radiolabeled amino acid (PerkinElmer Life Sciences) as described previously for l-[U-14C]histidine (11), l-[3,4,5-3H]leucine, l-[U-14C]valine, l-[U-14C]isoleucine (21), l-[2,3,4,5-3H]proline (19), and l-[3,4-3H]glutamate (45). The uptake was measured at 28 °C during 1 min and stopped by the addition of 50 mm (800 μl) of a cold solution of the corresponding unlabeled amino acid. Parasite pellets were washed twice with PBS (12,000 rpm, 2 min at 4 °C) and resuspended in a scintillation mixture, and incorporated radioactivity was measured using a scintillation counter (PerkinElmer Life Sciences Tri-Carb 2910 TR).
Western blot analysis
The presence of TcP5CDH (EC 1.5.1.12) and TcP5C-synthetase (2.7.2.11) proteins in the proliferative forms of T. cruzi was determined by Western blotting in parasite homogenates. Briefly, parasites were harvested by centrifugation (4,000 rpm for 10 min at 4 °C) and washed twice with cold PBS. Pellets were resuspended in lysis buffer containing 20 mm Tris-HCl, pH 7.9, 1 mm EDTA, pH 8.0, 0.25 m sucrose, 50 mm NaCl, 5% glycerol (v/v), 1% Triton X-100 (v/v), 1 mm PMSF, 10 μg/ml aprotinin, and 10 μm N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E-64), and 10 μg/ml of Nα-tosyl-l-lysine chloromethyl ketone. Samples were chilled on ice for 40 min and clarified by centrifugation (12,000 rpm for 15 min at 4 °C). Supernatants were recovered, and protein concentration was determined by the Bradford method using bovine serum albumin as standard (46). Samples were submitted to protein electrophoresis (SDS-PAGE) loading an equal amount (30 μg) of total protein per lane. Proteins were transferred to 0.2-μm PVDF membranes (GE Healthcare), blocked with PBS buffer plus 0.3% Tween 20 (v/v) (PBST) supplemented with 5% skim milk powder (w/v), and probed for 16 h at 4 °C against specific sera. TbP5CDH was probed with a polyclonal specific serum (1:4,000) raised against the recombinant TcP5CDH-His6 (TrytripDB accession number Tc00.1047053510943.50) (9). The enzyme P5C-synthetase was probed with a polyclonal serum (1:3,000) produced in mouse against its recombinant isoform of T. cruzi (TrytripDB accession number TCSYLVIO_005298).8 Membranes were washed three times and incubated with a secondary goat anti-mouse IgG horseradish peroxidase (Sigma) diluted in PBST (1:50,000). Signal was developed by using SuperSignal® West Pico Chemiluminescent ECL substrate (Thermo Scientific) following the manufacturer's instructions.
TcP5CDH and TcP5CR enzymatic activity assays
Enzymatic determinations for both P5C reduction to proline or P5C oxidation to glutamate were performed. The substrate used was a racemic mixture of dl-Δ1-pyrroline-5-carboxylate (dl-P5C) and its ring-open form γ-GS, which was synthesized from peroxidation with NaIO4 (Sigma) and purified by ion-exchange chromatography as described previously (9, 47). The steady-state activities for both TcP5CDH and TcP5CR were measured in cell-free homogenates from epimastigotes in exponential and stationary phases. The TcP5CDH reaction mixture contained 500 μm P5C/γGS (freshly prepared), 1 mm NAD+, 90 mm potassium phosphate buffer, pH 7.2, completed with distilled water up to 3 ml. The reaction was started after adding 200 μg of cell-free extracts, and the linear rate was determined by following the increase in absorbance (λ340 nm) over 5 min at 28 °C with constant stirring. A blank without P5C/γGS was used as a control. The P5C-reductase reaction mixture contained 600 μm P5C/γGS (freshly prepared), 30 μm NADPH, 100 mm Tris-HCl, pH 7.0, completed with distilled water up to 3 ml. The reaction was started after adding 200 μg of cell-free protein homogenates from parasites, and the linear rate was determined by following the decrease in absorbance (λ340 nm) over 3 min at 28 °C under constant stirring.
Author contributions
M. J. B., L. N. R., E. F. M., C. N., A. M. S., M. B. C. performed the conception and design, analysis and interpretation of the data, and drafting and revising the article; M. J. B., L. N. R., E. M. F. P., B. S. M., L. M., E. F. M. performed the acquisition and analysis of data. All authors read, corrected, and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Ana Rodriguez (University of New York, New York) for providing T. cruzi CL strain clone 14 epimastigotes used for part of these analyses in the Cassera laboratory. We thank Donna Huber for comments and corrections.
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grants 2013/18970-6 and 2016/06034-2 (to A. M. S.) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Grants 2013/18970-6 and 308351/2013-4 (to A. M. S.). Metabolomics analysis was supported by National Institutes of Health Grant AI108819 (to M. B. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. S1 and Tables S1 and S2.
Mantilla, B. S., Marchese, L., Casas-Sánchez, A., Dyer, N. A., Ejeh, N., Biran, M., Bringaud, F., Lehane, M. J., Acosta-Serrano, A., and Silber, A. M. (2017) Proline metabolism is essential for Trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog. 13, e1006158.
N. C. Manchola, A. M. Silber, and C. Nowicki, manuscript in preparation.
L. Marchese, B. S. Mantilla, K. Olavarria, and A. M. Silber, unpublished data.
- EPE
- exponential phase epimastigote
- SPE
- stationary phase epimastigote
- TCA
- tricarboxylic acid
- P5C
- Δ1-pyrroline-5-carboxylate
- P5CDH
- P5C dehydrogenase
- TSH
- trypanothione
- RTP
- reverse trans-sulfuration pathway
- MRM
- multiple-reaction monitoring
- PCA
- principal component analysis
- BCAA
- branched-chain amino acid
- γ-GS
- γ-glutamate semi-aldehyde
- CBS
- cystathionine β-synthase
- Pyr
- pyruvate
- α-KG
- α-ketoglutarate
- ESI
- electrospray ionization.
References
- 1. Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., and Werner-Washburne M. (2004) Sleeping beauty: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brener Z. (1971) Life cycle of Trypanosoma cruzi. Rev. Inst. Med. Trop. Sao Paulo 13, 171–178 [PubMed] [Google Scholar]
- 3. Barrett F. M., and Friend W. G. (1975) Differences in the concentration of free amino acids in the haemolymph of adult male and female Rhodnius prolixus. Comp. Biochem. Physiol. B 52, 427–431 [DOI] [PubMed] [Google Scholar]
- 4. Hernández R., Cevallos A. M., Nepomuceno-Mejía T., and López-Villaseñor I. (2012) Stationary phase in Trypanosoma cruzi epimastigotes as a preadaptive stage for metacyclogenesis. Parasitol. Res. 111, 509–514 [DOI] [PubMed] [Google Scholar]
- 5. de Godoy L. M., Marchini F. K., Pavoni D. P., Rampazzo Rde C., Probst C. M., Goldenberg S., and Krieger M. A. (2012) Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics 12, 2694–2703 [DOI] [PubMed] [Google Scholar]
- 6. Goldenberg S., and Avila A. R. (2011) Aspects of Trypanosoma cruzi stage differentiation. Adv. Parasitol. 75, 285–305 [DOI] [PubMed] [Google Scholar]
- 7. Cazzulo J. J. (1992) Energy metabolism in Trypanosoma cruzi. Subcell. Biochem. 18, 235–257 [DOI] [PubMed] [Google Scholar]
- 8. Shaw A. K., Kalem M. C., and Zimmer S. L. (2016) Mitochondrial gene expression is responsive to starvation stress and developmental transition in Trypanosoma cruzi. mSphere 1, e00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mantilla B. S., Paes L. S., Pral E. M., Martil D. E., Thiemann O. H., Fernández-Silva P., Bastos E. L., and Silber A. M. (2015) Role of δ1-pyrroline-5-carboxylate-dehydrogenase supports mitochondrial metabolism and host-cell invasion of Trypanosoma cruzi. J. Biol. Chem. 290, 7767–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paes L. S., Suárez Mantilla B., Zimbres F. M., Pral E. M., Diogo de Melo P., Tahara E. B., Kowaltowski A. J., Elias M. C., and Silber A. M. (2013) Proline dehydrogenase regulates redox state and respiratory metabolism in Trypanosoma cruzi. PLoS ONE 8, e69419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barison M. J., Damasceno F. S., Mantilla B. S., and Silber A. M. (2016) The active transport of histidine and its role in ATP production in Trypanosoma cruzi. J. Bioenerg. Biomembr. 48, 437–449 [DOI] [PubMed] [Google Scholar]
- 12. Lisvane Silva P., Mantilla B. S., Barisón M. J., Wrenger C., and Silber A. M. (2011) The uniqueness of the Trypanosoma cruzi mitochondrion: opportunities to identify new drug target for the treatment of Chagas disease. Curr. Pharm. Des. 17, 2074–2099 [DOI] [PubMed] [Google Scholar]
- 13. Contreras V. T., Salles J. M., Thomas N., Morel C. M., and Goldenberg S. (1985) In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 16, 315–327 [DOI] [PubMed] [Google Scholar]
- 14. Tonelli R. R., Silber A. M., Almeida-de-Faria M., Hirata I. Y., Colli W., and Alves M. J. (2004) l-Proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell. Microbiol. 6, 733–741 [DOI] [PubMed] [Google Scholar]
- 15. Magdaleno A., Ahn I. Y., Paes L. S., and Silber A. M. (2009) Actions of a proline analogue, l-thiazolidine-4-carboxylic acid (T4C), on Trypanosoma cruzi. PLoS ONE 4, e4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sayé M., Miranda M. R., di Girolamo F., de los Milagros Cámara M., and Pereira C. A. (2014) Proline modulates the Trypanosoma cruzi resistance to reactive oxygen species and drugs through a novel dl-proline transporter. PLoS ONE 9, e92028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silber A. M., Colli W., Ulrich H., Alves M. J., and Pereira C. A. (2005) Amino acid metabolic routes in Trypanosoma cruzi: possible therapeutic targets against Chagas' disease. Curr. Drug. Targets Infect. Disord. 5, 53–64 [DOI] [PubMed] [Google Scholar]
- 18. Martins R. M., Covarrubias C., Rojas R. G., Silber A. M., and Yoshida N. (2009) Use of l-proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect. Immun. 77, 3023–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silber A. M., Tonelli R. R., Martinelli M., Colli W., and Alves M. J. (2002) Active transport of l-proline in Trypanosoma cruzi. J. Eukaryot. Microbiol. 49, 441–446 [DOI] [PubMed] [Google Scholar]
- 20. Mancilla R., Naquira C., and Lanas C. (1967) Protein biosynthesis in trypanosomidae. II. The metabolic fate of dl-leucine-1-C14 in Trypanosoma cruzi. Exp. Parasitol. 21, 154–159 [DOI] [PubMed] [Google Scholar]
- 21. Manchola N. C., Rapado L. N., Barisón M. J., and Silber A. M. (2016) Biochemical characterization of branched chain amino acids uptake in Trypanosoma cruzi. J. Eukaryot. Microbiol. 63, 299–308 [DOI] [PubMed] [Google Scholar]
- 22. Nozaki T., Shigeta Y., Saito-Nakano Y., Imada M., and Kruger W. D. (2001) Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine β-synthase and serine acetyltransferase from Trypanosoma. J. Biol. Chem. 276, 6516–6523 [DOI] [PubMed] [Google Scholar]
- 23. Marciano D., Santana M., and Nowicki C. (2012) Functional characterization of enzymes involved in cysteine biosynthesis and H(2)S production in Trypanosoma cruzi. Mol. Biochem. Parasitol. 185, 114–120 [DOI] [PubMed] [Google Scholar]
- 24. Hunter K. J., Le Quesne S. A., and Fairlamb A. H. (1994) Identification and biosynthesis of N1,N9-bis(glutathionyl)aminopropylcadaverine (homotrypanothione) in Trypanosoma cruzi. Eur. J. Biochem. 226, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 25. Carrillo C., Cejas S., González N. S., and Algranati I. D. (1999) Trypanosoma cruzi epimastigotes lack ornithine decarboxylase but can express a foreign gene encoding this enzyme. FEBS Lett. 454, 192–196 [DOI] [PubMed] [Google Scholar]
- 26. González N. S., Huber A., and Algranati I. D. (2001) Spermidine is essential for normal proliferation of trypanosomatid protozoa. FEBS Lett. 508, 323–326 [DOI] [PubMed] [Google Scholar]
- 27. Monod J. (1949) The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394 [Google Scholar]
- 28. Dias Fde A., Guerra B., Vieira L. R., Perdomo H. D., Gandara A. C., Amaral R. J., Vollú R. E., Gomes S. A., Lara F. A., Sorgine M. H., Medei E., de Oliveira P. L., and Salmon D. (2015) Monitoring of the parasite load in the digestive tract of Rhodnius prolixus by combined qPCR analysis and imaging techniques provides new insights into the trypanosome life cycle. PLoS Negl. Trop. Dis. 9, e0004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cazzulo J. J., Franke de Cazzulo B. M., Engel J. C., and Cannata J. J. (1985) End products and enzyme levels of aerobic glucose fermentation in trypanosomatids. Mol. Biochem. Parasitol. 16, 329–343 [DOI] [PubMed] [Google Scholar]
- 30. Engel J. C., Franke de Cazzulo B. M., Stoppani A. O., Cannata J. J., and Cazzulo J. J. (1987) Aerobic glucose fermentation by Trypanosoma cruzi axenic culture amastigote-like forms during growth and differentiation to epimastigotes. Mol. Biochem. Parasitol. 26, 1–10 [DOI] [PubMed] [Google Scholar]
- 31. Adroher F. J., Osuna A., and Lupiáñez J. A. (1987) Fructose 1,6-bisphosphatase activity in two Trypanosoma cruzi morphological forms. J. Parasitol. 73, 438–441 [PubMed] [Google Scholar]
- 32. Maugeri D. A., Cannata J. J., and Cazzulo J. J. (2011) Glucose metabolism in Trypanosoma cruzi. Essays Biochem. 51, 15–30 [DOI] [PubMed] [Google Scholar]
- 33. Antunes L. C., Han J., Pan J., Moreira C. J., Azambuja P., Borchers C. H., and Carels N. (2013) Metabolic signatures of triatomine vectors of Trypanosoma cruzi unveiled by metabolomics. PLoS ONE 8, e77283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nogueira N. P., Saraiva F. M., Sultano P. E., Cunha P. R., Laranja G. A., Justo G. A., Sabino K. C., Coelho M. G., Rossini A., Atella G. C., and Paes M. C. (2015) Proliferation and differentiation of Trypanosoma cruzi inside its vector have a new trigger: redox status. PLoS ONE 10, e0116712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Henriques C., Miller M. P., Catanho M., de Carvalho T. M., Krieger M. A., Probst C. M., de Souza W., Degrave W., and Amara S. G. (2015) Identification and functional characterization of a novel arginine/ornithine transporter, a member of a cationic amino acid transporter subfamily in the Trypanosoma cruzi genome. Parasit. Vectors 8, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller-Fleming L., Olin-Sandoval V., Campbell K., and Ralser M. (2015) Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406 [DOI] [PubMed] [Google Scholar]
- 37. Brener Z., and Chiari E. (1965) Aspects of early growth of different Trypanosoma cruzi strains in culture medium. J. Parasitol. 51, 922–926 [PubMed] [Google Scholar]
- 38. Creek D. J., Nijagal B., Kim D. H., Rojas F., Matthews K. R., and Barrett M. P. (2013) Metabolomics guides rational development of a simplified cell culture medium for drug screening against Trypanosoma brucei. Antimicrob. Agents Chemother. 57, 2768–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Armenta J. M., Cortes D. F., Pisciotta J. M., Shuman J. L., Blakeslee K., Rasoloson D., Ogunbiyi O., Sullivan D. J. Jr., and Shulaev V. (2010) Sensitive and rapid method for amino acid quantitation in malaria biological samples using AccQ. Tag ultra performance liquid chromatography-electrospray ionization-MS/MS with multiple reaction monitoring. Anal. Chem. 82, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Birkler R. I., Støttrup N. B., Hermannson S., Nielsen T. T., Gregersen N., Bøtker H. E., Andreasen M. F., and Johannsen M. (2010) A UPLC-MS/MS application for profiling of intermediary energy metabolites in microdialysis samples–a method for high-throughput. J. Pharm. Biomed. Anal. 53, 983–990 [DOI] [PubMed] [Google Scholar]
- 41. Li Q., Zhang S., Berthiaume J. M., Simons B., and Zhang G. F. (2014) Novel approach in LC-MS/MS using MRM to generate a full profile of acyl-CoAs: discovery of acyl-dephospho-CoAs. J. Lipid Res. 55, 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia J., Sinelnikov I. V., Han B., and Wishart D. S. (2015) MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xia J., and Wishart D. S. (2011) Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinformatics Chapter 14, Unit 14.10 [DOI] [PubMed] [Google Scholar]
- 44. Xia J., Mandal R., Sinelnikov I. V., Broadhurst D., and Wishart D. S. (2012) MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 40, W127–W133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silber A. M., Rojas R. L., Urias U., Colli W., and Alves M. J. (2006) Biochemical characterization of the glutamate transport in Trypanosoma cruzi. Int. J. Parasitol. 36, 157–163 [DOI] [PubMed] [Google Scholar]
- 46. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 47. Mezl V. A., and Knox W. E. (1976) Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies. Anal. Biochem. 74, 430–440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.