Significance
The aryl hydrocarbon receptor (AHR) is unique among PAS family transcription factors in its diverse environmental and cellular ligands and broad array of biological endpoints. AHR dimerizes with AHR nuclear translocator (ARNT) and subsequently interacts with genomic enhancers, dioxin response elements (DREs). Our crystal structure of the AHR–ARNT heterodimer in complex with DRE provides structural insights into this transcriptional complex. Our studies reveal three-dimensional structural codes for specific engagement of DRE that discriminates it from the closely related hypoxia response elements; the highly intertwined dimerization and interdomain interfaces remotely control DRE-binding and ligand-induced exposure of nuclear localization signal. The structural similarity and ramifications of the AHR, HIF, and NPAS transcriptional complexes with ARNT provide general and unique insights into PAS family transcription factors and complex AHR signaling.
Keywords: AHR, ARNT, transcriptional complex, dimerization, DNA recognition
Abstract
The aryl hydrocarbon receptor (AHR) belongs to the PAS (PER-ARNT-SIM) family transcription factors and mediates broad responses to numerous environmental pollutants and cellular metabolites, modulating diverse biological processes from adaptive metabolism, acute toxicity, to normal physiology of vascular and immune systems. The AHR forms a transcriptionally active heterodimer with ARNT (AHR nuclear translocator), which recognizes the dioxin response element (DRE) in the promoter of downstream genes. We determined the crystal structure of the mammalian AHR–ARNT heterodimer in complex with the DRE, in which ARNT curls around AHR into a highly intertwined asymmetric architecture, with extensive heterodimerization interfaces and AHR interdomain interactions. Specific recognition of the DRE is determined locally by the DNA-binding residues, which discriminates it from the closely related hypoxia response element (HRE), and is globally affected by the dimerization interfaces and interdomain interactions. Changes at the interdomain interactions caused either AHR constitutive nuclear localization or failure to translocate to nucleus, underlying an allosteric structural pathway for mediating ligand-induced exposure of nuclear localization signal. These observations, together with the global higher flexibility of the AHR PAS-A and its loosely packed structural elements, suggest a dynamic structural hierarchy for complex scenarios of AHR activation induced by its diverse ligands.
The aryl hydrocarbon receptor (AHR) belongs to the PER-ARNT-SIM (PAS) family transcription factor that mediates broad responses to cellular and environmental cues. The AHR has been shown to be activated by diverse environmental toxicants and endogenous ligands, and play an important role in adaptive metabolism, dioxin toxicity, and normal vascular and immune development (1, 2), ever since it was identified four decades ago for mediating metabolic responses to aryl hydrocarbon toxicants (3, 4) and the acute toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (5). The AHR was more recently found to mediate diverse cellular and physiological responses and likely respond to unknown endogenous AHR ligands (1, 2, 6–8). Developmentally, the AHR plays a role in the normal development and function of both the vascular and immune systems (9–12), and has close links to cancer, metabolic, immune, and cardiovascular diseases (13–18).
Intense efforts in the past four decades have yielded important insights into the molecular processes governing AHR signaling. Newly synthesized AHR is located in cytosol and associated with the chaperones Hsp90 (19), P23 (20, 21), and AHR associated protein 9 (ARA9, also known as XAP2 or AIP) (22–24). Binding of ligands induces conformational changes in the AHR that lead to exposure of nuclear localization sequences (NLS) (25, 26). Following nuclear translocation, the AHR exchanges chaperones for a transcription partner, ARNT (1) and the AHR–ARNT heterodimer binds near the promoters of target genes at dioxin-response element (DRE) sequences, “GCGTG” (27, 28). Depending on the class of ligand, the up-regulated targets of the AHR–ARNT complex can vary from genes encoding enzymes of xenobiotic metabolism, to genes evoking the toxicity of TCDD or those required for normal vascular or immune function. As a negative feedback loop, AHR activation also induces the expression of a competitive inhibitor known as AHRR (repression of AHR) (29).
Both the AHR and ARNT harbor an internal PAS domain with two similar repeats, PAS-A and PAS-B, an N-terminal basic-helix–loop–helix (bHLH) domain and a C-terminal transcriptional activation domain (TAD) (Fig. 1A). The bHLH domain participates in DNA binding, and together with the PAS-A domain dictates AHR–ARNT dimerization (30–32). Distinctly different from other PAS family transcription factors, the PAS-B domain of AHR binds to diverse ligands, which trigger AHR nuclear translocation, ARNT dimerization, and transcriptional activation. Here, we determined the crystal structure of the AHR–ARNT heterodimer, encompassing the bHLH and PAS-A domains, in complex with a DRE. Our studies reveal unique structural malleability and hierarchy for controlling AHR signaling.
Fig. 1.
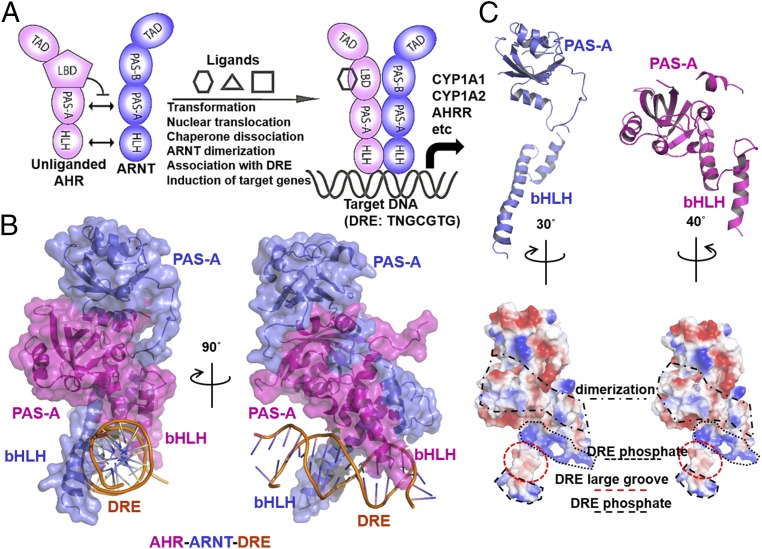
Overall architecture of the AHR–ARNT heterodimer in complex with DRE. (A) Schematic illustration of domain arrangements of AHR (magenta) and ARNT (blue), and ligand-induced AHR activation and assembly of the transcriptional complex. (B) Overall structure of the AHR–ARNT–DRE complex in two perpendicular views. (C) Individual structures and electrostatic contours of ARNT (blue) and AHR (magenta), illustrating surface features for DRE recognition and heterodimerization.
Quaternary Architecture of AHR–ARNT Heterodimer in Complex with DRE
In our initial attempts, we found that AHR encompassing the bHLH and PAS-A domains was poorly expressed in bacteria or insect cells, but coexpression with ARNT encompassing the same domains resulted in high levels of heterodimer expression in either host. This observation is consistent with the previous finding that the bHLH and PAS-A domains are crucial for AHR–ARNT dimerization (31). To gain insights into the structural basis of AHR–ARNT dimerization and DRE recognition, we performed crystallization screening for hundreds of AHR–ARNT–DRE complexes, encompassing varying bHLH and PAS-A domain boundaries and different lengths of DRE. From 1 of more than 40 different crystal forms, we solved the structure at a resolution of 4.0 Å (Fig. 1B and Table S1). Although AHR and ARNT have similar domain structures (Fig. 1A), we observed that ARNT curls around the AHR into a highly intertwined asymmetric architecture, similar to those of the CLOCK–BMAL1 and the HIF-α–ARNT complexes (33, 34). The bHLH and PAS-A domains of AHR and ARNT form extensive domain–domain and cross-domain interactions via highly conserved residues of AHR (Fig. 1B and Fig. S1). Consistent with the asymmetric nature of the architecture, the AHR has extensive interdomain contacts between the bHLH and PAS-A domains, but the two domains of ARNT barely contact each other (Fig. 1C). Despite the overall asymmetry, the bHLH domains of AHR and ARNT have a pseudosymmetric arrangement, with the N-terminal extended helix from each protein forming a pair of tweezer-like arms pinned to the major grooves of DRE from opposing sides (Fig. 1B and Fig. S1).
Table S1.
X-ray data collection and refinement statistics
| Crystallographic data | mAHR–hARNT–DRE complex |
| Data collection | |
| Beamline | LS-CAT ID-F and G |
| Wavelength, Å | 0.97872 |
| Space group | C121 |
| Cell dimension | |
| a, b, c, Å | 78.199, 64.364, 157.725, |
| α, β, γ, ° | 90, 100.136, 90 |
| Resolution, Å | 50–3.9 (3.97–3.90) |
| Total no. of reflections | 63,522 |
| No. of unique reflections | 6,690 (669) |
| I/σ (I) | 11.0 (1.5) |
| Multiplicity* | 7.9 (5.9) |
| Completeness, % | 99.50 (96.00) |
| Refinement | |
| Resolution, Å | 39.7–4.0 (4.14–4.0) |
| Rwork†/Rfree‡, % | 28.52/32.26 |
| No. of atoms* | 3,476/2,769/640/67 |
| B factor*, Å2 | 222.1/214.0/256.2/232.4 |
| rmsd bonds, Å | 0.010 |
| rmsd angles, ° | 1.30 |
| Ramachandran plot§, % | 91.0/8.1/0.9 |
The values in parentheses indicate the highest resolution shell (resolution range: 3.97–3.90 Å for data collection and 4.14–4.0 Å for refinement).
All atoms/protein/DNA/ligand.
Σhkl ||Fobs| − k|Fcalc||/ Σhkl |Fobs|.
Rfree was calculated by the same way as Rwork, but with the 5% of the reflections excluded from the refinement.
Preferred region/allowed region/disallowed region.
Fig. S1.
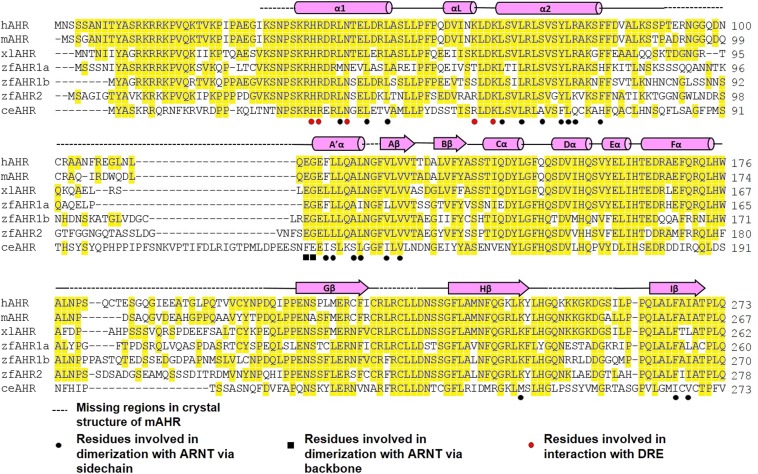
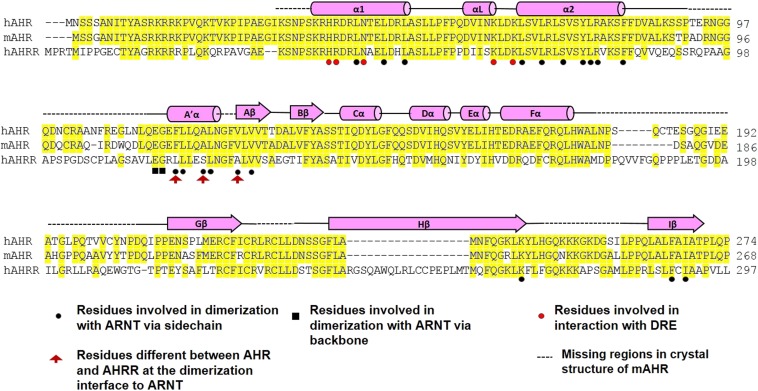
Sequence alignment of the AHR from human (h), mouse (m), Xenopus laevis (xl), zebra fish (zf), and C. elegans (ce). Secondary structural elements are indicated above the sequences. Conserved residues are highlighted in yellow. Residues that participate in dimerization with ARNT are indicated by black circles (via sidechain) and black squares (via backbone). Residues that participates in DRE binding are indicated by red circles.
The electrostatic potential of individual AHR and ARNT proteins reflect distinct surface features and patterns for dimerization and DRE binding (Fig. 1C). The surface inserted into the major groove of the DRE is sandwiched by two narrow strips of positively charged surfaces that contact the negatively charged rim of the DRE formed by phosphate groups. Right next to the DRE-docking sites is the extensive dimerization surface that is more hydrophobic than other surface areas, indicating that the dimerization interface of the AHR transcription complex is largely dictated by hydrophobic contacts.
Dimerization Interfaces Between AHR and ARNT
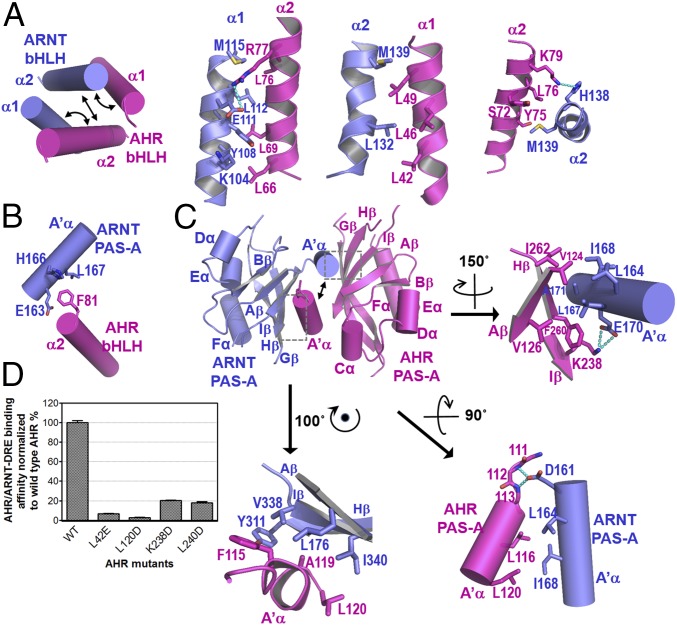
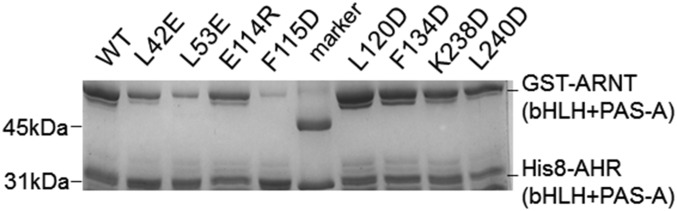
The highly intertwined domain–domain and cross-domain interactions between AHR and ARNT can be defined conveniently into six subcontact surfaces, three for bHLH domains (Fig. 2A), one for cross-domain interactions between the A′α helix of ARNT PAS-A and the α2 helix of AHR bHLH (Fig. 2B), and three for PAS-A domains (Fig. 2C). Up to 35 hydrophobic residues were found at the dimerization interfaces that form extensive hydrophobic contacts interspersed by merely a couple of H-bond and salt bridge interactions (Fig. 2 A–C). Consistent with the structural observations, AHR mutations, A119D, L120E, and F260D, were previously shown to disrupt AHR–ARNT dimerization in mammalian cells (35). Additionally, mutations to several other AHR residues at the interfaces were previously demonstrated to reduce AHR induction or dimerization with ARNT (35). Many of these single mutations and a few others can be tolerated when AHR and ARNT, encompassing both dimerization domains, were coexpressed in bacteria (Fig. S2). These observations suggest that the AHR single mutations at the dimerization interface tend to be tolerated by the broad contact surface areas when expressed in bacteria, but lead to a reduced ability to maintain the stability of its transcription complex in mammalian cells. Interestingly, the binding affinities of the mutant AHR–ARNT heterodimers to DRE were significantly reduced (Fig. 2D). This observation is intriguing because it indicates that changes at the heterodimerization interfaces remotely affect the spatial organization of the DNA reading head.
Fig. 2.
Heterodimerization interfaces of the AHR–ARNT complex. (A) Overall arrangements of α1 and α2 helices of AHR and ARNT bHLHs highlighting three subinterfaces for heterodimerization (Left), and close-up views of detailed interactions (right three images). (B) Cross-domain interactions involved in heterodimerization between the A′α helix of the ARNT PAS-A domain and the α2 helix of the AHR bHLH domain. (C) Overall pseudoasymmetric structure of the AHR–ARNT PAS-A domains highlighting three subinterfaces at the dimerization interface (Upper Left). A close-up view of the interactions between A′α of ARNT and Aβ, Iβ, and Hβ strands of AHR is shown at Upper Right, that between A′α of AHR and Aβ, Iβ, and Hβ strands of ARNT at Lower Left, and that between A′α helices of AHR and ARNT at Lower Right. For both A and C, H-bonds interactions are shown in cyan dash. (D) Effects of the AHR mutations at the dimerization interfaces to ARNT on the binding affinities of the AHR–ARNT heterodimer to an optimized DRE detailed in Fig. 3C.
Fig. S2.
Coexpression of GST-ARNT and His8-tagged AHR wild type and mutants in BL21DE3 followed by copurification over GS4B resin. The eluents were visualized on SDS/PAGE by Coomassie blue staining. AHR residues L53 and L115 were not located at the dimerization interfaces. Their mutations, L53E and F115D, likely affected the protein folding and interdomain interactions of AHR, and were used as control.
Specific Recognition of DRE and Comparison with Hypoxia Response Element
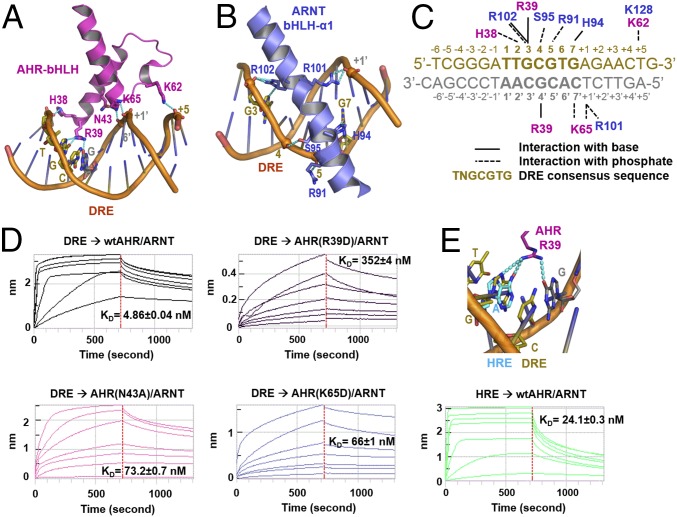
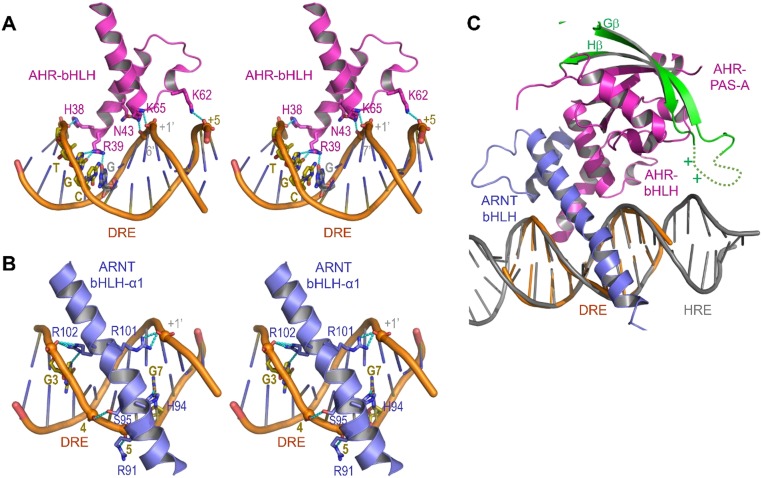
The bHLH domains of AHR and ARNT, in particular the α1 helix arms of the DRE-reading head tweezer, define an elegant 3D structural code for recognition of DRE sequences (Fig. 3 A–C and Fig. S3). These interactions include both specific recognition of base pairs in the DRE consensus sequence (TTGCGTG) and spatially well-positioned H-bond and salt bridge interactions to the phosphate groups both within the consensus DRE core and the neighboring sequence (Fig. 3 A–C and Fig. S3). Prominently, R39 of AHR forms three H-bond interactions with the base moieties of two base pairs GC/CG in the TNGC DRE half site (Fig. 3A and Fig. S3A). ARNT defines the recognition site for the GTG half site of the DRE sequence (Fig. 3B and Fig. S3B), which is almost identical to ARNT binding to the GTG half site in hypoxia response elements (TACGTG). Residues that interact with the phosphate groups of DNA are largely polar or positively charged, forming the properly spaced narrow strips of positively charged surfaces in both AHR and ARNT (Fig. 1C). Although not visible in the structure, several loops of AHR and ARNT PAS-A domains bear positively charged residues, which could be oriented for interaction with the target DNA outside the DRE consensus sequence (34) (Fig. S3C). This notion is corroborated by the crystal structures of the HIF–ARNT–hypoxia response element (HRE) complexes, in which, polar and positively charged residues in a HIF-2α PAS-A loop contact the nucleotides outside the HRE consensus sequence (34). Thus, similar to HREs, target DRE sequences can be modified by the sequence next to the DRE consensus core. The binding affinity of an optimized DRE target (Fig. 3C), selected based on a previous study (36), was measured at approximately 5 nM (Fig. 3D).
Fig. 3.
DRE recognition by the DRE-reading head formed by the bHLH domains of AHR and ARNT. (A) A close-up view of H bonds and salt bridge interactions of the AHR bHLH domain to the base moieties and phosphate groups of DRE. (B) A close-up view of H bonds and salt bridge interactions of ARNT bHLH to bases and phosphate groups of DRE. (C) Summary of interactions between AHR and ARNT residues and bases and phosphate groups of an optimized DRE. (D) Determination of binding affinities between the optimized DRE (as in C) and the AHR–ARNT deterodimers containing wild-type AHR or AHR bearing mutations to residues interacting with bases (R39) and phosphate groups (N43 and K65), respectively. The BLI signals for association at titrated concentrations and dissociation, and the calculated KD were shown. (E) A close-up view of H-bond interactions of R39 to the GC/CG base pairs of DRE overlaid with the adenine base of HRE in the HIF-2α–ARNT–HRE complex (Upper). The BLI association and dissociation signals of AHR–ARNT heterodimer to an HRE, with single G→A replacement to the optimized DRE (as in C) (Lower). The calculated KD was shown. The binding to the DRE control was the same as in D.
Fig. S3.
DRE recognition by the DRE-reading head formed by the bHLH domains of AHR and ARNT and predicted interaction with the AHR PAS-A domain. (A) A close-up stereoview of H bonds and salt bridge interactions of the AHR bHLH domain to the base moieties and phosphate groups of DRE. (B) A close-up stereoview of H bonds and salt bridge interactions of ARNT bHLH to bases and phosphate groups of DRE. (C) The invisible Gβ-Hβ loop (dashed curve) bearing positively charged residues is expected to be near the DNA (gray) outside the DRE consensus core (orange).
Consistent with the critical role of R39 in reading the AHR half site of DRE, a single mutation of R39 reduced the binding affinity of AHR–ARNT to the optimized DRE by more than 70-fold (Fig. 3D). AHR mutations that interrupt the H-bond and/or salt bridge interactions to the phosphate groups, N43A and K65E, reduced the binding affinity by more than 10-fold.
The consensus core sequences of the DRE used in our study and the HRE differ merely by a single nucleotide (TTGCGTG versus TACGTG). An overlay of our structure with that of HIF-2α–ARNT–HRE complex suggests that the G→A replacement interferes with at least one H-bond interaction with AHR R39. Introduction of this replacement to the optimized DRE reduced the binding affinity to the AHR–ARNT heterodimer by fivefold (Fig. 3E). Because the neighboring sequence had been optimized for AHR–ARNT binding, we expect that the HRE targets in the genome would largely disfavor binding of the AHR–ARNT heterodimer.
Structural Comparison with HIF-2α–ARNT–HRE and NPAS3–ARNT–HRE
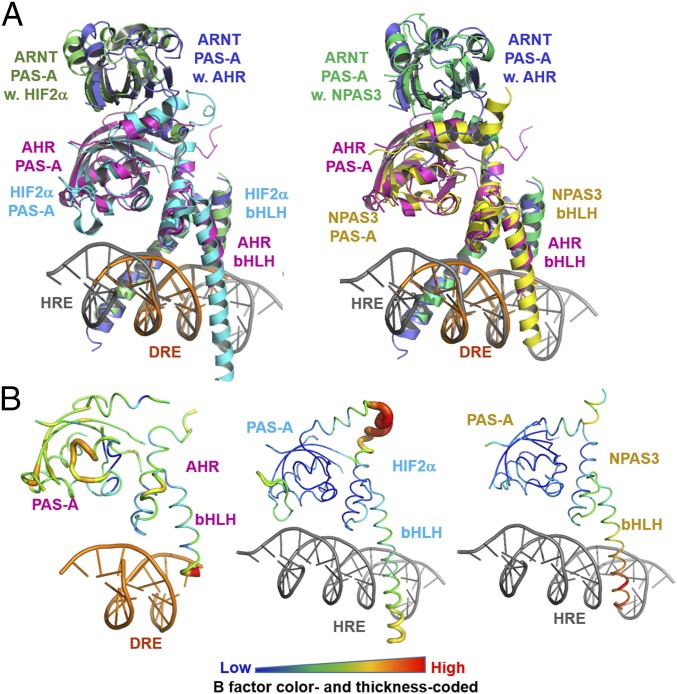
Besides HIF-1α, HIF-2α, and AHR, many other PAS family transcription factors heterodimerize with ARNT to form transcriptionally active complexes, among which the crystal structures of the transcriptional complexes for NPAS1 and NPAS3 were recently determined (37). Intriguingly, although the NPAS–ARNT complexes recognize HREs similar to the HIF–ARNT complexes (37), the overall architectures of the AHR–ARNT–DRE and NPAS3–ARNT–HRE complexes exhibit a much higher similarity than the HIF-2α–ARNT–HRE complex, with obvious structural ramifications in domain positioning between the AHR–ARNT–DRE and HIF-2α–ARNT–HRE complexes (Fig. 4A). Although the bHLH domains are similar to each other, the positions of PAS-A domains of both AHR and ARNT are shifted up to 5 Å to different positions (Fig. 4A). This structural shift might lead to different orientation of PAS-B and transactivation domains and cofactor recruitment, which likely contributes to different scenarios of downstream gene activation.
Fig. 4.
Comparison of structures of AHR–ARNT–DRE, HIF–2α-ARNT–HRE, and NPAS3–ARNT–HRE complexes. (A) Overall view of the overlay of the HIF-2α–ARNT–HRE and AHR–ARNT–DRE complexes (Left) and NPAS3–ARNT–HRE and AHR–ARNT–DRE complexes (Right). AHR is colored magenta; HIF-2α, cyan; NPAS3, yellow; DRE, orange; and HRE, gray. ARNT in complex AHR, HIF-2α, and NPAS3 are colored slate, lime, and green, respectively. (B) Structures of AHR, HIF-2α, and NPAS3 in worm with the color and thickness reflecting the scale of the B factors.
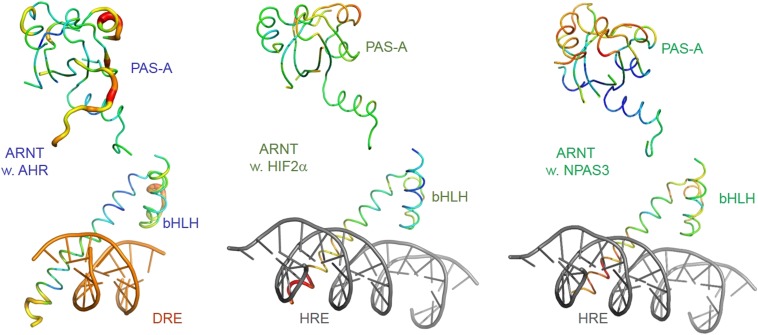
Similar to a majority of domain structures, both HIF-2α and NPAS3 have generally low B factors in several spatially proximate and stably packed structural elements, including the PAS-A domain, the A′α helix, and the second helix of the bHLH domain (Fig. 4B). In contrast, the corresponding domain and structural elements in AHR have globally higher B factors than HIF-2α and NPAS3 (Fig. 4B), whereas the B factors of ARNT in the three complexes are not significantly different (Fig. S4A). These structural observations suggest a globally higher flexibility and loose domain packing of the AHR PAS-A domain, the A′α helix, and the second helix of the bHLH domain. The PAS-B ligand-binding domain of AHR was previously suggested to have a higher structural flexibility than other PAS family transcription factors, providing a structural basis for AHR to adopt different conformations for accommodating diverse AHR ligands (38). The structural flexibility of the AHR bHLH and PAS-A domains might provide an extended allosteric structural pathway for DNA reading head to sense structural variations of the PAS-B ligand-binding domain induced by different ligands and modify target DNA recognition.
Fig. S4.
Structures of ARNT in the AHR–ARNT–DRE (Left) HIF-2α–ARNT–HRE (Center), and NPAS3–ARNT–HRE (Right) in worm with the color and thickness reflecting the scale of the B factors as in Fig. 4B.
Interdomain Interactions in AHR
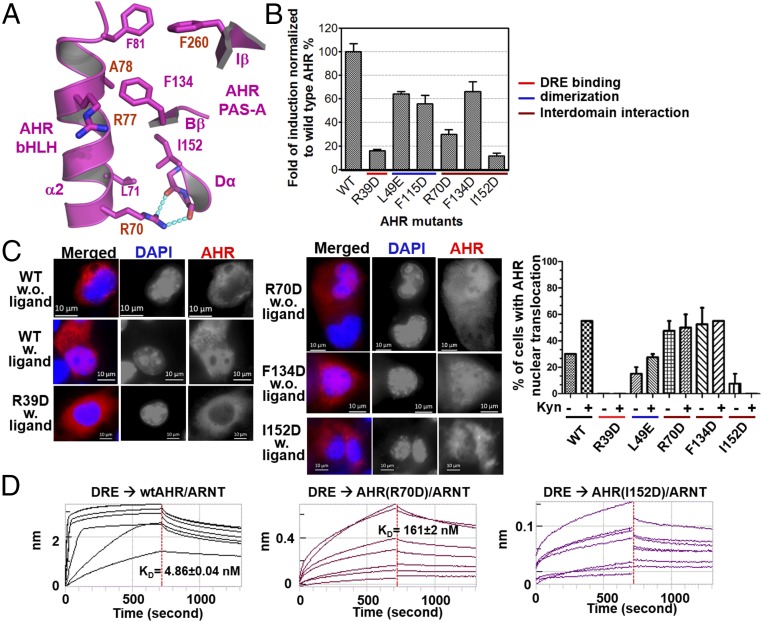
The broad interdomain interface in AHR harbors significant hydrophobic contacts and H-bond interactions between bHLH α2 helix and several structural elements in PAS-A (Fig. 5A). Mutation of residues at this interface revealed intriguingly different functions for the AHR transcriptional complex. Perturbing the H-bond interactions by the AHR mutation R70D drastically reduced the activity of AHR induction in COS-1 cells by 2 nM FICZ (6-formylindolo [3,2-b] carbazole, a potent AHR ligand generated by UV irradiation of tryptophan; refs. 39–41) (Fig. 5B). Intriguingly, the R70D mutation led to a high level of constitutive AHR nuclear translocation in the absence of ligands (Fig. 5C), but the binding affinity for the DRE was reduced by more than 30-fold in vitro (Fig. 5D); the latter likely contributes to its drastically reduced AHR induction activity (Fig. 5B). Mutations to residues at the interdomain interfaces appear to give distinctly different effects. AHR I152D completely abolished the AHR induction activity (Fig. 5B), the nuclear translocation in response to ligands (Fig. 5C), and drastically reduced DRE-binding in vitro (Fig. 5D), similar to the DNA-binding mutation R39D (Figs. 3D and 5 B and C). The failure of ligand-induced nuclear translocation of AHR R39D (Fig. 5B) is consistent with the previous observation that R39 is part of a key nuclear localization signal of AHR, R37H38R39 (42). AHR F134D reduced the AHR induction activity by ∼50%, similar to some of the single mutations at the heterodimerization interface, L49E and F115D (Fig. 5B). However, unlike L49E, which responds to ligands for increased nuclear translocation, AHR F134D is localized in the nucleus at a high level regardless of the presence or absence of FICZ (Fig. 5C).
Fig. 5.
AHR interdomain interactions and hierarchical control of AHR induction, AHR localization and DRE recognition. (A) A close-up view of interdomain interactions between the α2 helix of the AHR bHLH domain and the Iβ/Bβ strands and Dα helix of the AHR PAS-A domain. H-bonds interactions are shown in cyan dash. (B) The effects of AHR mutations on the induction of AHR activity by 2 nM FICZ, normalized to wild-type AHR–ARNT heterodimer (Upper). The mutations at the interdomain interfaces were compared with those at the DRE reading head and the dimerization interfaces to ARNT. (C) Immunofluorescence staining to detect nuclear translocation of AHR WT and mutants. Cells with ligand were treated with 10 nM FICZ. AHR was stained red, and DNA was stained with DAPI (blue). The percentages of cells with AHR nuclear translocation with and without ligand induction were calculated by counting the transfected cells (Right). (D) The effects of mutations at the interdomain interfaces on DRE binding, measured as in Fig. 3D.
These observations demonstrated that perturbing of interdomain interactions would cause either constitutive exposure of NLS or failure to expose NLS upon ligand binding, and affect target DNA binding. Our results suggest that the AHR interdomain interactions are crucial for establishing ligand-induced nuclear translocation and activation, and are the critical part of the allosteric structural pathway that senses the structural changes at the ligand-binding domain for exposure of NLS and for proper positioning of DNA reading head for target DNA recognition.
Discussion
The ability of the AHR to respond to diverse environmental and cellular ligands is believed to primarily reside in the ligand-binding PAS-B domain of AHR, which possesses distinct structural determinants and flexibility for controlling ligand-binding promiscuity and specificity (38). How AHR activation by different ligands leads to distinct and broad biological consequences remains unclear. Our studies here reveal a highly intertwined and flexible structural hierarchy dictating AHR–ARNT dimerization and DRE recognition in the AHR activation complex. The structure of the complex reveals spatially well-defined structure codes for specific recognition of the DRE (Fig. 3), which provides a simple and elegant mechanism for discriminating the closely related HRE that differs by merely a single nucleotide (Fig. 3E). Intriguingly, the complex dimerization and interdomain interfaces remotely control target DNA binding and the induction of AHR activity (Figs. 2 and 5), which, together with the overall more flexible nature of the AHR PAS-A domain and its loosely packed A′α and bHLH helices (Fig. 4 and Fig. S4), suggests an allosteric structural pathway for mediating changes from the ligand-binding PAS-B domain to the DNA-reading head, or reciprocally, from the DNA-reading head to the ligand-binding domain or farther to the transactivation domain. Besides affecting target DNA binding, we showed that the AHR interdomain interactions are crucial for shielding NLS before ligand binding and mediating ligand-induced exposure of NLS (Fig. 5 C and D). Thus, the allosteric structural pathway of AHR are also crucial for sensing and transducing the structural changes of the ligand-binding domain to the N-terminal NLS.
It is important to mention that such structural allostery is not unique to AHR. Similar observations had been made to the hormone nuclear receptors, which have sharply different structures of their DNA-binding and ligand-binding domains (43). Changes in the dimer interfaces of glucocorticoid receptor (GR), one of the hormone nuclear receptors, was found to allosterically affect DNA sequence-specific signaling (44); allosteric changes are crucial for communications between domains and alters the GR structure in response to target DNA binding (45, 46). Structural allostery is virtually broadly recognized as an important mechanism for signaling switches of transcription factors and has been used to engineer transcription factors for versatile response to diverse signaling cues (47, 48), suggesting that structural allostery might be a general mechanism for nuclear receptors to respond to their ligands. It is plausible that broad AHR ligands might induce diverse structural allostery that might modify target DNA recognition and the C-terminal TAD and lead to distinctly different biological consequences. Further characterization of the AHR transcriptional machinery to better define the allosteric structural pathway for transmitting changes of the ligand-binding domain to the DNA reading head and TAD and to examine ligand-specific structural allostery is expected to reveal key structural basis for controlling the diverse and distinct signaling mediated by different AHR ligands.
It is intriguing that the heterodimerization interfaces of CLOCK–BMAL1, HIF-α–ARNT, and AHR–ARNT transcriptional complexes are all dominated by hydrophobic contacts (33, 34, 49) (Fig. 2). This mode of interaction might contribute to the overall higher malleability of the complexes and might be a common feature for the PAS family transcription factors. CLOCK, HIF-α, and AHR are also common in harboring interdomain interactions, distinctly different from their transcriptional partners. Our observations on the role of AHR interdomain interfaces in remote control of the target DNA-binding, in establishing ligand-induced nuclear translocation, and in the induction of AHR activity (Figs. 2 and 5) suggest that the interdomain interactions might be important for the function of CLOCK, HIF-α, and other PAS family transcription factors.
Importantly, the AHR residues involved in DRE recognition and the majority of residues at the dimerization interfaces with ARNT are identical in AHRR (Fig. S5), underlying a structural mechanism for AHRR to competitively block AHR interactions with ARNT and DREs. A few residues at the dimerization interfaces with ARNT are different between AHR and AHRR (Fig. S5). In particular, A119 of AHR is converted to serine in AHRR, which may affect the mode of interaction at the dimerization interface. Given the strong impact of the dimerization interfaces on DRE binding (Fig. 3), AHRR might have varied preferences in target DNA recognition comparing to AHR. Whether this altered mode of interaction contributes remotely to the transcriptional repression activity of AHRR is also intriguing to test. Collectively, our insights into the structural hierarchy of AHR transcriptional complex provides an important platform for understanding the complex AHR signaling and regulation, and signaling ramifications of PAS family transcription factors.
Fig. S5.
Sequence alignment of the human and mouse AHR with human AHRR. Similar to Fig. S1, secondary structural elements are indicated above the sequences; conserved residues are highlighted in yellow. Residues that participate in dimerization with ARNT are indicated by black circles (via sidechain) and black squares (via backbone). Residues that participates in DRE binding are indicated by red circles. Residues that are different between AHR and AHRR at the dimerization interface with ARNT are indicated by red arrowhead.
Methods
Molecular Cloning and Protein Preparation.
Various constructs of mouse and human AHR and ARNT encompassing the bHLH and PAS-A domains were cloned in pQlink vector harboring an N-terminal His8-tag and a TEV cleavage site using standard PCR-based molecular cloning procedures. AHR mutants were generated by using site-directed mutagenesis. AHR and ARNT were expressed in BL21 (DE3) Rosetta (Novagen) after assembly of their expression cassettes into the same pQlink vector. Bacteria was lysed in a buffer containing 25 mM Tris, 500 mM NaCl, 5 mM MgCl2, 300 mM KCl, 5 mM ATP, 0.5% Triton X-100, and the AHR–ARNT heterodimer was purified over Ni-NTA resin. Upon removal of the His8-tag, the complexes were further purified by cation exchange chromatography (GE Healthcare). For assembly of the AHR–ARNT–DRE complexes, recombinant AHR–ARNT heterodimers were mixed with DRE oligonucleotides in 1:1.1–1.2 molar ratio and purified by gel filtration chromatography (Superdex 200, GE Healthcare).
Crystallization and Data Collection.
Crystals of the AHR–ARNT–DRE complex were grown at 4 °C by using the sitting-drop vapor-diffusion method by mixing 200–250 nL of 4 mg/mL AHR–ARNT heterodimer in complex with DRE, with an equal volume of reservoir solution containing 10–12% PEG 20000, 4–6% Tacsimate, pH 7.0, or 0.1 M Bis-Tris, pH 6.5. The crystals appeared in 2 d and grew to full in a week. The crystals were flash frozen in liquid nitrogen after dehydration and being equilibrated in the reservoir buffer with 25% glycerol (vol/vol). The X-ray diffraction datasets were collected at Advanced Photon Source Life Sciences Collaborative Access Team (APS LS-CAT); different datasets in the same space group were combined and processed to 4.0 Å by using HKL2000 (50).
Structure Determination.
The structure of the AHR/ARNT/DRE complex was solved by molecular replacement using the structure of HIF-2α–ARNT–HRE complex (34) encompassing the bHLH and PAS-A domains (PDB ID code: 4ZPK) as the searching model. One complex was found per asymmetric unit. The structure was built by using Coot (51) and refined by using Phenix (52). Repeated model building and refinement were performed, during which ensemble refinement with diffraction-restricted molecular dynamic simulation (53), followed by refinement of individual models was used to constructively remove model errors. The structure was refined to 4.0 Å, with the free and working R factors of 32.3% and 28.5%, respectively.
Determination of Binding Affinities of AHR–ARNT Heterodimer to DRE or HRE.
The interaction between WT and mutant AHR–ARNT heterodimers and DRE or HRE was tested by biolayer interferometry (BLI) on the FortéBio Octet System (FortéBio). The biotinylated DRE or HRE were immobilized on the streptavidin (SA) sensors and signals for association of the AHR–ARNT heterodimer in a series of titrated concentrations (varied from 10 nM to 2 μM) followed by dissociation were recorded at 25 °C. Data analysis was performed by using the FortéBio data analysis software to determine the on- and off-rates and binding affinities.
Luciferase Reporter Gene Assay.
COS-1 cells were cultured in 96-well plates (1.5–2 × 104 per well) overnight and transfected with pTarget vector containing wild-type or mutant mAHR (3 ng), together with pGudLu6.1 DRE-driven luciferase reporter vector (14 ng) (54) and TK-renilla luciferase vector (3 ng). Six hours after transfection, cells were treated with 2 nM FICZ or vehicle alone (0.1% DMSO) for 4 h, and assayed with dual luciferase reporter assay system (Promega). The expressed luciferase activity was measured by ENSPIRE plate reader (Perkin-Elmer). Data analysis was performed by using GraphPad Prism 5 (GraphPad Software). The experiments were performed in triplicate and repeated three times. Representative results are shown in mean ± SEM.
Immunofluorescence Staining.
COS-1 cells were cultured on 18-mm coverslips. pTarget vector (1.6 μg) harboring the expression cassette for WT or mutant mouse AHR (mAHR) was transfected into COS-1 cells by using Lipofectamine 2000 (Invitrogen Life Technologies) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were treated with 10 nM FICZ or vehicle alone (0.1% DMSO) for 2 h. Cells were fixed with 4% PFA for 15 min at room temperature and permeabilized for 10 mins with PBS containing Triton X-100. The antibody that specifically recognize the mAHR (bear-4) was used for staining and Alexa Fluor 594-conjugated goat anti-rabbit (Invitrogen) was used for visualization. Nuclei were counterstained with DAPI. Sections were subsequently dehydrated, mounted, and observed under Zeiss AxioObserver Z1 (Zeiss).
Coexpression and Copurification of AHR/ARNT Heterodimer.
See SI Methods for additional information.
SI Methods
Lysates of bacteria coexpressing GST-ARNT and His8-tagged AHR wild type and mutants harboring the bHLH and PAS-A domains from 1 L of culture was load into 1 mL of glutathione resin via GST tag. The resins were washed with 10 mL of washing buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 3 mM DTT four times to remove the excess unbound protein). Proteins were eluted with 7 mL of elution buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 20 mM glutathione, 3 mM DTT) before being examined by SDS/PAGE, and visualized by Coomassie blue staining.
Acknowledgments
We thank David Smith and Elena Kondrashkina (APS LS-CAT) for assistance on X-ray diffraction data collection, Kenneth A Satyshur (University of Wisconsin, Madison) for assistance on data processing, and Ed Glover and Hui Chen (University of Wisconsin, Madison) for assistance on AHR antibodies and immunofluorescence. The work is supported by University of Wisconsin and McArdle new direction research funds R01 ES020668 (to C.A.B.) and R01 GM096060-01 (to Y.X.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5V0L).
See Commentary on page 5330.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617035114/-/DCSupplemental.
References
- 1.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas PE, Hutton JJ. Genetics of aryl hydrocarbon hydroxylase induction in mice: Additive inheritance in crosses between C3H-HeJ and DBA-2J. Biochem Genet. 1973;8:249–257. doi: 10.1007/BF00486177. [DOI] [PubMed] [Google Scholar]
- 4.Thomas PE, Kouri RE, Hutton JJ. The genetics of aryl hydrocarbon hydroxylase induction in mice: A single gene difference between C57BL-6J and DBA-2J. Biochem Genet. 1972;6:157–168. doi: 10.1007/BF00486400. [DOI] [PubMed] [Google Scholar]
- 5.Poland AP, Glover E, Robinson JR, Nebert DW. Genetic expression of aryl hydrocarbon hydroxylase activity. Induction of monooxygenase activities and cytochrome P1-450 formation by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mice genetically “nonresponsive” to other aromatic hydrocarbons. J Biol Chem. 1974;249:5599–5606. [PubMed] [Google Scholar]
- 6.Oesch-Bartlomowicz B, et al. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: Divergent signaling pathways. Proc Natl Acad Sci USA. 2005;102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hestermann EV, Brown M. Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol Cell Biol. 2003;23:7920–7925. doi: 10.1128/MCB.23.21.7920-7925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Rowlands C, Safe S. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicol Appl Pharmacol. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savouret JF, Berdeaux A, Casper RF. The aryl hydrocarbon receptor and its xenobiotic ligands: A fundamental trigger for cardiovascular diseases. Nutr Metab Cardiovasc Dis. 2003;13:104–113. doi: 10.1016/s0939-4753(03)80026-1. [DOI] [PubMed] [Google Scholar]
- 10.Korashy HM, El-Kadi AO. The role of aryl hydrocarbon receptor in the pathogenesis of cardiovascular diseases. Drug Metab Rev. 2006;38:411–450. doi: 10.1080/03602530600632063. [DOI] [PubMed] [Google Scholar]
- 11.Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: A perspective on potential roles in the immune system. Immunology. 2009;127:299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 14.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 15.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 16.Mulero-Navarro S, Fernandez-Salguero PM. New trends in aryl hydrocarbon receptor biology. Front Cell Dev Biol. 2016;4:45. doi: 10.3389/fcell.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polyzos KA, Ketelhuth DF. The role of the kynurenine pathway of tryptophan metabolism in cardiovascular disease. An emerging field. Hamostaseologie. 2015;35:128–136. doi: 10.5482/HAMO-14-10-0052. [DOI] [PubMed] [Google Scholar]
- 18.Carreira VS, et al. Disruption of Ah receptor signaling during mouse development leads to abnormal cardiac structure and function in the adult. PLoS One. 2015;10:e0142440. doi: 10.1371/journal.pone.0142440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem. 1988;263:13802–13805. [PubMed] [Google Scholar]
- 20.Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J Biol Chem. 1999;274:13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 21.Cox MB, Miller CA., 3rd The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol Lett. 2002;129:13–21. doi: 10.1016/s0378-4274(01)00465-9. [DOI] [PubMed] [Google Scholar]
- 22.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol. 1998;18:978–988. doi: 10.1128/mcb.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem. 1998;273:33580–33587. doi: 10.1074/jbc.273.50.33580. [DOI] [PubMed] [Google Scholar]
- 24.Meyer BK, Perdew GH. Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry. 1999;38:8907–8917. doi: 10.1021/bi982223w. [DOI] [PubMed] [Google Scholar]
- 25.Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J Biol Chem. 1998;273:2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- 26.Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH. The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J Biol Chem. 2003;278:2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- 27.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 28.Chan WK, Chu R, Jain S, Reddy JK, Bradfield CA. Baculovirus expression of the Ah receptor and Ah receptor nuclear translocater. Evidence for additional dioxin responsive element-binding species and factors required for signaling. J Biol Chem. 1994;269:26464–26471. [PubMed] [Google Scholar]
- 29.Watanabe T, et al. Human arylhydrocarbon receptor repressor (AHRR) gene: Genomic structure and analysis of polymorphism in endometriosis. J Hum Genet. 2001;46:342–346. doi: 10.1007/s100380170070. [DOI] [PubMed] [Google Scholar]
- 30.Dolwick KM, Swanson HI, Bradfield CA. In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc Natl Acad Sci USA. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 32.Lindebro MC, Poellinger L, Whitelaw ML. Protein-protein interaction via PAS domains: Role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang N, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 35.Wu D, Potluri N, Kim Y, Rastinejad F. Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol Cell Biol. 2013;33:4346–4356. doi: 10.1128/MCB.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 37.Wu D, Su X, Potluri N, Kim Y, Rastinejad F. NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. eLife. 2016;5:5. doi: 10.7554/eLife.18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing Y, et al. Identification of the Ah-receptor structural determinants for ligand preferences. Toxicol Sci. 2012;129:86–97. doi: 10.1093/toxsci/kfs194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rannug A, et al. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J Biol Chem. 1987;262:15422–15427. [PubMed] [Google Scholar]
- 40.Helferich WG, Denison MS. Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol Pharmacol. 1991;40:674–678. [PubMed] [Google Scholar]
- 41.Rannug U, et al. Structure elucidation of two tryptophan-derived, high affinity Ah receptor ligands. Chem Biol. 1995;2:841–845. doi: 10.1016/1074-5521(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 42.Bunger MK, et al. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 43.Chandra V, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson LC, et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol. 2013;20:876–883. doi: 10.1038/nsmb.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas-Chollier M, et al. A naturally occurring insertion of a single amino acid rewires transcriptional regulation by glucocorticoid receptor isoforms. Proc Natl Acad Sci USA. 2013;110:17826–17831. doi: 10.1073/pnas.1316235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor ND, et al. Engineering an allosteric transcription factor to respond to new ligands. Nat Methods. 2016;13:177–183. doi: 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raman S, Taylor N, Genuth N, Fields S, Church GM. Engineering allostery. Trends Genet. 2014;30:521–528. doi: 10.1016/j.tig.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao N, Whitelaw ML, Shearwin KE, Dodd IB, Chapman-Smith A. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burnley BT, Afonine PV, Adams PD, Gros P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife. 2012;1:e00311. doi: 10.7554/eLife.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han D, Nagy SR, Denison MS. Comparison of recombinant cell bioassays for the detection of Ah receptor agonists. Biofactors. 2004;20:11–22. doi: 10.1002/biof.5520200102. [DOI] [PubMed] [Google Scholar]