Significance
Mammalian cells have evolved to develop multiple mechanisms for tumor suppression. It remains challenging to dissect the dynamic mechanism by which signaling proteins are engaged in inhibiting aberrant cell proliferation. Here, we propose a comprehensive network model composed of three signaling pathways and show how this network responds to mitogenic and oncogenic signals. This modeling approach elucidates how a precise temporal control of signaling pathways modulates the cellular response. We reveal the underlying principle for cell-fate decision and identify the roles of the different network components in tumor inhibition. This study sheds light on how the tumor-suppressive function can be modulated by the dynamics of the signaling proteins.
Keywords: cell-fate determination, oncogene activation, signal transduction
Abstract
Intrinsic tumor-suppressive mechanisms protect normal cells against aberrant proliferation. Although cellular signaling pathways engaged in tumor repression have been largely identified, how they are orchestrated to fulfill their function still remains elusive. Here, we built a tumor-suppressive network model composed of three modules responsible for the regulation of cell proliferation, activation of p53, and induction of apoptosis. Numerical simulations show a rich repertoire of network dynamics when normal cells are subject to serum stimulation and adenovirus E1A overexpression. We showed that oncogenic signaling induces ARF and that ARF further promotes p53 activation to inhibit proliferation. Mitogenic signaling activates E2F activators and promotes Akt activation. p53 and E2F1 cooperate to induce apoptosis, whereas Akt phosphorylates p21 to repress caspase activation. These prosurvival and proapoptotic signals compete to dictate the cell fate of proliferation, cell-cycle arrest, or apoptosis. The cellular outcome is also impacted by the kinetic mode (ultrasensitivity or bistability) of p53. When cells are exposed to serum deprivation and recovery under fixed E1A, the shortest starvation time required for apoptosis induction depends on the terminal serum concentration, which was interpreted in terms of the dynamics of caspase-3 activation and cytochrome c release. We discovered that caspase-3 can be maintained active at high serum concentrations and that E1A overexpression sensitizes serum-starved cells to apoptosis. This work elucidates the roles of tumor repressors and prosurvival factors in tumor repression based on a dynamic network analysis and provides a framework for quantitatively exploring tumor-suppressive mechanisms.
Keeping cell proliferation in check is essential for tumor suppression (1). Although the activation of oncogenes tends to induce cell transformation, a variety of innate mechanisms are engaged to protect against neoplastic progression by triggering cell-cycle arrest, senescence, or apoptosis while retaining the proliferative capacity of normal cells (2). Dysfunction of these mechanisms usually promotes tumorigenesis. Tumor repression is achieved via concerted actions of sensors, transducers, modulators, and effectors, which constitute a tumor-suppressive network. This complex network is interconnected to the machinery responsible for cell-cycle arrest and apoptosis. Although the operating mechanism of tumor suppression is a focus of intensive research (2), unraveling it has been a challenge.
As a tumor sensor, ARF is induced by activated oncogenes such as Myc and E1A (3). ARF transduces oncogenic signals by stabilizing and activating p53 (4). p53 is a potent tumor suppressor capable of inducing cell-cycle arrest, senescence, and apoptosis (5). Contrary to the ARF–p53 axis, environmental trophic factors such as growth factors and cytokines tend to trigger intracellular programs that buffer or repress proapoptotic signals. They do so mainly by activating receptor tyrosine kinases and signaling through the Ras/PI3K cascade, in which the Akt kinase acts as a key mediator and promotes cell survival (2, 6). On the other hand, the E2F family regulates the entry into and progression through the S phase of the cell cycle. Specifically, deregulated E2F1 promotes both cell proliferation and apoptosis (7).
Generally, the cell-fate decision depends on the competition between proapoptotic signals induced by tumor suppressors and prosurvival signals evoked by trophic factors. Consistent with this notion, cells exposed to oncogene activation are more susceptible to apoptosis upon serum withdrawal than in serum-rich situations (8–11). Despite this qualitative understanding, we still need a quantitative and comprehensive characterization of how cells respond to these competing signals.
Here, we construct a three-module network model to unravel the mechanism of tumor suppression following E1A activation. The modules detect and process physiological and oncogenic stimuli for cellular decision-making. We explore how cell fate is controlled by competing factors under various conditions and examine the impact of growth factors on apoptosis induction after serum starvation. Simulation results agree well with multiple experimental observations, and testable predictions are presented. This work provides an insight into how different signaling pathways are orchestrated dynamically to repress aberrant proliferation.
Models and Methods
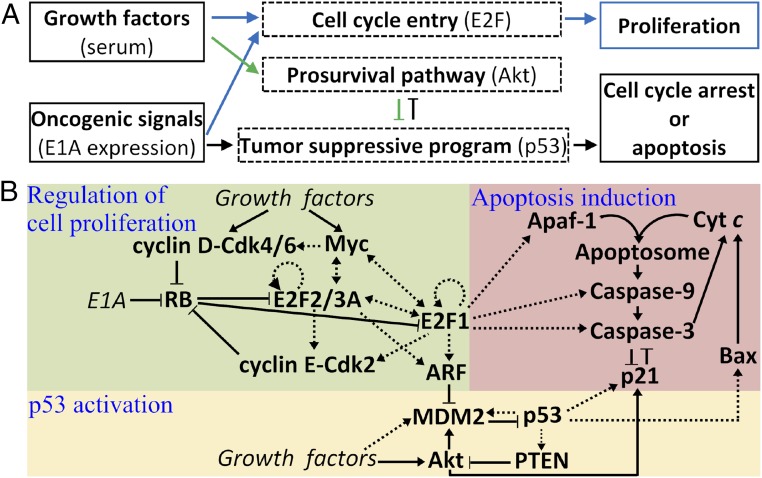
The network model characterizes the cellular response to both growth factors and oncogenic signals derived from the overexpression of adenovirus E1A. Growth factors stimulate two pathways that separately promote cell proliferation and survival (Fig. 1A); E2F and Akt are the main regulators, respectively. Oncogenic signals not only promote cell proliferation but also activate the tumor-suppressive program that is mainly mediated by p53. Consequently, normal cells may undergo proliferation, cell-cycle arrest, or apoptosis. To dissect the dynamics and function of the network, we divided it into three functional modules (Fig. 1B). Notably, these modules are endowed with positive feedback loops, and thus some components exhibit switch-like behaviors when activated. Detailed descriptions of these modules are available in SI Appendix, Methods (see also SI Appendix, Fig. S1).
Fig. 1.
A three-module model of the tumor-suppressive network. (A) Outline of the model. Growth factors stimulate two signaling pathways separately regulating cell-cycle progression and promoting cell survival. Oncogenic signals promote cell proliferation and activate tumor-suppressive programs via the canonical ARF–p53 axis. (B) The three-module network model characterizing the regulation of cell proliferation, p53 activation, and apoptosis induction. Dotted arrows, including two-way ones, represent transcriptional regulation; solid arrows and lines denote the promotion and inhibition, respectively, of production, transition, or activation.
The framework of the module regulating cell proliferation is derived from ref. 12, with the incorporation of recent advances and the characterization of more interactions and processes (see SI Appendix, Method S1, for details). As the central node, E2F activators (E2F1, E2F2, and E2F3A; hereafter referred to as “E2F” for simplicity) are normally regulated by four positive feedback loops, i.e., E2F–cyclin E/Cdk2–RB, E2F–Myc, E2F–Myc–cyclin D/Cdk4/6–RB, and E2F self-activation. E1A can disrupt the delicate regulation of E2F by inactivating RB, resulting in ectopic expression of E2F and ARF induction (13, 14). E2F1 is distinguished from E2F2 and E2F3A because of their different functions and interactions with RB and E1A.
Two opposing factors affect the stability and activity of p53 via modulating MDM2. ARF forms a complex with MDM2 to inhibit its E3 ligase activity toward p53 (15), whereas growth factors not only facilitate the transcription of mdm2 but also induce the activation of Akt (16). Activated Akt phosphorylates MDM2 to promote its nuclear accumulation and weaken its interaction with ARF (16–18). On the other hand, p53 induces the transcription of mdm2 and pten, and PTEN dephosphorylates PIP3 to inhibit Akt activation (19, 20). Thus, the interactions between p53, PTEN, Akt, and MDM2 constitute a positive feedback loop (21).
The backbone of the apoptosis module is the caspase-activation cascade, which involves Bax activation, the release of cytochrome c (Cyt c) from mitochondria, apoptosome formation, and the activation of caspases (see SI Appendix, Method S2, for details). p53 initiates apoptosis by inducing Bax (22), which triggers the release of Cyt c, and E2F1 promotes apoptosis via inducing Apaf-1, procaspase-9, and procaspase-3 (23). Once in the cytosol, Cyt c binds to and activates Apaf-1, forming the apoptosome. Procaspase-9 forms dimers and becomes active; caspase-9 (Casp9), either free or bound to the apoptosome, leads to the conversion of procaspase-3 into caspase-3 (Casp3). The Casp3-mediated amplification loop of Cyt c release is responsible for switch-like activation of Casp3 (24). It was shown experimentally that a small pool of soluble Cyt c in the intermembrane space and Cyt c bound to the inner membrane of mitochondria are released sequentially during cell apoptosis (25, 26); this two-wave release of Cyt c is considered in the model. On the other hand, p53 induces p21, which promotes cell-cycle arrest and inhibits apoptosis. Akt-mediated phosphorylation of p21 enhances its accumulation in the cytosol. A mutual inhibition exists between Casp3 and p21: Cytosolic p21 can form a complex with procaspase-3, inhibiting its activation (27, 28), whereas Casp3 mediates the cleavage of p21 (29). Here, the persistent activation of Casp3 and p21 is taken as the indicator of apoptosis and cell-cycle arrest, respectively.
The concentration of each protein in the network, denoted by square brackets, is represented by a state variable. Their temporal evolution is governed by ordinary differential equations (ODEs) (SI Appendix, Method S3). The gene transcription mediated by transcription factors is characterized by the Hill function, whereas the (de)phosphorylation and (de)activation processes are taken as enzyme-catalyzed reactions and are assumed to follow the Michaelis–Menten kinetics (30–33). The definition of the variables and their initial concentrations, the description of reaction kinetics, and a set of standard parameter values are listed in SI Appendix, Tables S1–S3. The ODEs were numerically solved using MTALAB ode15s. The bifurcation diagrams were plotted using Oscill8.
Results
Differentiation Between Normal Mitogenic and Oncogenic Signals by ARF.
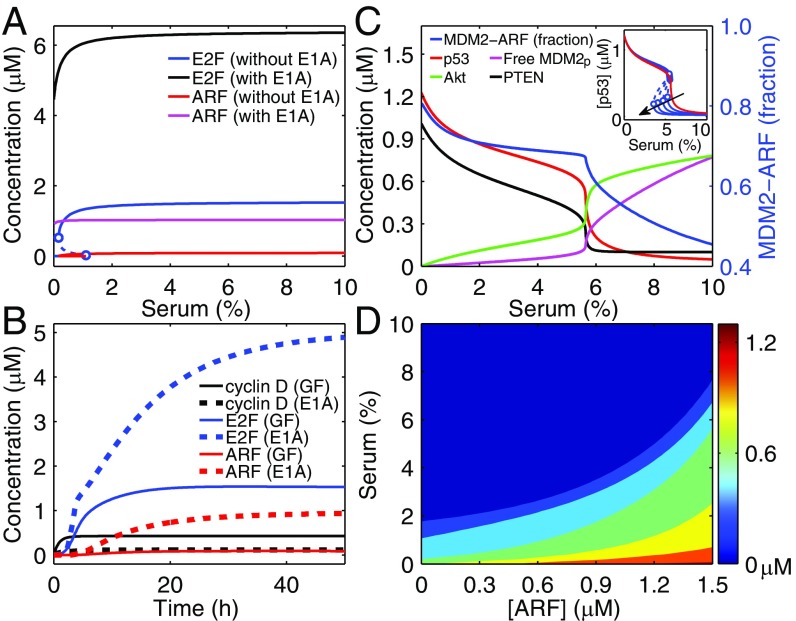
E2F activators are well-known for the ability to govern the R-point in the cell cycle together with RB. It was shown that E2F behaves as a bistable switch in response to serum stimulation (12); this feature is reproduced here. A bistable regime exists in the bifurcation diagram of the steady-state level of E2F versus serum concentration (C), and the lower bifurcation point occurs at C = 1% (Fig. 2A). Of note, [E2F] equals the total concentration of all active forms of E2F. In contrast, the delicate R-point regulation is disrupted upon E1A expression; the steady-state E2F level rises toward saturation with increasing C. E1A induces ectopic expression of E2F even without serum. Consequently, ARF is actively induced by E2F, in contrast to its basal expression under normal growth conditions. This result agrees with the experimental observations that ARF becomes active only when the levels of its inducers exceed aberrantly high thresholds (14, 34, 35). Collectively, ARF is induced in an all-or-none manner to transduce oncogenic signals. The bifurcation diagrams for cyclin D and cyclin E are presented in SI Appendix, Fig. S2A.
Fig. 2.
Responses of activator E2F, ARF, and p53 to serum stimulation and E1A expression. (A) Bifurcation diagrams of the steady-state levels of E2F and ARF versus serum concentration with or without E1A expression. (B) Temporal evolution of the levels of cyclin D, E2F, and ARF in response to 10% serum (solid lines) or E1A expression with 0.1% serum (dashed lines). (C) Bifurcation diagrams for components in the p53 activation module with fixed E1A expression. The Inset shows the transition of the p53 kinetic mode from ultrasensitivity to bistability. Following the direction of the arrow, the rate constant and Michaelis constant for Akt dephosphorylation take the following values: kDP3/KAkt = 9.6/0.2, 8.8/0.16, 8.01/0.12, 7.17/0.08, and 6.51/0.05. (D) Steady-state p53 level as a function of independent serum and ARF concentrations. Here, [ARF] denotes the total amount of ARF, including free ARF, the MDM2–ARF (MA) and MDM2P–ARF (MpA) complexes. The dynamics of MA and MpA obey SI Appendix, Method 3, Eqs. S19 and S20, whereas [free ARF] = [ARF] − [MA] − [MPA] replaces SI Appendix, Method 3, Eq. S11(b). The colored scale denotes the p53 level.
Cyclin D is an early-response gene, reaching its steady level within 6 h after serum stimulation, whereas E2F becomes fully activated around 13 h (12); these features are recapitulated here (Fig. 2B). On the contrary, E1A alone fails to activate the production of cyclin D but induces high expression of E2F; as a consequence, the levels of cyclin E and ARF are markedly up-regulated. The activation of E2F exhibits biphasic kinetics. Over the initial short period (0–5 h), [E2F] rises quickly (rapid phase); subsequently, it rises more slowly, and a long time is required for the saturation of [E2F] (slow phase). This biphasic feature is attributed mainly to the conversion of dominant regulators of E2F activity between the two phases: The rapid phase is triggered mainly by E1A-mediated RB suppression and E2F release, whereas the slow phase is associated with the transcriptional activity of E2F and E1A/RB-mediated inhibition of E2F degradation (see SI Appendix, Fig. S2 B and C, for details). Taken together, the dynamics of E2F and ARF expression differ remarkably between the two cases, so that normal signaling can be distinguished from aberrant proliferative signaling.
Dynamics of p53 Activation.
p53 activation by ARF is essential to the initiation of tumor-suppressive programs. Because of the p53–PTEN–Akt–MDM2 positive feedback loop, the dependence of steady-state concentrations of these proteins on the serum concentration C can be ultrasensitive (Fig. 2C). The p53 level depends on C as follows: For C ≤5.5% [p53] remains at relatively high levels (ON state), but it drops steeply to low levels (OFF state) around 6% and remains there thereafter. A similar transition mode has been reported previously (21, 36). The transition point is set around C = 5.5% to match the experimental observation that high levels of p53 can be induced when cells are supplied with 5% serum (37, 38) (Of note, this value is cell-type specific). Moreover, changing the feedback strength can switch the dependence of p53 levels on C from ultrasensitivity to bistability, with the upper bifurcation point changing slightly (Fig. 2C, Inset). Unless otherwise specified, p53 always operates in the ultrasensitive mode in the following analysis.
ARF and Akt are two opposing factors regulating p53 levels via MDM2. Unlike p53, the concentrations of free phosphorylated MDM2 (MDM2p) and active Akt switch to relatively high levels after C = 6%. Consistently, MDM2 in complex with ARF is the dominant form of MDM2 for C <6%, leading to the sequestration of MDM2 in the nucleolus and p53 activation; for C >6%, Akt phosphorylates MDM2 to promote its nuclear entry and weaken its interaction with ARF, resulting in MDM2p up-regulation and p53 inactivation.
When both the concentrations of serum and ARF are controllable inputs, the steady-state level of p53 is represented by a contour map (Fig. 2D). Because growth factors facilitate both mdm2 expression and the accumulation of MDM2p via Akt, Fig. 2D is significantly different from SI Appendix, Fig. S2D, which shows [p53] as a function of [ARF] and [Akt]. The contour map in Fig. 2D can be roughly divided into three regions: (i) the deep blue region represents the inactive state of p53, in which the impact of serum on p53 expression suppresses that of ARF; (ii) the light blue and green regions correspond to moderate levels of p53, with the effects of serum and ARF being comparable; and (iii) the yellow and red regions denote the highly active state of p53, in which the influence of ARF is predominant. Together, the p53 level depends on the relative strength of pro- versus antiproliferative signals.
Cell-Fate Determination Under Diverse Conditions.
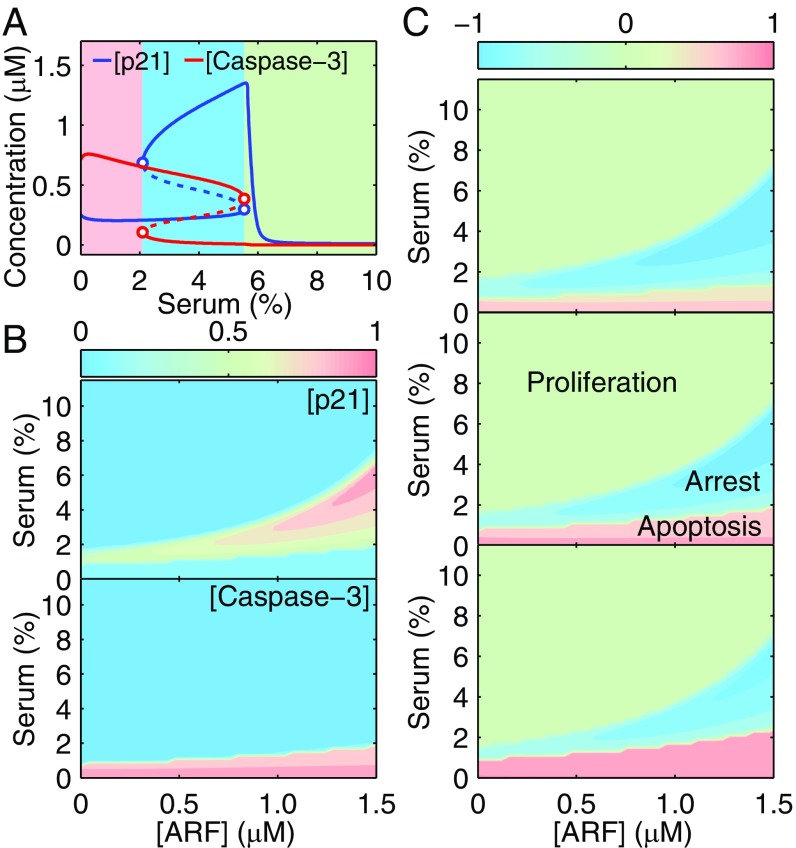
Activated p53 induces p21 and Bax, which inhibit and promote the activation of Casp3, respectively. With fixed E1A expression, the steady-state levels of p21 and Casp3 can exhibit bistability over a range of serum concentrations (Fig. 3A). Two saddle-node bifurcation points divide the serum concentration into three ranges: low (0–2%), intermediate (2–5.5%), and high (5.5–10%). Given that the initial concentrations of all species are their steady-state levels at 10% serum without E1A, the cell will sequentially undergo proliferation, cell-cycle arrest, and apoptosis when C is decreased from 10 to 0%.
Fig. 3.
Cell-fate decisions among proliferation, cell-cycle arrest, and apoptosis. (A) Steady-state levels of p21 (including phosphorylated and nonphosphorylated forms) and Casp3 versus serum concentration. (B) Dependence of the steady-state levels of p21 and Casp3 on independent ARF and serum concentrations. Protein levels are normalized by their respective maxima and are shown in contour maps. (C) Distribution of cell fates. By subtracting [p21] from [Casp3] in B, we roughly depict the distribution of cell fates. Compared with those in the middle panel, the E2F1-dependent expression rates of Apaf1, procaspase-9, and procaspase-3 drop (in the top panel) or rise (in the bottom panel) by 12.5%. The notation for [ARF] is the same as in Fig. 2D.
At high serum levels, p53 remains inactive, and both free p21 and Bax are kept at low levels (SI Appendix, Fig. S3). Cyt c is maintained in mitochondria, and little apoptosome is formed. Thus, little Casp3 is produced despite the high expression of procaspase-3 by E2F1. Consequently, cell proliferation is permitted. At moderate serum levels, p21 and Bax are induced by p53; although Bax promotes Cyt c release, Akt-mediated phosphorylation of p21 enhances its localization in the cytosol to inhibit procaspase-3 activation. Thus, Casp3 is still at low levels, and cell-cycle arrest is induced. Conversely, p21 phosphorylation becomes weak at low serum levels, and Casp3 is activated to trigger apoptosis.
More generally, the steady-state levels of p21 and Casp3 are calculated when the concentrations of both serum and ARF are independent inputs (Fig. 3B). Because p21 and Casp3 antagonize each other, they either are predominant in different regions or are simultaneously inactive. By subtracting the p21 level from the Casp3 level (both levels normalized by their respective maxima), we got a new plot reflecting the distribution of three cell fates (Fig. 3C, Middle). The comparison between Figs. 2D and 3C reveals that activated p53 inhibits cell proliferation, whereas the relative strength of prodeath versus antideath signals guides the decision between cell-cycle arrest and apoptosis.
The distribution of cell fates is also affected by E2F1 activity, p21/Casp3 interaction, and the kinetic mode of p53. First, increasing the transcriptional rates of Apaf-1, procaspase-9, and procaspase-3 by E2F1 results in the expansion of the region corresponding to apoptosis (compare the three panels in Fig. 3C). This effect agrees with the notion that E2F1 contributes to p53-dependent apoptosis under oncogenic conditions (7, 39). This role is highlighted when cellular responses to E1A expression and to ARF overexpression alone are compared. Without E1A, overexpression of ARF facilitates cell-cycle arrest rather than apoptosis in the presence of serum stimulation (SI Appendix, Fig. S4A), in agreement with experimental observations (40, 41). Second, as the rate constant for forming the p21p/procaspase-3 complex rises, the region corresponding to apoptosis shrinks (SI Appendix, Fig. S4B), indicating the antiapoptotic role of p21. Third, if the kinetic mode of p53 changes from ultrasensitivity to bistability, and the bistable regime enlarges, the right-side OFF-to-ON transition point of p21, which is governed by p53, moves leftward in the bifurcation diagram (SI Appendix, Fig. S4C). Thus, the region corresponding to cell-cycle arrest contracts in SI Appendix, Fig. S4D, whereas the region corresponding to apoptosis remains unchanged. Collectively, the determination of cell fate is controlled largely by p53 activity and is fine-tuned by multiple factors, thus allowing flexible control of cellular outcome.
Time Delay in Apoptosis Induction.
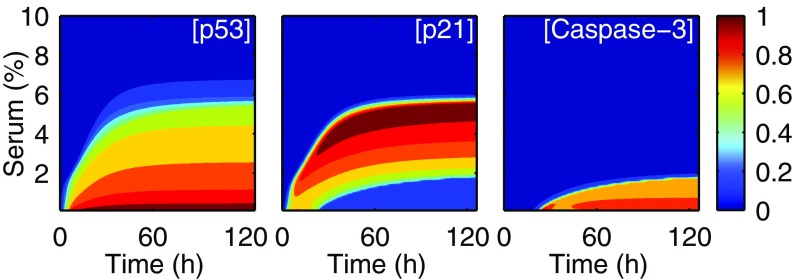
Fig. 4 shows the temporal evolution of p53, p21, and Casp3 levels, with the initial concentrations being their steady-state levels at 10% serum without E1A. As C rises from 0.1 to 5.5%, the timing for full activation of proteins is increasingly delayed; for p53 and p21, it rises from hours to days with increasing C, because increasing the amount of growth factors enhances the expression of nuclear MDM2 and thus decreases the accumulation rate of p53 (17, 42). Casp3 activation depends on the slow accumulation of cytosolic Cyt c, which is promoted by Bax and Casp3, on rate-limiting steps such as apoptosome formation, and on antagonism between p21 and Casp3. Thus, the full activation of Casp3 is retarded more prominently, with the delay ranging from one to a few days. This delay has been well manifested experimentally (38).
Fig. 4.
Temporal evolution of protein levels. Time courses of the levels of p53, p21, and Casp3 in response to different serum stimuli (0–10%). Protein levels are normalized by their respective maxima.
Of note, cells first undergo cell-cycle arrest before commitment to apoptosis under low serum conditions; this sequence has physiological implications. This feature was observed previously in experiments with virus infection (38) or p53 overexpression (43). With E1A expression in serum-free systems, however, the initial p21-induced cell-cycle arrest at G2/M phase was not so remarkable (44). Possibly, the concentration of p21 preceding Casp3 activation does not reach a sufficiently high level, or p21 is rapidly cleaved by subsequently activated Casp3 before inducing a prominent arrest.
Two Stages in Apoptosis Induction upon Serum Starvation and Recovery.
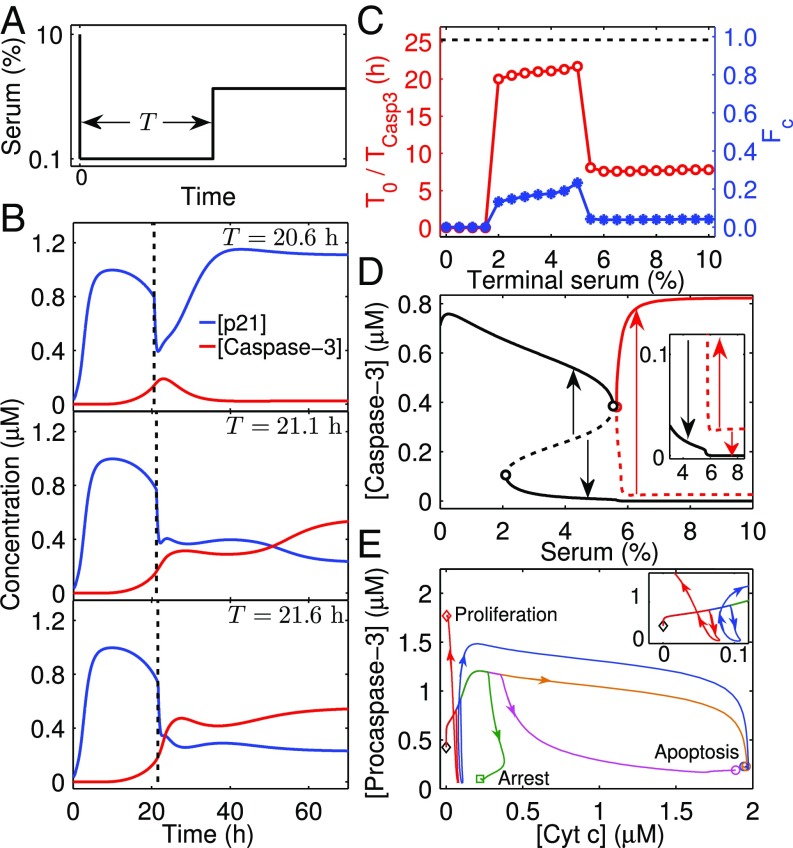
To unravel the influence of growth factors on apoptosis induction and the dynamics of Casp3 activation further, we devised a special case in which serum concentration is increased from a low to high level. We assume that the cell is first kept in a medium with 0.1% serum for a certain period (T) and then is supplied with 4% serum (Fig. 5A). Proapoptotic proteins accumulate during the starvation period, and there may exist a particular time beyond which Casp3 is always activated irrespective of serum. Indeed, if T < T0 (∼21.1 h), the activation of Casp3 is readily halted after serum recovery (Fig. 5B). As T approaches T0, the activation process persists until its completion. For T > T0, the activation process is more resistant to serum restoration. These results imply that apoptosis induction may involve the transition from a quenchable stage to an unquenchable stage.
Fig. 5.
Impact of serum recovery on apoptosis induction with fixed E1A expression. (A) Illustration of the simulation protocol. The cell is exposed to 0.1% serum for a period of T and is then to higher serum concentrations (e.g., 4%). (B) Critical T0 for irreversible activation of Casp3. Time courses of [p21] and [Casp3] are shown for different starvation times: T = 20.6, 21.1, and 21.6 h. T0 is ∼21.1 h. (C) Dependence of T0 (red circles) and FC (blue stars) on terminal serum concentration. The dashed line denotes the required time (TCasp3) for Casp3 activation when the cell is constantly exposed to 0.1% serum. (D) Full bifurcation diagram for Casp3 with the new branches denoted in red. An enlarged view is shown in the Inset. Arrows schematically denote the tendency of [Casp3] to progress to steady state when initiated from an unstable steady state. (E) Phase portraits of [Cyt c] and [procaspase-3] for different cases with C/T = 0.1%/+∞ (yellow), 3%/20.1 h (green), 3%/21.1 h (magenta), 8%/6.7 h (red), or 8%/8.7 h (blue). The Inset shows an enlarged view. The system begins with the steady state at 10% serum without E1A and ends at the steady state corresponding to proliferation, cell-cycle arrest, or apoptosis.
Such a critical timing exists for other terminal serum concentrations; the T0–C curve is roughly divided into three parts: left (0–2%), middle (2–5.5%), and right (5.5–10%). T0 remains at 0 in the left region, indicating that cells always commit to apoptosis. T0 rises slightly as C is increased from 2 to 5%. Surprisingly, T0 is nearly a constant for 5.5 ≤ C ≤ 10%, markedly less than that in the middle region. This result seems to be incompatible with the bifurcation diagram in Fig. 3A showing Casp3 is inactivated for C > 5.5%.
We found that another branch of stable fixed points exists for 5.5 ≤ C ≤ 10% when the bifurcation diagram is plotted with different initial conditions (e.g., with a high Casp3 level) (Fig. 5D). This new branch still results from the positive feedback loops in the apoptosis module. Here, maintenance of high Casp3 levels does not require the sustained activation of p53/Bax, and activation of the caspase cascade is self-maintainable once being turned on (SI Appendix, Fig. S5). Notably, the threshold of Casp3 activation for C ≥ 5.5% is markedly lower than that for 2 ≤ C < 5.5% because p21 is expressed only at basal levels; thus, Casp3 activation can be initiated more easily.
Given that Casp3 levels depend largely on the amounts of free procasepse-3 and Cyt c in the cytosol, the dependence of T0 on C can be interpreted as follows. If serum is restored to moderate concentrations, p21p will form a complex with procaspase-3 to inhibit Casp3 production. Only when Cyt c has accumulated sufficiently during a long starvation period can Casp3 activation persist, leading to cell apoptosis; otherwise, Casp3 decays to low levels, and cell-cycle arrest is induced (Fig. 5E). If serum is restored to high concentrations, p21 will be expressed at basal levels, and a large amount of procaspase-3 can be activated; thus, a relatively short starvation time is required for Cyt c elevation to induce apoptosis (otherwise, a very low amount of Cyt c leads to cell proliferation).
Two Waves of Cyt c Release.
As shown above, sufficient accumulation of cytosolic Cyt c is essential for apoptosis induction. The release of Cyt c from mitochondria to the cytoplasm is assumed to follow biphasic dynamics (SI Appendix, Method S3, Eq. S30), so there exists a two-wave release during apoptosis, consistent with the experimental results (25, 26). The first wave engages a small pool of soluble Cyt c in the intermembrane space of mitochondria, whereas the second releases a large pool of Cyt c, which is evoked by the Casp3-mediated cleavage of mitochondrial complexes. The first wave is a slow process, whereas the second wave, Casp3-mediated amplification of Cyt c release, is a fast process associated with Casp3 activation (SI Appendix, Fig. S3).
The fraction of Cyt c released to the cytosol at t = T0 (FC) versus C parallels the T0–C curve (Fig. 5C). FC denotes the minimal fraction required for apoptosis induction. For 2 ≤ C ≤ 5%, FC remains around 0.14, which is the fraction of soluble Cyt c that resides in mitochondria (45); the transition between two waves of Cyt c release roughly corresponds to the conversion between the two stages of apoptosis induction. In contrast, for 5.5 ≤ C ≤ 10%, FC is less than 0.14, because a small amount of Cyt c is enough for apoptosis induction at a later time. Nevertheless, a long time is required for the full activation of Casp3 because free procaspase-3 is restored to high levels only after p21 is degraded to basal levels, and a sharp increase in Casp3 levels still corresponds to the initiation of the second wave of Cyt c release (see SI Appendix, Figs. S6 and S7 and analyses therein in terms of activation of the p21–Casp3 and Casp3–Cyt c release positive feedback loops). Of note, FC would not be so small if more antiapoptotic proteins such as Bcl-2 were included in the model.
The left boundary of the new branch in Fig. 5D is determined by the OFF-to-ON transition point of p53 in the bifurcation diagram (SI Appendix, Fig. S8A). When the kinetic mode of p53 converts from ultrasensitivity to bistability and the bistable regime enlarges, the region covered by this new branch expands leftward. Accordingly, the right parts of the T0–C and FC–C curves expand leftward (SI Appendix, Fig. S8B). Additionally, when each parameter is increased or decreased by 10% with respect to its default value, phenomena similar to those shown in Figs. 4 and 5 can be observed (SI Appendix, Fig. S9), suggesting that the main results described above are insensitive to parameter variations in a limited range.
Collectively, critical T0 and FC serve as a timer and a biomarker, respectively, for dictating the irreversibility of apoptosis induction; that is, E1A-expressing cells commit to apoptosis if the starvation time is longer than T0 or if the fraction of released Cyt c exceeds FC. The dependence of T0 and FC on terminal serum concentration reflects the extent to which apoptosis induction is resistant to serum readdition.
Discussion
The tumor-suppressive network enables appropriate cellular responses to mitogenic and oncogenic signals. We identify how key network components can dynamically contribute to tumor suppression. The discrimination between normal and oncogenic proliferative signaling is made via the all-or-none expression of ARF. Thus, cell-fate decisions can be reached by the competition of the proapoptotic and prosurvival signals through downstream functional modules, such as activated p53 in the cases of cell-cycle arrest and apoptosis.
Our results indicate that, in addition to p53, other regulators such as E2F1, p21, and Akt can affect cellular output. (i) Because E2F1 has a critical proapoptotic role, E1A-expressing cells tend to suppress uncontrolled proliferation by inducing apoptosis. Alternatively ARF overexpression alone leads mostly to cell-cycle arrest (41). (ii) In the presence of p21/procaspase-3 interaction, cells may undergo apoptosis, cell-cycle arrest, or proliferation. Otherwise, cells could commit only apoptosis or proliferation. This inference is consistent with experimental observations (46). (iii) It is well-known that Akt promotes cell survival by engaging multiple pathways (6). For simplicity, we have limited our analysis to two loops, Akt–MDM2–p53–PTEN and p21–Casp3. They act either upstream or downstream of p53. Accordingly, Akt can modulate p53 levels via MDM2 and apoptosis induction via p21. Such a hierarchical organization of Akt signaling mediates its antiapoptotic function. Modulating the dynamics of and interactions between those proteins effectively controls the cellular outcome and may be exploited to find mechanisms to reactivate the tumor-suppressive functions in cancer cells.
Adding growth factors to serum-starved cells may evoke distinct cellular outcomes that will depend on the dynamics of serum recovery. Cells undergo cell-cycle arrest or apoptosis at moderate concentrations and proliferation or apoptosis at high concentrations. Apoptosis can be induced only when the starvation time is longer than a certain threshold, depending on the terminal serum concentration. These results depend on the antiapoptotic mechanism in the model: p21 can be phosphorylated by Akt to form a complex with procasepase-3, inhibiting its activation. Incorporating additional mechanisms, such as adding Bcl-2, may enrich our understanding of the impact of growth factors on apoptosis induction.
The proposed model provides insights into the role of the tumor-suppressive network in processing normal and oncogenic signals and making cell-fate decisions. Despite the multifaceted regulation of ARF at the transcriptional, translational, and posttranslational levels (47), the model focuses on the induction of ARF expression, because it is the main outcome of oncogenic signaling. The model could be extended to incorporate additional ARF regulation, so that we could gain more insights into delicate modulation of p53 and ARF activities.
Supplementary Material
Acknowledgments
We thank Drs. Zhaohui Feng and Guang Yao for helpful discussions. This work was supported by 973 Program of China Grant 2013CB834104 and National Natural Science Foundation of China Grants 31361163003, 81421091, 11574139, and 11175084. J.N.O. was supported by National Science Foundation Grants PHY-1427654 and NSF-CHE 1614101 and by Cancer and Prevention Institute of Texas Grant R1110.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702412114/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 7.Polager S, Ginsberg D. p53 and E2f: Partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 8.Lowe SW, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 9.Harrington EA, Bennett MR, Fanidi A, Evan GI. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canman CE, Gilmer TM, Coutts SB, Kastan MB. Growth factor modulation of p53-mediated growth arrest versus apoptosis. Genes Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- 11.Deng J, Xia W, Hung MC. Adenovirus 5 E1A-mediated tumor suppression associated with E1A-mediated apoptosis in vivo. Oncogene. 1998;17:2167–2175. doi: 10.1038/sj.onc.1202148. [DOI] [PubMed] [Google Scholar]
- 12.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 13.de Stanchina E, et al. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K. Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J. 2005;24:3724–3736. doi: 10.1038/sj.emboj.7600836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stott FJ, et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ries S, et al. Opposing effects of Ras on p53: Transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103:321–330. doi: 10.1016/s0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 18.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 20.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 21.Wee KB, Aguda BD. Akt versus p53 in a network of oncogenes and tumor suppressor genes regulating cell survival and death. Biophys J. 2006;91:857–865. doi: 10.1529/biophysj.105.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 23.Nahle Z, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 24.Kirsch DG, et al. Caspase-3-dependent cleavage of Bcl-2 promotes release of cytochrome c. J Biol Chem. 1999;274:21155–21161. doi: 10.1074/jbc.274.30.21155. [DOI] [PubMed] [Google Scholar]
- 25.Garrido C, et al. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q, Gong B, Almasan A. Distinct stages of cytochrome c release from mitochondria: Evidence for a feedback amplification loop linking caspase activation to mitochondrial dysfunction in genotoxic stress induced apoptosis. Cell Death Differ. 2000;7:227–233. doi: 10.1038/sj.cdd.4400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou BP, et al. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 28.Child ES, Mann DJ. The intricacies of p21 phosphorylation: Protein/protein interactions, subcellular localization and stability. Cell Cycle. 2006;5:1313–1319. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Fujita N, Tsuruo T. Caspase-mediated cleavage of p21Waf1/Cip1 converts cancer cells from growth arrest to undergoing apoptosis. Oncogene. 1999;18:1131–1138. doi: 10.1038/sj.onc.1202426. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XP, Liu F, Wang W. Two-phase dynamics of p53 in the DNA damage response. Proc Natl Acad Sci USA. 2011;108:8990–8995. doi: 10.1073/pnas.1100600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Wang J. Quantifying the underlying landscape and paths of cancer. J R Soc Interface. 2014;11:20140774. doi: 10.1098/rsif.2014.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C, Wang J. A physical mechanism and global quantification of breast cancer. PLoS One. 2016;11:e0157422. doi: 10.1371/journal.pone.0157422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Wang J. Quantifying the landscape for development and cancer from a core cancer stem cell circuit. Cancer Res. 2015;75:2607–2618. doi: 10.1158/0008-5472.CAN-15-0079. [DOI] [PubMed] [Google Scholar]
- 34.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 35.Iaquinta PJ, Aslanian A, Lees JA. Regulation of the Arf/p53 tumor surveillance network by E2F. Cold Spring Harb Symp Quant Biol. 2005;70:309–316. doi: 10.1101/sqb.2005.70.050. [DOI] [PubMed] [Google Scholar]
- 36.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 37.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 38.Sabbatini P, McCormick F. Phosphoinositide 3-OH kinase (PI3K) and PKB/Akt delay the onset of p53-mediated, transcriptionally dependent apoptosis. J Biol Chem. 1999;274:24263–24269. doi: 10.1074/jbc.274.34.24263. [DOI] [PubMed] [Google Scholar]
- 39.Hershko T, Chaussepied M, Oren M, Ginsberg D. Novel link between E2F and p53: Proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–383. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- 40.Rao L, et al. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 42.Ashcroft M, et al. Phosphorylation of HDM2 by Akt. Oncogene. 2002;21:1955–1962. doi: 10.1038/sj.onc.1205276. [DOI] [PubMed] [Google Scholar]
- 43.Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013;20:576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabbatini P, Lin J, Levine AJ, White E. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 45.Scorrano L, et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 46.Gorospe M, et al. p21Waf1/Cip1 protects against p53-mediated apoptosis of human melanoma cells. Oncogene. 1997;14:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 47.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.