Significance
Little is known about the molecular mechanisms by which the microbiota influences T-cell responses. We have determined that a secreted protein, Erdr1, is highly upregulated in germfree T cells and that microbial products can suppress Erdr1 through MyD88. Erdr1 functions to induce apoptosis through a Fas-dependent pathway that can impact the process of immunization and dictate the clinical course of autoimmunity. Thus, microbial colonization functions to regulate T-cell survival through suppression of the cytokine Erdr1, revealing a potentially new T-cell factor that can be therapeutically targeted.
Keywords: microbiota, T cells, apoptosis, Fas, Toll-like receptors
Abstract
Symbiotic microbes impact the severity of a variety of diseases through regulation of T-cell development. However, little is known regarding the molecular mechanisms by which this is accomplished. Here we report that a secreted factor, Erdr1, is regulated by the microbiota to control T-cell apoptosis. Erdr1 expression was identified by transcriptome analysis to be elevated in splenic T cells from germfree and antibiotic-treated mice. Suppression of Erdr1 depends on detection of circulating microbial products by Toll-like receptors on T cells, and this regulation is conserved in human T cells. Erdr1 was found to function as an autocrine factor to induce apoptosis through caspase 3. Consistent with elevated levels of Erdr1, germfree mice have increased splenic T-cell apoptosis. RNA sequencing of Erdr1-overexpressing cells identified the up-regulation of genes involved in Fas-mediated cell death, and Erdr1 fails to induce apoptosis in Fas-deficient cells. Importantly, forced changes in Erdr1 expression levels dictate the survival of auto-reactive T cells and the clinical outcome of neuro-inflammatory autoimmune disease. Cellular survival is a fundamental feature regulating appropriate immune responses. We have identified a mechanism whereby the host integrates signals from the microbiota to control T-cell apoptosis, making regulation of Erdr1 a potential therapeutic target for autoimmune disease.
The incidence of autoimmune diseases, such as type 1 diabetes, multiple sclerosis, and rheumatoid arthritis, is increasing in Western countries at confounding rates. These diseases are thought to occur through a combination of genetic and environmental factors. Research over the past decade has provided compelling evidence that resident commensal microbes act as one environmental feature that influences the induction of autoimmunity (1–3). The effects of the microbiota on the development of autoimmunity could be due in part to the known contribution to steady-state immune system development (4, 5). A vast majority of the microbiota are found within the gastrointestinal tract; however, many autoimmune diseases occur at extraintestinal sites. Therefore, understanding the mechanisms by which resident gut microbes regulate the immune response outside of the gut might identify novel therapeutic targets for autoimmunity.
Regulation of T-cell apoptosis is an essential characteristic of appropriate immunity. Survival of antigen-specific T cells is required to eradicate infectious agents whereas induction of cellular death is key to resolution of inflammation (6). Apoptosis also impacts steady-state T-cell responses during development in the thymus and homeostatic turnover at mucosal surfaces (7, 8). Dysregulation of T-cell death can have serious consequences to health that can lead to autoimmunity or immunodeficiency. Thus, the ability to modulate the persistence of T cells has clear medicinal potential. One of the most well-studied death receptor pathways in T cells is Fas, which functions through activation of caspases 8 and 3 to induce apoptosis (9). Although pathogens are known to exploit cell death or survival pathways to infect hosts (10), little is known about whether cues from symbiotic bacteria function to influence T-cell survival. Here we have identified that the microbiota suppresses the expression of a secreted factor, erythroid differentiation factor 1 (Erdr1), which functions to control T-cell apoptosis in a Fas-dependent manner.
Erdr1 is highly conserved between mice and humans, and its expression has been reported to be dysregulated in human cancer (11, 12). Erdr1 was originally identified in supernatants of B-cell leukemic cells and named for its hemoglobin-inducing activity on erythroid lines, but now has implicated roles in cellular survival, NK cytotoxicity, and metastasis; however, its function within T cells remains completely unexplored (11–15).
Results
Erdr1 Expression in T Cells Is Suppressed by the Microbiota.
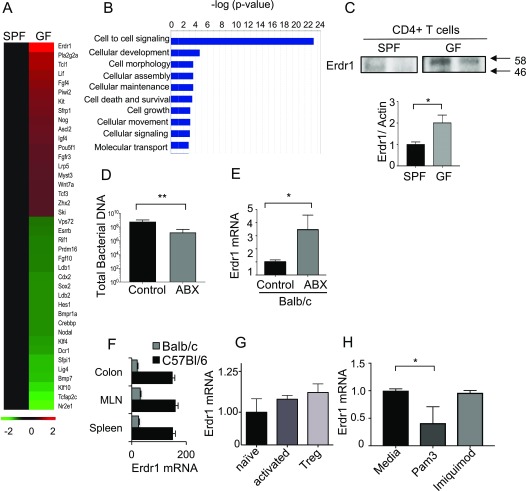
To broadly characterize how the microbiota might affect T-cell responses outside of the gut and potentially identify novel genes regulated by the microbiota, we compared transcriptional profiles from splenic, sort-purified CD4+ T cells from germfree or specific pathogen-free (SPF) animals. Using Ingenuity Pathway Analysis, we observed that genes involved in cellular maintenance, death, and survival were among the most influenced by the microbiota, implying that commensal organisms can affect T-cell homeostasis (Fig. S1 A and B). One gene in particular, Erdr1, was one of the most highly up-regulated genes in germfree T cells in the Kyoto Encyclopedia of Genes and Genomes (KEGG) category “cellular maintenance,” suggesting that the microbiota suppressed expression of Erdr1 in T cells (Fig. S1A). We verified the results of our transcriptional profiling by qRT-PCR of Erdr1 in either germfree or SPF splenic CD4+ T cells and found that T cells from germfree mice consistently had elevated levels of Erdr1 transcripts and protein (Fig. 1A and Fig. S1C). Antibiotics are known to cause perturbations to the microbiota that are associated with harmful consequences to the host (16, 17). To determine whether Erdr1 expression is sensitive to changes in microbiota composition, adult SPF animals were treated with a mixture of antibiotics to deplete commensal microbes. As with germfree mice, Erdr1 expression became elevated within splenic CD4+ T cells from antibiotic-treated SPF animals from two different genetic backgrounds, demonstrating that the microbiota continuously regulates Erdr1 (Fig. 1B and Fig. S1 D and E). Expression levels of Erdr1 in CD4+ T cells isolated from multiple tissues including the colon, mesenteric lymph node, and spleen remained similar, indicating that proximity to the microbiota does not affect expression of Erdr1 in T cells (Fig. S1F). Additionally, Erdr1 expression is not significantly different in naive, activated, or Treg subsets sort-purified directly from the spleen of an animal (Fig. S1G), suggesting that Erdr1 expression is similarly expressed during a variety of T-cell activation states.
Fig. S1.
Erdr1 is up-regulated in germfree T cells and regulated by microbial signals. (A) Heat map of the most significantly changed genes in germfree mice compared with SPF in splenic T cells in the KEGG category of cellular maintenance. n = 2/group. (B) Ingenuity Analysis on microarray data from A showing the most significantly different pathways when comparing splenic germfree (GF) versus SPF CD4+ T cells. (C) CD3+CD4+ T-cell lysate from GF or SPF spleens was collected and analyzed for Erdr1 protein levels. (D) Total bacteria from fecal samples of animals after 2 wk of antibiotic treatment. (E) BALB/c mice were treated with antibiotics for 2 wk. CD3+CD4+ T cells were isolated from the spleen, and Erdr1 was measured by qRT-PCR. n = 4–5/group. (F) CD3+CD4+ T cells were isolated from the indicated compartments, and levels of Erdr1 were measured by qRT-PCR. n = 3/group. (G) RNA was collected from naive (CD3+CD4+CD44−CD62L+), activated (CD3+CD4+CD44+CD62L−), or Treg (CD3+CD4+CD25hi) T cells, and levels of Erdr1 were analyzed. (H) CD3+CD4+ T cells from GF spleens were collected and incubated with either Pam3 or Imiquimod, and Erdr1 was measured by qRT-PCR. n = 3/group. *P < 0.05; **P < 0.01. Statistical significance was determined by Student’s t test.
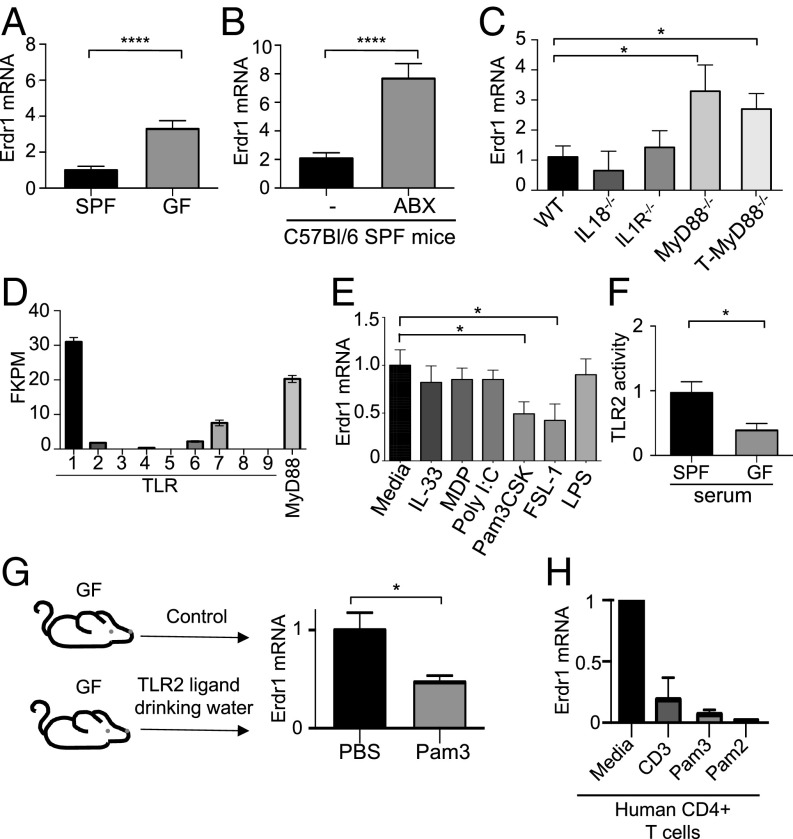
Fig. 1.
Erdr1 is a conserved T-cell factor that is dynamically regulated by the microbiota through MyD88. (A) RNA was collected from CD3+CD4+ T cells from indicated animals and Erdr1 was analyzed by qRT-PCR. n = 8/group compiled from three experiments. (B) qRT-PCR was performed for Erdr1 from CD4+ T cells in antibiotic-treated animals. The n = 10/group was compiled from three experiments. (C) qRT-PCR was performed for Erdr1 from CD4+ T cells isolated from the indicated animals. n = 3–6/group. (D) RNA was collected from the CD3+CD4+ splenic T cells isolated and used for RNA-seq. n = 4/group. (E) Germfree CD4+ T cells were stimulated with anti-CD3 either alone or in combination with indicated stimuli. n = 5–8/group. (F) HEK293T NF-κB reporter cells activated by TLR2 were incubated with serum isolated from either SPF or germfree animals. Performed from the sera of six independent animals. (G) qRT-PCR was performed for Erdr1 from CD4+ T cells from germfree mice provided Pam3CysK in their drinking water. Data were compiled from two experiments; n = 6–8/group. (H) Primary human CD4+CD3+ T cells were isolated from whole blood and stimulated with anti-CD3 either alone or in combination with the indicated TLR2 ligands. Representative of three experiments. *P < 0.05 and ****P < 0.0001 were determined by a Student’s t test.
Given that Erdr1 levels are sensitive to cues provided by the microbiota, we sought to determine the mechanism by which commensal bacteria can influence T-cell expression of Erdr1. The microbiota is an abundant source of Toll-like receptor (TLR) ligands; therefore, we tested whether regulation of Erdr1 was dependent on MyD88, an adaptor molecule that controls signaling through most TLRs. Erdr1 expression was elevated in splenic CD4+ T cells isolated from MyD88 total body knockout animals (MyD88−/−) as well as from mice with a T-cell–specific ablation of MyD88 (T-MyD88−/−) (18, 19) (Fig. 1C). As MyD88 can also signal through IL-1 and IL-18 receptors, we tested the involvement of these cytokines in Erdr1 regulation as well. Erdr1 expression levels in splenic CD4+ T cells from IL-1R– and IL-18–deficient animals were similar to those of wild-type (WT) controls, indicating that Erdr1 is not regulated by these cytokines (Fig. 1C) (12). It is possible that microbiota composition could be different in these various knockout strains and account for the differential expression of Erdr1. Therefore, we tested whether TLR ligands could directly suppress Erdr1 in T cells in vitro. To identify the most relevant T-cell intrinsic TLRs, we analyzed RNA sequencing data from splenic SPF CD4+ T cells and observed that TLR1, -2, -6, and -7 were most highly expressed under homeostatic conditions (Fig. 1D). Innate immune signaling agonists including Pam3CysK (TLR1/2), FSL-1 (TLR2/6), IL-33, Poly I:C (TLR3), Muramyl Dipeptide (NOD2), LPS (TLR4), and Imiquimod (TLR7) were used to stimulate T cells in vitro in the presence of anti-CD3. T cells treated with TLR2 ligands suppress Erdr1 expression compared with media controls (anti-CD3 alone), whereas other stimuli do not affect Erdr1 transcripts (Fig. 1E and Fig. S1H). These data suggest that T-cell–intrinsic TLR2 signaling is most relevant for suppression of Erdr1 within T cells.
Previous reports have demonstrated that TLR4 ligands from the microbiota are present in the serum and are able to enhance neutrophil function within the bone marrow (20). Consistent with this, elevated TLR2 activity could be detected from the serum of SPF animals compared with germfree mice (Fig. 1F). This suggests that TLR2 agonists can escape from the intestine and into the blood to potentially modulate Erdr1 expression. Based on this, we introduced Pam3CysK into the gut of germfree mice and observed that Erdr1 expression in splenic T cells was significantly reduced (Fig. 1G). This pathway for Erdr1 suppression is conserved in human T cells. Primary CD4+ T cells isolated from healthy donors and treated with the TLR2 ligands Pam3CysK and Pam2CysK had significantly reduced Erdr1 expression compared with media-treated controls (Fig. 1H). These data indicate that Erdr1 can be regulated under homeostatic conditions by microbial cues from the gut by TLR signals in an MyD88-dependent fashion.
Erdr1 Functions to Induce T-Cell Apoptosis.
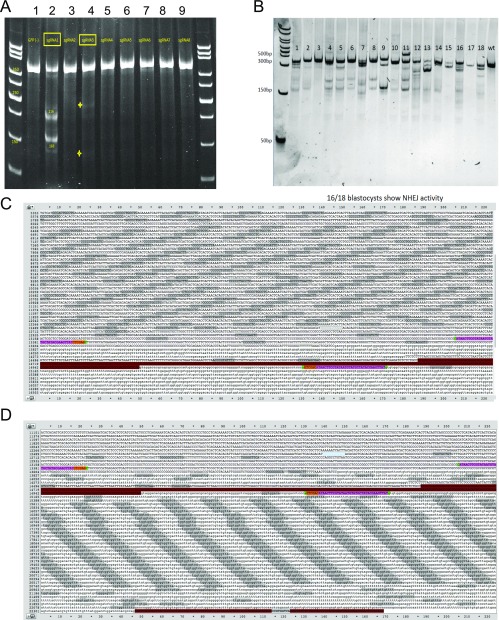
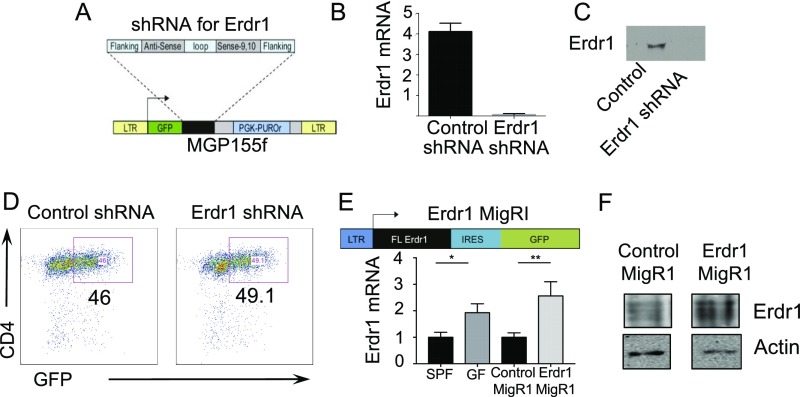
Erdr1 is a vesicle-associated secreted protein that is ubiquitously expressed (11, 15). It has reported functions in keratinocytes and NK cells, although there are currently no known roles for Erdr1 in T lymphocytes (13, 14). A knockout mouse model does not exist to study the role of Erdr1, and our attempts to make a whole-body knockout using Crisper/Cas9 technology revealed that loss of this gene is not tolerated in utero (Fig. S2 A and B). Additionally, two independent attempts to insert LoxP sites to create a conditional allele were not successful due to the highly repetitive nature of the locus surrounding Erdr1 (Fig. S2 C and D). Therefore, we manipulated Erdr1 expression using overexpression and depletion approaches in primary mouse T cells. We used retroviral constructs encoding GFP to express an Erdr1-specific shRNA for silencing endogenous expression or to overexpress the Erdr1 cDNA. We validated that these constructs either suppressed or enhanced Erdr1 expression in splenic mouse T cells at both the mRNA and the protein levels (Fig. S3 A–F). Importantly, overexpression of Erdr1 using this construct causes elevated levels of Erdr1 that are similar to those found in germfree and antibiotic-treated animals (Fig. S3E).
Fig. S2.
Erdr1 knockout or floxed animals cannot be generated. (A) Two of eight guide RNAs (gRNAs) had activity at the Erdr1 locus in vitro. Numbers indicate the various gRNAs. gRNA 1 and gRNA 3 had activity. (B) Sixteen of 18 blastocysts showed Crisper/Cas9 activity at the Erdr1 locus. However, 711 embryos injected with these blastocysts could not produce viable pups, indicating that this gene is necessary during development. (C and D) A high degree of sequence repetition inhibits efficient targeting of Erdr1. The first exon of Erdr1 is shown highlighted in red, and flanking LoxP sites shown in pink indicate the target sites for CRISPR genome editing. Dark-gray highlights indicate repetitive sequencing surrounding either (C) the 5′ site or (D) the 3′ site.
Fig. S3.
Validation of shRNA and overexpression of Erdr1 in primary T cells. (A–F) The 293T cells are transfected with either the Erdr1 MigRI or Erdr1 shRNA or with the respective control plasmids along with pCLECO packaging vector to obtain virus. Erdr1-modified virus is then used to infect CD4+CD3+ splenic mouse T cells as described in SI Materials and Methods. (A) Schematic of the Erdr1 shRNA vector used throughout the study. (B and C) Validation that the Erdr1 shRNA construct functions to reduce levels of Erdr1 at the RNA and protein levels in T cells. (D) Efficiency of delivery of Erdr1 and control shRNA into primary CD4+CD3+ T lymphocytes. Cells are sorted by GFP+ before use. (E) Schematic of vector used for Erdr1 overexpression. RNA was collected from CD3+CD4+ splenic mouse T cells from SPF, GF, control infected, or Erdr1overexpressing T cells, and levels of Erdr1 were assayed by qRT-PCR. (F) Validation that the Erdr1 MigRI results in overexpression of the Erdr1 in primary splenic CD4+ T cells at the protein levels. *P < 0.05 and **P < 0.01. Statistical significance was determined by Student’s t test.
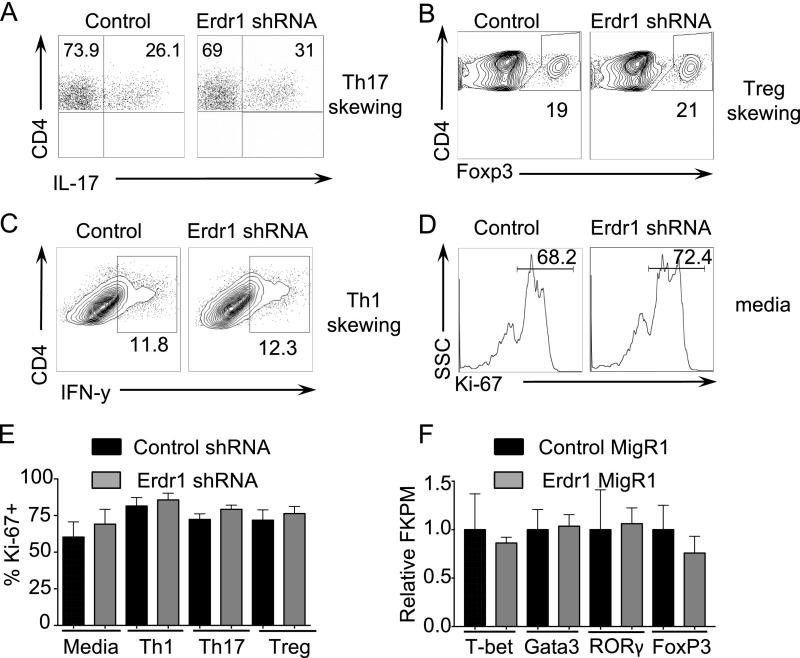
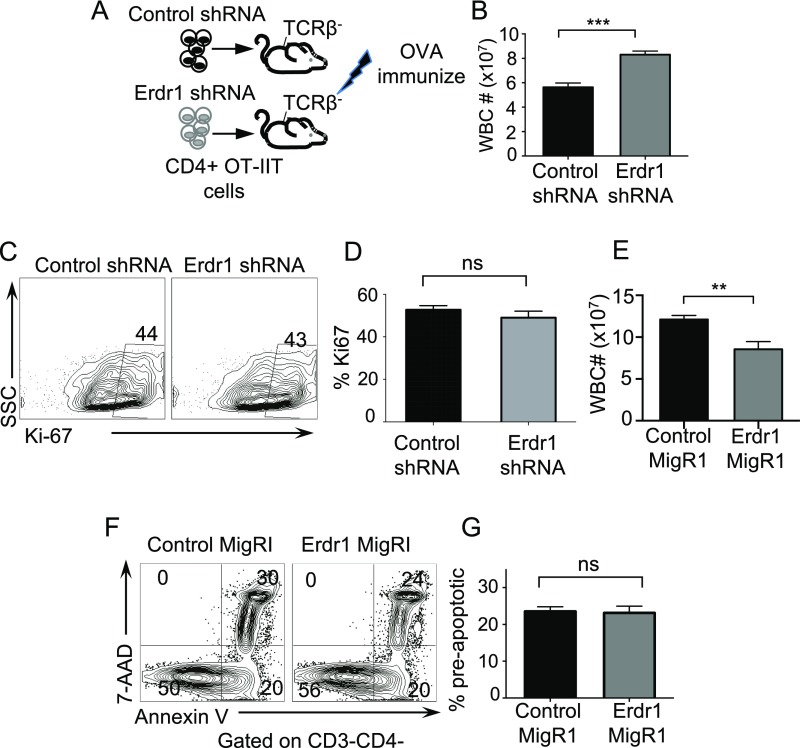
The microbiota has important impacts on the differentiation of CD4+ T-helper-cell subsets in both the gut and the spleen (4, 21, 22). To address whether Erdr1 directly influences T-helper-cell responses, we performed in vitro T-cell–skewing assays during Erdr1 shRNA knockdown. Control or Erdr1 shRNA-treated T cells were placed in media (anti-CD3/CD28 only) and in Th1-, Th17-, or Treg-skewing conditions, and Ki-67 was used to measure proliferation. Reduction of Erdr1 expression levels in primary T cells did not result in differences in the percentage of in vitro-skewed T-helper-cell populations (Fig. S4 A–C), nor did it result in any differences in proliferation (Fig. S4 D–E). Additionally, transcriptional profiling of primary T cells overexpressing Erdr1 did not reveal any changes in the expression of the transcriptional regulators known to be important for differentiation of these T-cell lineages including Gata3, Tbet, Foxp3, and Rorc (Fig. S4F). These data suggest that Erdr1 does not directly influence the differentiation of any particular T-helper-cell subset.
Fig. S4.
Erdr1 does not influence T-helper-cell differentiation or proliferation. (A–C) Representative flow cytometry plots of sort-purified CD4+CD3+ T cells from the spleens of SPF animals after transfection with control or Erdr1-specific shRNA. T cells were incubated under the designated skewing conditions (A) Th17, (B) Treg, and (C) Th1 and analyzed for differentiation. (D) Representative flow cytometry plot of Ki-67 staining from sort-purified CD4+CD3+ T cells in response to anti-CD3/CD28 stimulation. All experiments were performed from cells isolated from at least four mice, and each experiment was replicated at least twice independently. (E) Ki-67 staining of sort-purified CD4+CD3+ T cells under indicated skewing conditions. n = 6–8/group compiled from two independent experiments. (F) CD3+CD4+ GFP+ sort-purified T cells from cells overexpressing either control MigRI or Erdr1 MigRI were stimulated with anti-CD3 and anti-CD28 in vitro for 2 d, and RNA was collected for RNA-seq. Analysis of fragments per kilobase of transcript per million mapped reads (FPKM) of master regulators of T-helper-cell differentiation. n = 4/group.
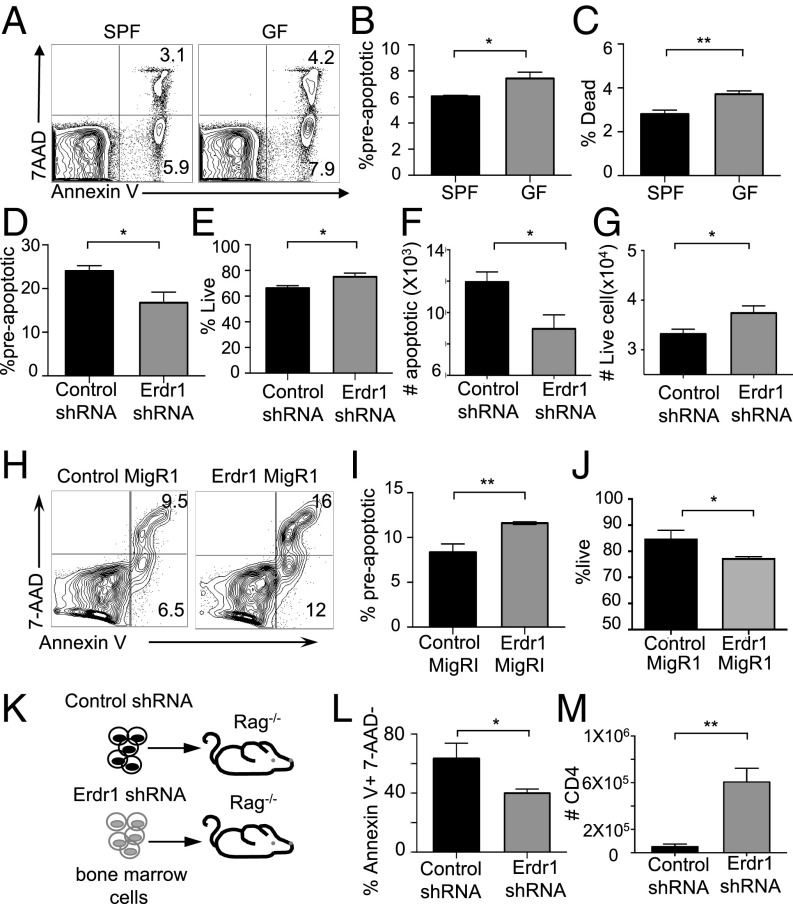
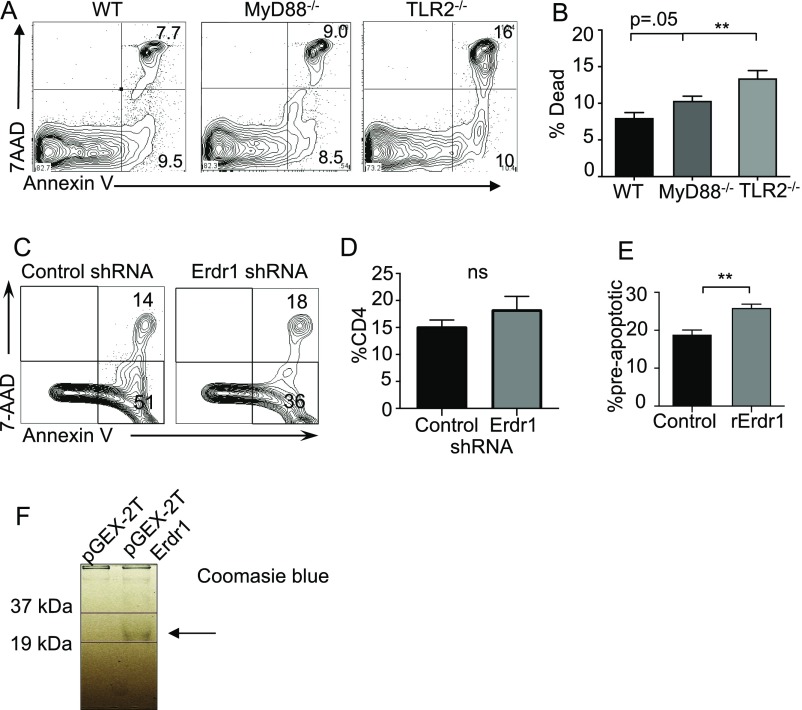
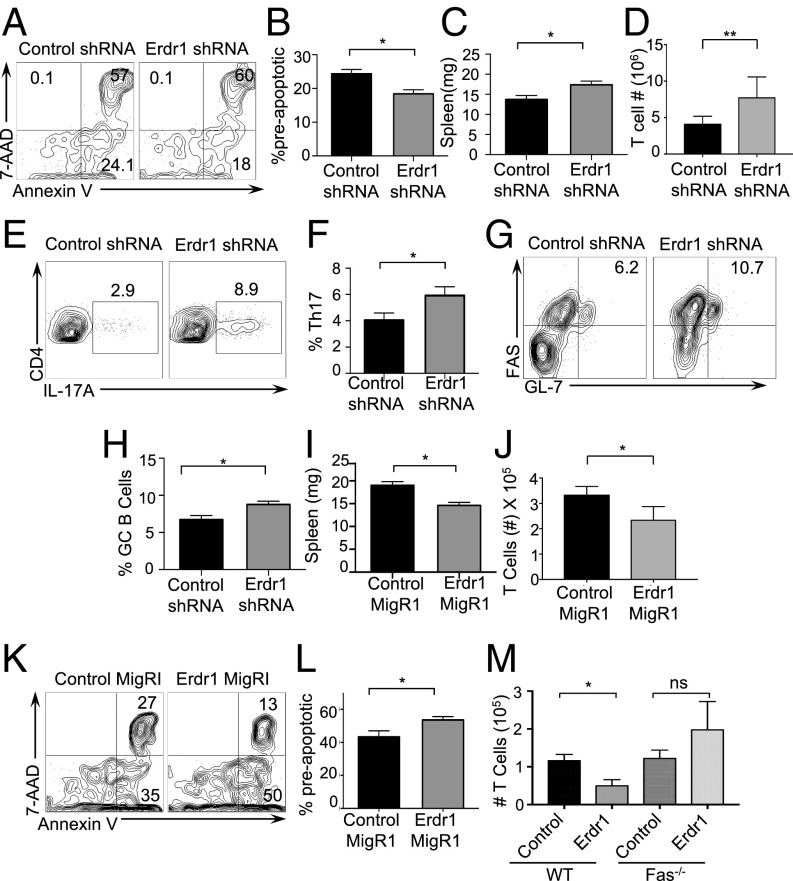
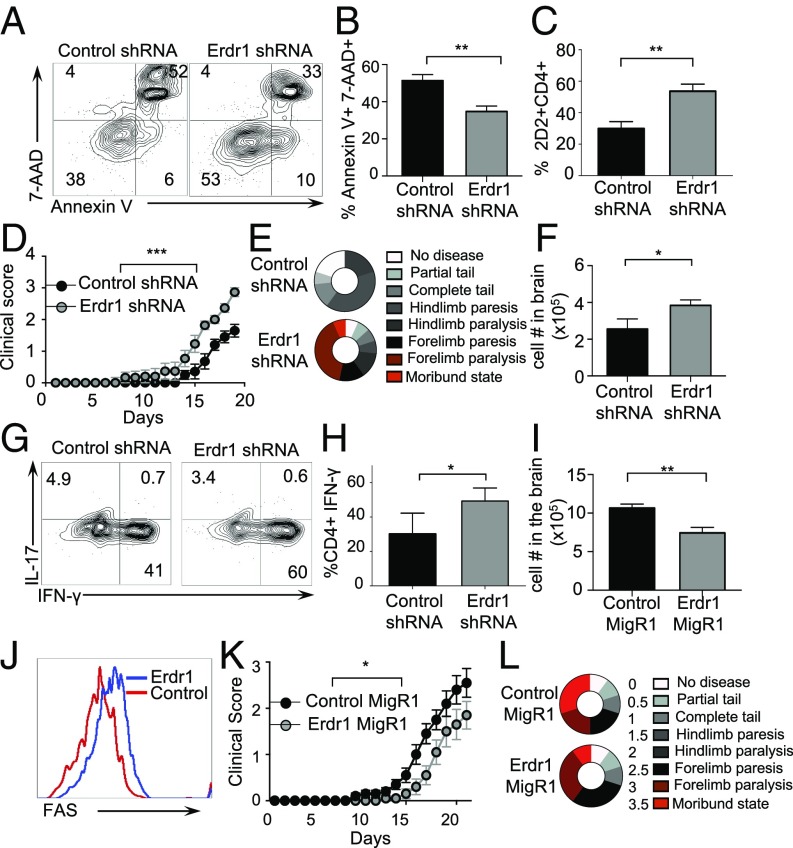
Throughout these in vitro experiments, we consistently observed changes in the number of cells upon Erdr1 manipulation despite equal proliferation rates. One of the many functions suggested for Erdr1 is regulation of cell death; however, this has not been well studied. Consistent with this, genes involved in cellular survival and death are significantly regulated by the microbiota in our microarray analysis (Fig. S1B). Using Annexin V, a well-known marker for cells about to undergo apoptosis and 7AAD, a dead-cell stain, we observed that splenic CD4+ T cells isolated from germfree mice had a small, but significant, increase in preapoptotic (AnnexinV+7AAD−) and dead cells (Annexin V+7AAD+) (Fig. 2 A–C). Additionally, splenic CD4+ T cells isolated from animals deficient in MyD88 or TLR2 also have significant increases in cellular death (Fig. S5 A and B). As macrophages rapidly engulf dead cells, it might be difficult to appreciate differences in actively dying cells under homeostatic conditions in vivo. To address a role for Erdr1 in T-cell apoptosis, we used overexpression and shRNA in primary splenic T cells in vitro. Suppression of Erdr1 by shRNA in cultured splenic CD4+ T cells led to an ∼10–15% reduction in the proportion of preapoptotic cells. This rate of apoptosis results in a twofold decrease in the absolute numbers of cells undergoing apoptosis and significantly more live cells within the culture compared with control (Fig. 2 D–G). The opposite phenotype is observed in cells overexpressing Erdr1. Overexpression of Erdr1 increases the percentage of preapoptotic T cells and reduces the number of live cells observed in culture (Fig. 2 H–J). Erdr1 influences T-cell survival during homeostatic cellular development in vivo. Indeed, adoptive transfer of control or Erdr1 shRNA-expressing bone marrow into Rag−/− recipients reveals a significant reduction in the proportion of apoptotic cells (Fig. 2 K and L). This modest, but significant, decrease in the proportion of T-cell apoptosis has substantial effects on the absolute number of CD4+ T cells. Although there is no difference in the percentage of CD4+ T cells in control versus Erdr1 shRNA-expressing T cells (Fig. S5 C and D), there is a sixfold increase in the absolute numbers of T cells in the spleen of animals expressing Erdr1 shRNA (Fig. 2M). Further supporting a role for Erdr1 as a mediator of T-cell death, T cells treated with recombinant Erdr1 (rErdr1) also induced an increase in the percentage of preapoptotic T cells (Fig. S5 E and F). This result demonstrates that modulation of Erdr1 expression can alter T-cell survival and have profound impacts on the number of functionally available T cells.
Fig. 2.
Erdr1 induces T-cell apoptosis. (A–C) Cell apoptosis was analyzed by Annexin V and 7AAD in CD4+ T cells from germfree mice by flow cytometry. n = 7–8/group compiled from two experiments. (D–G) Indicated CD4+ T cells were stimulated with anti-CD3/CD28 and analyzed for cell death. Preapoptotic are Annexin V+7AAD−, live are Annexin V−7AAD−. The data represent two pooled independent experiments. n = 4/group. (H–J) Indicated CD4+ sort-purified T cells were stimulated with anti-CD3/CD28 and analyzed for cell death as above. The data represent two pooled independent experiments. n = 4/group. (K–M) Total bone marrow was infected with either control shRNA or Erdr1 shRNA-containing virus. GFP+ cells were flow-sorted and injected into Rag−/− animals. Eight weeks postreconstitution animals were analyzed for cellular death. n = 3/group. *P < 0.05 and **P < 0.01 were determined by a Student’s t test.
Fig. S5.
Cell apoptosis is modulated by Erdr1 expression. (A and B) CD3+CD4+ splenic T cells isolated from the indicated knockout animals and analyzed for cell death. (C) Representative flow cytometry plot of Annexin V staining in bone marrow chimeras from Fig. 2K. n = 3/group. (D) Percentage of CD4 in the spleens from animals receiving indicated bone marrow transplant. n = 3/group. (E) CD4+CD3+ T cells were treated with recombinant Erdr1 for 3 d in the presence of anti-CD3/CD28 and stained for Annexin V and 7AAD. n = 3/group. (F) Coomassie-stained gel of rERDR1 protein purification. **P < 0.01. ns, not significant. Statistical significance was determined by Student’s t test.
Erdr1-Mediated Apoptosis Is Dependent on Fas.
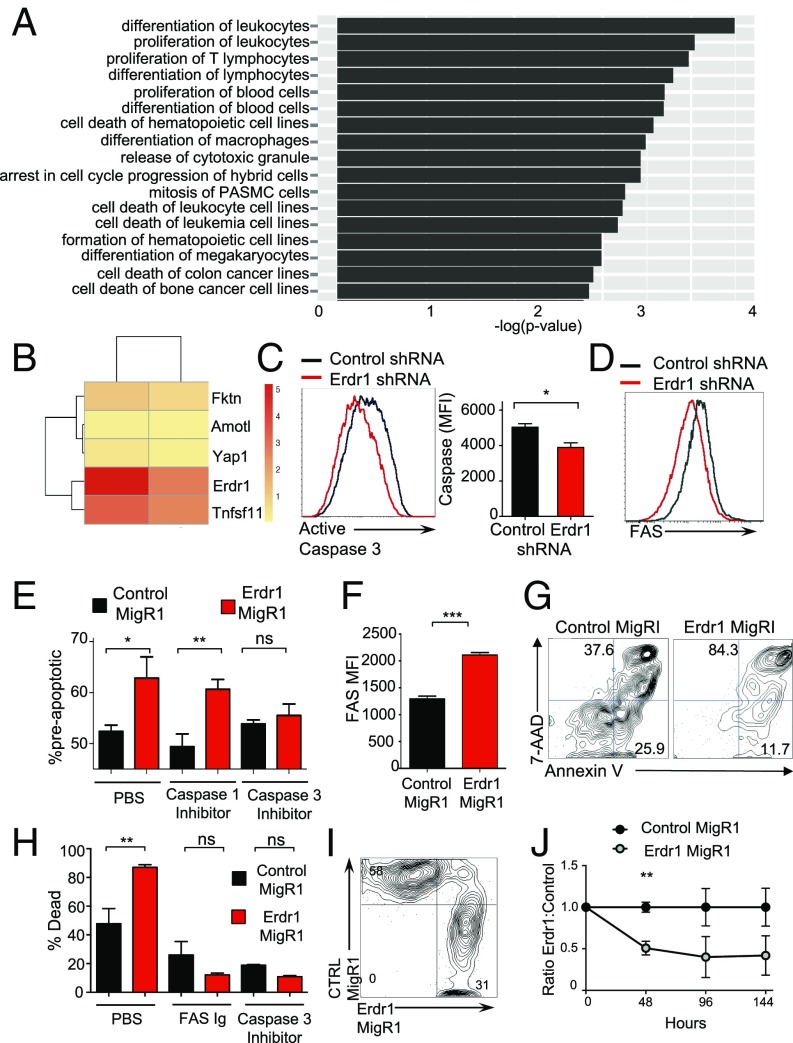
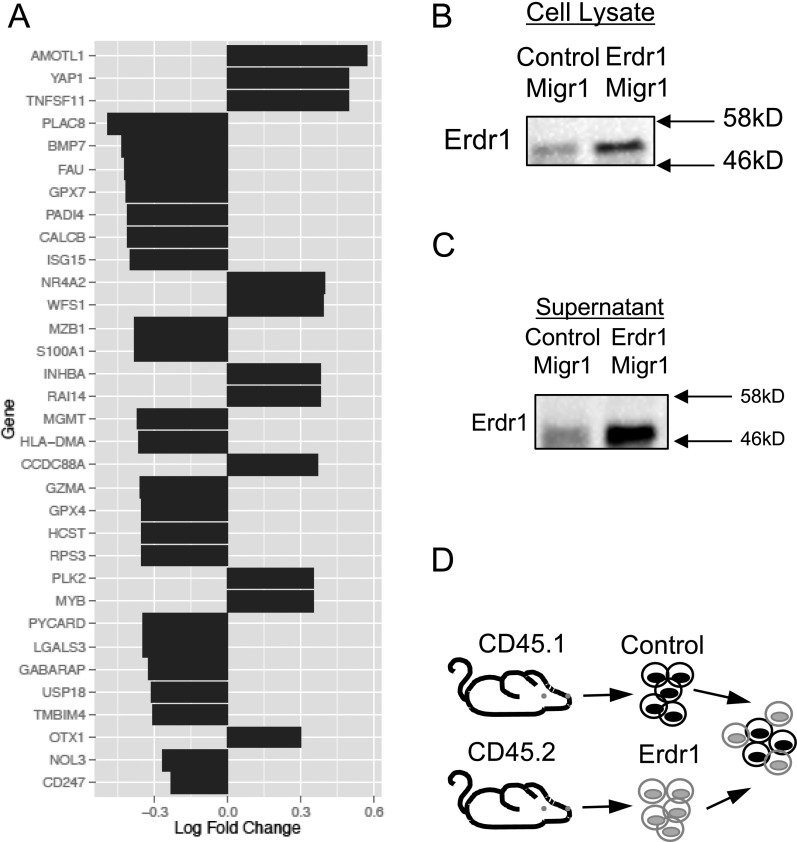
To better understand the mechanism by which Erdr1 influences T-cell apoptosis, RNA-seq was performed on primary splenic Erdr1-overexpressing CD4+ T cells. Consistent with enhanced cell apoptosis mediated by Erdr1, the most significantly different genes and pathways affected by Erdr1 were those involved in cell death and survival (Fig. 3A and Fig. S6A). Three of the five genes that were most highly up-regulated by forced Erdr1 overexpression were Amotl1, Tnfsf11, and Yap1, all of which are known to be involved in cell death (Fig. 3B) (23–26). The most significantly up-regulated gene was Tnfsf11, also known as RankL. RankL is highly expressed in activated T cells and has a myriad of functions (27). However, RankL has been reported to enhance the growth of T cells and therefore is not likely to mediate T-cell death through Erdr1 on its own (26). Interestingly, RankL can up-regulate apoptosis in dendritic cells and enhance cell death only in collaboration with Fas/FasL signaling (28). Fas/FasL interactions are a primary, well-studied mechanism leading to apoptosis through caspase 8 and caspase 3 within T cells (29–31). To determine whether Fas expression and caspase 3 activation are influenced by Erdr1, we analyzed these molecules during Erdr1 knockdown by shRNA. Reduced Erdr1 expression in primary splenic CD4+ T cells led to a reduction in the level of cleaved and activated caspase 3 (Fig. 3C) and a significant reduction in the surface expression of Fas (Fig. 3D). Whereas caspase-3 is associated with the induction of apoptosis, caspase 1 is known to influence the cleavage and activation of innate cytokines such as IL-1. To demonstrate that Erdr1 functions within the caspase-3–dependent cell death pathway, control or Erdr1-overexpressing primary T cells were incubated with caspase 1 or 3 inhibitors. Enhanced T-cell death induced by Erdr1 is blocked by a caspase 3, but not by a caspase 1 inhibitor (Fig. 3E). To further validate these findings, we constructed an Erdr1-overexpressing DO11.10 hybridoma T-cell line (32) (Fig. S6 B and C). Importantly, overexpression of Erdr1 in this cell line is only three- to fourfold higher than in control lines and therefore has similar levels of overexpression as is observed in germfree mice. Consistent with our findings in primary mouse T cells (Fig. 3D), forced overexpression of Erdr1in this cell line significantly elevated the levels of Fas on the cell surface (Fig. 3F). More importantly, Erdr1 overexpression in this cell line led to a dramatic increase in the percentage of dead cells found in the culture (Fig. 3 G and H), and the addition of a Fas-blocking antibody or a caspase 3 inhibitor completely ameliorates Erdr1mediated cell death (Fig. 3H).
Fig. 3.
Erdr1-induced death is dependent on Fas and caspase 3. (A and B) Indicated CD4+ T cells were used for RNA-seq and analyzed using Ingenuity Pathway Analysis. n = 4/group. (B) The five most significantly changed genes between Erdr1 MigRI and controls are shown. (C and D) Indicated CD4+ T cells were analyzed for active caspase 3 or Fas by flow cytometry. Three experiments were performed with n = 2–3/group. (E) Indicated CD4+ T cells stimulated with anti-CD3/CD28 in vitro in the presence of indicated inhibitors. n = 4/group from two trials. (F and G) Fas expression was measured by flow cytometry in control MigR1- or Erdr1 MigR1-expressing DO11.10 hybridoma T cells or (H) incubated with indicated inhibitors or Fas-blocking antibody and subsequently measured for cell death. Representative of four trials. (I and J) Equal numbers of Erdr1- and control MigRI-expressing T cells were mixed and stimulated with anti-CD3 to anti-CD28 for 6 days. n = 3/group. *P < 0.05, **P < 0.01, and ***P < 0.001 were determined by Student’s t test. ns, not significant.
Fig. S6.
Characterization of gene expression in Erdr1-overexpressing T cells. (A) List of the most significantly changed genes involved in cell death (up- or down-regulated). (B and C) Western blot of supernatant from cultured cells that were overexpressing Erdr1 or control cells to demonstrate that Erdr1 can be detected in the culture media. Western blot of cell lysate from Erdr1-overexpressing DO11.10 cells. (D) Schematic of experimental design for Fig. 3I.
Erdr1 is secreted and can function in both an autocrine and a paracrine fashion (15). To establish whether Erdr1 represents a mechanism of autoregulation, we forced expression of Erdr1 in CD45.2 T cells and incubated them with CD45.1-marked control cells (Fig. S6D). If Erdr1 functioned as a paracrine factor, there would be an equal representation of CD45.1 and CD45.2 cells in the culture. However, although Erdr1 protein was verified to be secreted into the T-cell media (Fig. S6C), within 48 h of cell culture, splenic CD4+ T cells overexpressing Erdr1 were rapidly out-competed by control cells (Fig. 3 I and J). Collectively, these data demonstrate that Erdr1 acts in an autocrine manner to induce apoptosis through induction of Fas and caspase 3.
Erdr1-Mediated Cell Death Influences the Immune Response.
Regulation of cellular survival is a critical component to the maintenance of appropriate immunity. Our data suggest that signals from the microbiota can function to promote cellular survival through Erdr1, and this could have important consequences for T-cell reactions in vivo. To test this, we manipulated Erdr1 expression by shRNA or overexpression in OT-II T cells. Use of OT-II TCR transgenic cells allows analysis of a T-cell population responding to the same antigen, ovalbumin peptide (OVA) (SI Materials and Methods). CD4+OT-II+Erdr1 shRNA or control T cells were adoptively transferred into TCRβ-deficient mice (animals that lack T cells) and subsequently immunized with OVA (Fig. S7A). Consistent with our findings in vitro, suppression of Erdr1 led to a reduced percentage of antigen-specific preapoptotic CD4+ T cells (Fig. 4 A and B). Suppression of Erdr1 led to moderate 15–20% decreases in the proportion of preapoptotic cells; however, this rate of cellular death has significant impacts on the immune response. Indeed, animals that received T cells with Erdr1 shRNA had marked increases in spleen size with greater cellularity and increased numbers of OT-II T cells (Fig. 4 C and D and Fig. S7B). There was also a significant increase in the percentage of inflammatory OT-II Th17 cells and enhanced germinal center B cells (Fig. 4 E–H). Of note, there was equal Ki-67 staining of cells within these experiments, emphasizing that Erdr1 does not influence proliferation (Fig. S7 C and D).
Fig. S7.
Erdr1 influences cellular survival during immunization. (A) Schematic of experimental design shown in Fig. 4. TCRβ−/− animals were provided with Erdr1-overexpressing, Erdr1 shRNA, or control OT-II cells and subsequently immunized with OVA. (B) Quantification of total splenic cells after RBC lysis from mice in A and Fig. 4. (C and D) Representative FACS plot and quantification of Ki-67 from splenic T cells isolated from animals in A and Fig. 4. Plots are gated on CD4+CD3+ cells. (E) Quantification of total splenic cells after RBC lysis from mice in Fig. 4. (F and G) There is no difference in non-CD4 cell apoptosis during overexpression of Erdr1 in T cells from Fig. 4, indicating that cell death is specific to the cells overexpressing Erdr1. **P < 0.01; ***P < 0.001. ns, not significant. Statistical significance was determined by Student’s t test.
Fig. 4.
Erdr1 regulates apoptosis in vivo during immunization. (A–D) Indicated OT-II CD4+ T cells were transferred into TCRβ−/− animals and immunized with OVA and CFA. Representative plots are from the draining lymph node stained for Annexin V and 7AAD, spleen size in milligrams, number of live OT-II T cells in the spleen after red blood cell (RBC) lysis. n = 9/group compiled from two experiments. (E and F) Representative plots of cells from the draining lymph node of experiments in A stained for CD3+CD4+IL-17A+. (G and H) Representative plots of cells stained for Fas and GL-7 and gated on B220+ IgD− cells. n = 9/group. Data were compiled from two experiments. (I–L) Indicated OT-II CD4+ T cells were transferred into TCRβ−/− animals and subsequently immunized with OVA and CFA. Seven days postimmunization, spleen size in milligrams (I), number of OT-II T cells (J), and apoptotic cells (K and L) were analyzed. n = 4–5/group. Data were compiled from two independent experiments. (M) Splenic CD4+ T cells from B6 or FAS−/− mice immunized with OVA-overexpressing Erdr1 or the control vector were transferred into TCRβ−/− mice and immunized with OVA. T-cell numbers were determined in the draining lymph nodes. n = 3–6/group. *P < 0.05 and **P < 0.01 were determined by Student’s t test. ns, not significant.
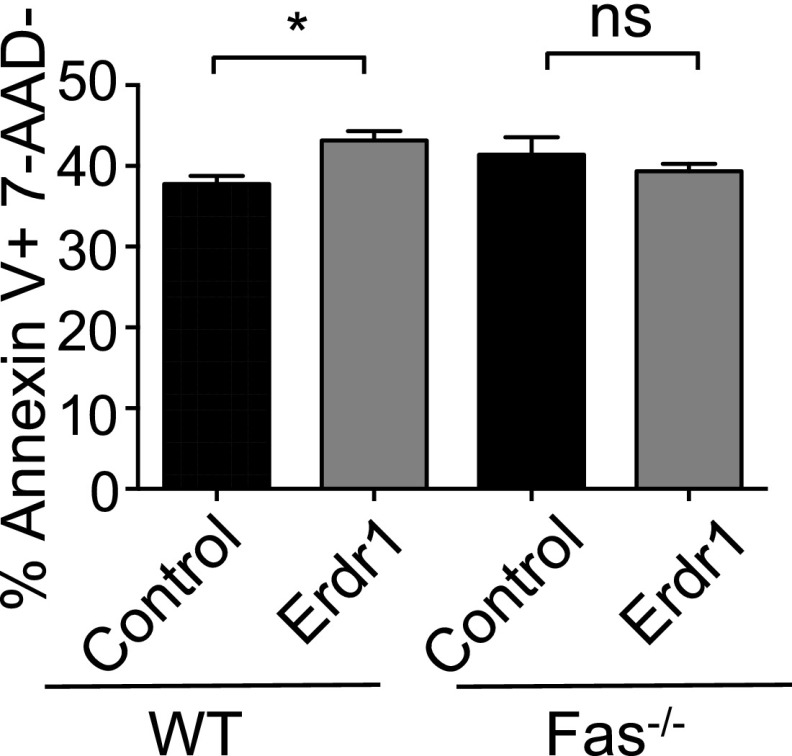
Conversely, the opposite phenotype is observed in animals receiving T cells that overexpress Erdr1. Indeed, elevated expression of Erdr1 led to enhanced T-cell apoptosis and significantly smaller, less cellular spleens and reduced OT-II T-cell numbers compared with control animals (Fig. 4 I and J and Fig. S7E). Notably, there was no difference in non-CD4 cellular apoptosis within the spleen, supporting an intrinsic role for Erdr1 (Fig. S7 F and G). To further investigate the dependence of Erdr1-mediated cell death on Fas signaling in vivo, Fas-deficient or WT animals were immunized with OVA, and these cells were used to manipulate Erdr1 expression. This was done to expand the number of antigen-specific T cells in our assay. Fas−/− or WT T cells were transferred into TCRβ−/− mice, and these animals were subsequently immunized to measure cell death of OVA-specific T cells. Mice receiving Erdr1-overexpressing cells had increased apoptosis of T cells and fewer numbers of T cells compared with controls (Fig. 4M and Fig. S8). However, when mice received T cells from Fas KO mice that overexpressed Erdr1, there were no differences in apoptosis or live T-cell counts in the spleen (Fig. 4M and Fig. S8). Thus, modulation of Erdr1 expression in vivo can impact the immune response through regulation of cellular survival, and this activity is dependent on induction of Fas.
Fig. S8.
Erdr1-induced cell death is dependent on Fas signaling. Percentage of preapoptotic T cells from mice receiving WT or Fas−/− T cells. *P < 0.05. ns; ns, not significant. Statistical significance was determined by Student’s t test.
Erdr1-Mediated Cellular Death Dictates the Severity of Neuro-Inflammatory Autoimmunity.
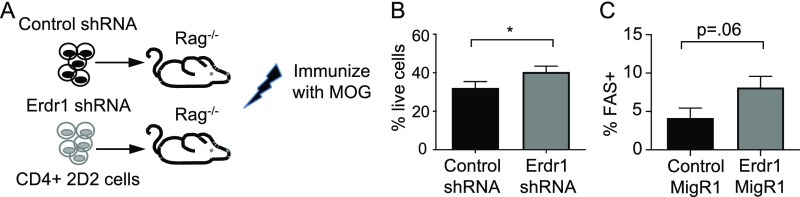
The microbiota is known to influence autoimmune diseases outside of the intestine. Indeed, germfree mice are reported to be resistant to Experimental Autoimmune Encephalomyelitis (EAE), an animal model of multiple sclerosis (2, 33). Although the mechanisms are not fully understood for this resistance, the loss of inflammatory T-cell lineages in germfree mice is thought to be involved in part. Our data demonstrate that microbiota-mediated suppression of Erdr1 promotes antigen-specific T-cell survival during immune responses; therefore, this could have critical implications for the development of autoimmune diseases. We therefore sought to test whether variation in Erdr1 expression levels could influence autoimmunity using an EAE model. Erdr1 expression levels were reduced by shRNA in 2D2 TCR transgenic T cells that recognize the myelin oligodendrocyte protein (MOG35–55) (Fig. S9A). Upon immunization with MOG35–55, animals develop ascending paralysis that mimics many aspects of human disease. The 2D2 T cells expressing either Erdr1 shRNA or control shRNA were adoptively transferred into Rag−/− animals and subsequently immunized with MOG35–55 to induce disease. Consistent with our findings, suppression of Erdr1 enhanced cellular survival of MOG-specific T cells during EAE (Fig. 5 A–C). Reduction of Erdr1 expression caused decreased T-cell apoptosis, increased live cells, and promoted an accumulation of MOG-specific T cells during disease (Fig. 5 A–C and Fig. S9B). Whereas control animals developed symptoms of EAE that ranged from mild tail paralysis to hind-limb paresis, animals with reduced Erdr1 expression in 2D2 T cells developed disease faster and had worsened clinical scores compared with controls (Fig. 5 D and E). Animals with Erdr1 shRNA-expressing T cells presented with complete forelimb paralysis and a ruffled, hunched appearance (Fig. 5E), indicating that differences in T-cell survival can have critical consequences on host health. Consistent with worsened symptoms of EAE and enhanced T-cell survival, there is a twofold increase in infiltrating cells within the brains of animals (Fig. 5F) and greater inflammatory Th1 cells within the CNS (Fig. 5 G and H). Conversely, Erdr1 overexpression in MOG35–55–specific T cells leads to fewer cells infiltrating into the brain (Fig. 5I). Moreover, Erdr1 overexpression results in an increased expression of Fas on T cells, corroborating our findings that overexpression of Erdr1 leads to up-regulation of programmed cell death through Fas (Fig. 5J and Fig. S9C). Importantly, animals with elevated Erdr1 developed a milder form of disease that extended to partial loss of their hind limbs, whereas control animals experienced complete loss of their hind limbs and showed signs of forelimb paralysis (Fig. 5 K and L). Collectively, these findings identify Erdr1 as an autocrine factor that controls T-cell survival and has critical functions during homeostasis, inflammation, and autoimmunity.
Fig. S9.
Erdr1 influences cell survival during EAE. (A) Schematic of experimental design shown in Fig. 5. Rag−/− animals were provided with Erdr1-overexpressing, Erdr1 shRNA, or control 2D2 cells and subsequently immunized with MOG + CFA. (B) Percentage of live (Annexin V−7AAD−) total cells in the spleens of animals after RBC lysis. (C) Fas-expressing cells from the spleens of animals from Fig. 5 H–L as measured by flow cytometry. Data are compiled from three independent experiments with a total of 9–10/group. *P < 0.05. Statistical significance was determined by Student’s t test.
Fig. 5.
Erdr1 effects on T-cell survival influence extraintestinal autoimmunity. (A–H) The indicated 2D2 T cells were transferred into Rag−/− animals and EAE-induced. (A) Representative flow cytometry plots of Annexin V and 7AAD. (B) Compiled plots from splenic cells. n = 7–9/group from three experiments. (C) Percentage of CD4+ T cells isolated from spleens. n = 9/group from three experiments. (D and E) Clinical symptoms of disease are shown. ***P < 0.005 by two-way ANOVA statistical test. n = 9/group. Data were compiled from three experiments. (F) Total number of infiltrating lymphocytes in the brains of indicated animals. n = 4–5/group representative of three experiments. (G and H) Representative plots of brain infiltrating IL-17 and IFN-γ from T cells. n = 4–5/group representative of three trials. (I–L) The indicated 2D2 T cells were transferred into Rag−/− animals, and EAE was induced. (I) Number of total lymphocytes isolated from the brains of animals. n = 9–10/group. (J) Representative plot of Fas staining from the brains of animals. Data were compiled from two experiments. (K and L) Animals were monitored for signs of paralysis for 21 d. n = 9–10/group representative of two experiments. *P < 0.05 by two-way ANOVA statistical test. Data were compiled from two experiments. *P < 0.05, **P < 0.01, and ***P < 0.005 by Student’s t test unless otherwise indicated.
SI Materials and Methods
Mice.
C57BL/6J, 2D2, Rag1−/−, and T-MyD88−/− mice were reared in the SPF facility at the University of Utah. Germfree C57BL/6J and BALB/c mice were reared in gnotobiotic chambers in the germfree facility at the University of Utah. IL-1−/−, IL-18−/− (B6.129P2-IL-18tm1Aki/J), MyD88−/−, OT-II [B6.Cg-Tg(TcraTcrb)425 Cbn/J], TCRβ−/− (B6.Tcrbtm/Mom/J), FAS−/− (B6.MRL-Faslpr/J), and TLR2−/− were purchased from Jackson Laboratory. Mice used in this study were 6–10 wk old except for the bone marrow transfer mice, which were 12 wk old. All experiments were performed in accordance with US regulations as well as the guidelines for animal use set forth by the University of Utah Institutional Animal Care and Use Committee.
Microarray.
Total RNA was collected from sort-purified splenic CD4+ T cells from age-matched SPF and germfree mice using the miRNeasy Mini Kit (Qiagen). Agilent 1 color gene expression array was used to measure gene expression and run by the University of Utah microarray core facility.
Retroviral Knockdown.
After 15 min of incubation, this mixture was added to 5 mL of DMEM in a tissue culture flask containing ∼5 × 106 293T cells. After 3 d of incubation, supernatant from cells was collected and passed through a 0.45-μm syringe tip. A total of 150–200 μL of retroviral containing media was then added to 96-well plates containing CD4+ splenic cells isolated from mice using a CD4+ T-cell Isolation Kit (Miltenyi) with plate-bound anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) (eBioscience) and spun at 2,500 × g for 90 min at 37 °C. DMEM (100 μL) was removed and replaced with complete DMEM containing anti-CD28, and plates were incubated for 3 d at 37°. After incubation, cells were sort-purified for GFP+CD3+ using the BD FACSAria.
Retroviral Overexpression.
Erdr1 was amplified, and the Erdr1 PCR product and MIGR1 plasmid were digested using the restriction enzymes EcoRI and Bgl2 (New England Biolabs) and ligated using T4 DNA Ligase (New England Biolabs). Plasmid was introduced into Mix & Go Competent Escherichia coli (Zymo Research) and plated on LB + ampicillin overnight. After 15 min of incubation, this mixture was added to 5 mL of DMEM in a tissue culture flask containing ∼5,000,000 293T cells. After 3 d of incubation, supernatant from cells was collected and passed through a 0.45-μm syringe tip. A total of 150–200 μL of retroviral-containing media was then added to 96-well plates containing 200 μL of RPMI containing CD4+ splenic cells isolated from mice using the CD4+ T-cell Isolation Kit (Miltenyi) with anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) (eBioscience) and spun at 2,500 × g for 90 min at 37 °C. One hundred microliters of RPMI was removed and replaced with complete RPMI containing anti-CD28, and plates were incubated for 3 d at 37°. After incubation, cells were sort-purified for GFP+CD3+ using FACSAria.
qPCR Analysis.
RNA was collected from cells using a Ribozol (Amresco) extraction method. RNA was treated with DNaseI (Sigma-Aldrich) per supplier protocol. cDNA was synthesized using qScript cDNA SuperMix (Quantigene). Amplification of cDNA was measured using Lightcycler 480 SYBR Green I Master (Roche) with the cycle threshold (CT) method and normalized to L32. Primers used were L32F 5-AAGCGAAACTGGCGGAAAC-3, L32R 5-TAACCGATGTTGGGCATCAG-3, ERDR1F 5-ATAGGATCCATGCCCACGGGACGC-3, ERDR1R 5-GGGGAATTCTTATTGAGGGGGGGCATTT-3, FASF 5-ATGCACACTCTGCGATGAAG-3, and FASR 5-CAGTGTTCACAGCCAGGAGA-3.
TLR2 Reporter Cell.
HEK Blue mTLR2 cells (hkb-mtlr2, InvivoGen) were used per supplier directions. Briefly, 50,000 HEK-mTLR2 cells were incubated in a 96-well round-bottom plate with serum collected from age-matched SPF and germfree mice in HEK Blue Detection media (InvivoGen) overnight at 37 °C. Sixteen hours after incubation, media absorbance was measured at 620 nm using a Synergy 2 Microplate Reader (BioTek). Media controls were subtracted from samples and normalized to SPF values.
Antibiotic and TLR Treatment.
SPF mice were treated with an antibiotic mixture of ampicillin, erythromycin, neomycin, and gentamycin (0.5 g/L) supplemented with Equal (4 g/L). For in vivo experiments, Pam3CSK4 (10 μg/mL) was added to the drinking water of germfree mice for 2 wk. In vitro CD4+ T cells were plated in 200 μL RPMI treated with Pam3CSK4, LPS, MDP, IL-33, FSL-1, Imiquimod, or Poly I:C (InvivoGen) (0.1 μg/mL).
B6 and FAS KO Ova Immunizations.
B6 mice or FAS deficient mice were immunized with 100 μL of albumin from chicken egg white (0.5 mg/mL) (Sigma Aldrich) and emulsified in 0.5 mg/mL CFA (Difco) subcutaneously. After 10 d, splenic CD4+ T cells were collected from immunized mice and transfected with Erdr1 overexpression vector. Control or overexpressing T cells were then transferred into TCRβ/- mice. After 1 d, TCRβ/- mice receiving Ova-reactive cells were immunized with 100 μL of albumin from chicken egg white (0.5 mg/mL) (Sigma Aldrich) and emulsified in 0.5 mg/mL CFA (Difco) subcutaneously. After 10 d, spleens and inguinal lymph nodes were collected from each animal and stained with CD4(RM45, Biolegend), CD3 (145-2C11, Biolegend), IL-17a (eBio17B7, eBioscience), IFN-γ (XMG1.2, Biolegend), FoxP3 (FJK-16s, eBioscience), IL-10 (JES5-16E3, Biolegend), B220 (RA3-6B2, Biolegend), CXCR5 (SPRCL5, Biolegend), IgD (11-26c.2a, Biolegend), IgG (Poly4053, Biolegend), Ki-67 (16A8, Biolegend), PD-1 (RMP1-30, Biolegend), FAS (JO2, BD Pharmagen) Annexin V (Biolegend), or 7-AAD (Biolegend) and analyzed using BD LSRFortessa. For quantification of OVA-specific IgG, serum from immunized TCRβ/- mice was collected, and plates were coated overnight at 4 °C with 4 µg/mL ovalbumin in 1 × PBS. Ova-specific IgG was measured the next day using mouse IgG quantification ELISA (Affymetrix).
Experimental Autoimmune Encephalomyelitis.
The 2D2 splenic CD4+ T cells from retroviral knockdown were retroorbitally injected into Rag1−/− mice in 50 μL of HBSS. The next day, mice were injected subcutaneously with 200 μL with 100 μg/mL of MOG35–55 peptide (Difco) emulsified in CFA. Mice were then injected with 200 ng of pertussis toxin on days 0 and 1. Mice were next scored daily as follows: 0, no symptoms; 0.5, partially limp tail; 1, totally limp tail; 1.5, partial hind limb paresis; 2, hind-limb paralysis; 2.5, partial forelimb paresis; 3, complete forelimb paresis; and 4, death. Spleen, inguinal lymph node, and brain were collected from mice at the end of experimental time points and stained with CD4 (RM45, Biolegend), CD3 (145-2C11, Biolegend), IL-17A (eBio17B7, eBioscience), IFN-γ (XMG1.2, Biolegend), FoxP3 (FJK-16s, eBioscience), IL-10, (JES5-16E3, Biolegend), Annexin V, or 7-AAD (Biolegend) and analyzed using BD LSRFortessa.
Recombinant ERDR1.
Full-length ERDR1 was introduced into the pGEX-2T GST Expression Vector (GE Healthcare) and purified. Briefly, ERDR1 was PCR-amplified. ERDR1 PCR product was gel-purified using a QIAquick gel extraction kit (Qiagen). ERDR1 and pGEX-2T GST were cut using restriction enzymes EcoRI and XbaI (New England Labs), ligated with T4 DNA ligase (New England Biolabs), introduced into Mix & Go Competent E. coli (Zymo Research), and plated on LB + ampicillin overnight. After confirming successful Erdr1 cloning, E. coli was grown in 500 mL LB + ampicillin broth to OD0.45. IPTG (100 mM) was added and allowed to incubate for 2.5 h. Bacteria were collected and sonicated with complete protease inhibitor tablets (Roche). Sonicate was then incubated with Glutathione Sepharose 4B (GE Healthcare) after washing beads were incubated with 10 units of thrombin (GE Healthcare). Sonicate + thrombin was then incubated with benzamidine Sepharose (GE Healthcare) to remove thrombin. Beads were spun-down and supernatant was collected. Purity was assessed using SDS/PAGE and RAPIDStain (G Biosciences) to view the ∼17-kDA product. Splenic CD4+ T cells from B6 mice were collected and treated with 20 ng of rErdr1 for 48 h in 200 μL of RPMI with anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) at 37 °C.
CD4+ T-Cell Lineage Skewing.
Isolated splenic CD4+ T cells were collected and plated with anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) (eBioscience). Th17-skewing conditions were 2 μg/mL anti–IL-4 (eBioscience), 2 μg/mL anti–IFN-γ (eBioscience), 20 ng/mL IL-6 (eBioscience), 5 ng/mL TGF-β (eBioscience), and 20 ng/mL IL-2 (eBioscience). Treg-skewing conditions were 5 ng/mL TGF-β (eBioscience) and 20 ng/mL IL-2 (eBioscience).
Erdr1 Antibody.
Erdr1 antibody was generated using the 36–51 amino acids of ERDR1 (N-RAPRPPRHTRHTRHTR-C) generated at the University of Utah Peptide Core. This peptide was conjugated to KLH and used to immunize rabbits (Harlan Laboratories). Rabbits were boosted with ERDR1 peptide three times and bled. IgG was purified using Protein G purification to a final concentration of 3.27 mg/mL.
Western Blots.
Cell lysate was collected from cells lysed in Nonidet P-40 cell lysis buffer with protease inhibitors (Sigma-Aldrich). Protein samples were run on 10.5% SDS/PAGE gels and transferred to activated PVDF membrane. Membrane was incubated overnight in anti-ERDR1 (Harlan Laboratories) at 1:200 in 2.5% nonfat milk. Membrane was then incubated in anti-rabbit IgG HRP (Promega) at 1:5,000 for 1 h in 5% nonfat milk. Membrane was imaged using LumiGLO Chemiluminescent Substrate (KPL) using the ChemiDoc Imaging System (Bio-Rad) and analyzed with Image Lab Software (Bio-Rad). For HA-tagged Erdr1, Erdr1 was inserted into the pCMV-HA vector (BD Biosciences/Clontech) using BglII and XhoI to create a fusion of Erdr1 to HA on the N terminus. HA-Erdr1 was cotransfected with the Erdr1 shRNA construct into 293T cells to determine the level of knockdown for Erdr1. Equal amounts of lysates as determined by Bradford assay were run on an SDS/PAGE gel, and membrane was probed with an anti-HA antibody.
Human T Cell.
Whole blood was collected from healthy donors. CD4+ T cells were negatively selected from peripheral blood mononuclear cells and plated for 24 h in the absence of stimulation or with anti-CD3/CD28, or Pam3CsK, or Pam2CsK. RNA was collected from cells using a Ribozol (Amresco) extraction method and treated with DNaseI (Sigma-Aldrich) per supplier protocol. cDNA was synthesized using qScript cDNA SuperMix (Quantigene). Amplification of cDNA was measured using Lightcycler 480 SYBR Green I Master (Roche) with the CT method and normalized to L32 or GAPDH for human samples. Primers used were hGAPF 5-GGGCTCTCCAGAACATCATCC-3, hGAPR 5-GTCCACCACTGACAC GTTGG-3, ERDR1F 5-ATAGGATCCATGCCCACGGGACGC-3, ERDR1R 5-GGGGAATTCTTATTGAGGGG GGGCATTT-3, L32F 5-AAGCGAAACTGGCGGAAAC-3, and L32R 5-TAACCGATGTTGGGCATCAG-3.
T-Cell Competition.
CD4+CD3+ splenic T cells from CD45.1 or CD45.2 mice were retrovirally infected with Erdr1 overexpression as described above. GFP+CD4+CD3+ T cells were then sorted out and plated in equal numbers in 200 μL of RPMI with anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) (eBioscience) and incubated at 37 °C. Cells were stained with anti-CD45.1 (Tonbo Biosciences) and CD45.2 (Tonbo Biosciences) and analyzed using BD LSRFortessa.
RNA-Isolation and RNA-Sequencing of Purified T Cells.
mRNA was collected from 150,000 FACS sort-purified splenic CD4+ T cells using the Qiagen miRNeasy Mini Kit (Qiagen) following kit instructions. mRNA was prepared following quality control via an Illumina TruSeq Stranded RNA Sample Prep with RiboZero treatment (human, mouse, rat, etc.) and analyzed using Illumina HiSeq Sequencing.
Caspase Inhibitor Splenic CD4+ T cells were collected via negative magnetic selection (Miltenyi) and plated in equal numbers in 200 μL of RPMI with anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL) (eBioscience) and incubated at 37 °C with Caspase-3 Inhibitor Z-DEVD-FMK (100 μM) (R&D Systems) or Caspase-1/ICE Inhibitor Z-WEHD-FMK (100 μM) (R&D Systems) for 96 h.
Cell Culture.
D.10 mouse T cells were transfected with ERDR1 in the MIGR1 plasmid as previously mentioned (Addgene). After incubation, cells were sort-purified for GFP+CD3+CD4+ using FACSAria. Cells were then cultured in complete RPMI. Cells were incubated with PBS, Caspase-3 Inhibitor Z-DEVD-FMK (100 μM) (R&D Systems), Fas antibody (GeneTex), or RANKL antibody (eBioscience).
OT-II Immunizations.
OT-II splenic CD4+ T cells from overexpression or knockdown experiments were retroorbitally injected into TCRβ/- mice in 50 μL of HBSS. The next day, mice were immunized with 100 μL of OVA 323–339 peptide (0.5 mg/mL) (InvivoGen) emulsified in (0.5 mg/mL) CFA (Difco) subcutaneously. After 7 d, spleens and inguinal lymph nodes were collected from each animal and stained with CD4 (RM45, Biolegend), CD3 (145-2C11, Biolegend), IL-17a (eBio17B7, eBioscience), IFN-γ (XMG1.2, Biolegend), FoxP3 (FJK-16s, eBioscience), IL-10 (JES5-16E3, Biolegend), B220 (RA3-6B2, Biolegend), CXCR5 (SPRCL5, Biolegend), IgD (11-26c.2a, Biolegend), IgG (Poly4053, Biolegend), Ki-67 (16A8, Biolegend), PD-1 (RMP1-30, Biolegend), FAS (JO2, BD Pharmagen), Annexin V (Biolegend), or 7-AAD (Biolegend) and analyzed using BD LSRFortessa.
Discussion
Survival of antigen-specific T-cell populations is necessary for appropriate inflammation to control pathogens, whereas induction of apoptosis in self-reactive T cells is needed to prevent autoimmunity. Thus, regulation of T-cell survival is a cardinal feature of healthy immunity, making the identification of factors that function to control apoptosis important therapeutic targets. The microbiota is emerging as a critical component of T-cell maturation and function. The complex interactions that take place between commensal microbes and host cells are only beginning to be defined. Many important immune genes and processes have been discovered through the study of how pathogens subvert immunity to cause disease, yet little is known about the mechanisms by which commensal bacteria influence homeostasis of host tissues. We therefore hypothesized that studying how commensal bacteria can influence host immunity might result in identification of unappreciated pathways of immune system regulation. Indeed, this strategy has led us to study the role of a secreted autocrine factor, Erdr1, which we show functions within T cells to regulate apoptosis.
It remains unclear which signals function to maintain Erdr1 levels; however, we demonstrate that suppression is dependent on MyD88. Microbial molecules that activate TLR2 in particular appear to be relevant, although it is likely that other pathways exist. Even potent bacterial molecules, such as LPS or muramyl dipeptide, did not repress Erdr1 in T cells, despite reported expression of these receptors on T cells (34). NF-κB has long been known to promote T-cell survival and activation. Because most MyD88-dependent signals activate an NF-κB–dependent program, it is likely that Erdr1 suppression is dependent on NF-κB signaling. Given the complexity and multiple pathways leading to activation of NF-κB, we did not evaluate this within this study. Future experiments should be aimed at addressing a role for NF-κB signaling, as NF-κB could be acting to induce an intermediate factor that represses Erdr1 or this could be an example of how NF-κB functions as a transcriptional repressor (35). TLR2 is able to pair with other receptors including TLR1 and -6 that increase the array of agonists that can bind to TLR2. Thus, multiple molecules from the microbiota might function to stimulate the immune system through TLR2.
Host–microbiota interactions are often thought to have beneficial consequences for the host. Regulation of T-cell death through suppression of Erdr1 by microbial products would promote T-cell survival and enhance host fitness by poising the immune system to fend off pathogens. Supporting this, germfree animals, which have elevated levels of Erdr1 and reduced T cells, are susceptible to a myriad of pathogenic infections (36, 37). In contrast, excessive reduction of Erdr1 may have pathological consequences by promoting the survival of auto-reactive T lymphocytes and consequently setting the stage for autoimmunity. A role for Erdr1 in immune cells of the intestine should be examined because Erdr1 levels are impacted by the presence of microbial products. Given that the intestine has a greater diversity and increased abundance of microbial molecules, it is likely that Erdr1 is highly relevant within gut resident immune cells and might be differentially expressed in distinct T-cell types, including CD8 T cells, that were not examined within this study. Additionally, differences in sensitivity to microbial signaling may exist in various T-cell populations of the gut through differential TLR expression, making the study of Erdr1 within select T-cell subsets an important future direction. Collectively, our results suggest that regulation of Erdr1 by the microbiota could provide flexibility to fine-tune T-cell responses against various antigens, whether self or foreign, and to poise T cells for future insults and highlight the extensive cross-talk that has evolved between commensal microorganisms and host to promote homeostasis.
Materials and Methods
All experiments were performed in accordance with US regulations as well as the guidelines for animal use set forth by the University of Utah Institutional Animal Care and Use Committee. Retroviral knockdown was performed using a Murine Stem Cell Virus backbone. EAE was induced by injection of MOG35–55 peptide (Difco) emulsified in Complete Freund’s Adjuvant. Two-tailed Student’s t test or two-way ANOVA statistical tests were used where indicated (GraphPad Software).
Acknowledgments
We thank the Flow Cytometry Core and James Marvin for assistance with flow sorting necessary throughout these studies. The Flow Cytometry Core is supported by NIH National Center for Research Resource Grant 1S10RR026802-01. Some of the germfree mice used in this publication were provided by the University of North Carolina’s Gnotobiotic Facility (Grants 5-P39-DK034987 and 5-P40-OD010995). C.P. is supported by a T32 Fellowship (AI-055434). R.S. was supported by T32 Training Grant GM007464. This project was also supported by National Institute of Allergy and Infectious Diseases Grant R21 AI109122, the Pew Scholars Program, NSF Career Award IOS-1253278, and a Packard Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619336114/-/DCSupplemental.
References
- 1.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Dobber R, Hertogh-Huijbregts A, Rozing J, Bottomly K, Nagelkerken L. The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Dev Immunol. 1992;2:141–150. doi: 10.1155/1992/57057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann S, Krueger A, Kirchhoff S, Krammer PH. Regulation of T cell apoptosis during the immune response. Curr Mol Med. 2002;2:257–272. doi: 10.2174/1566524024605671. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Hartig H, Dzhagalov I, Draper D, He YW. The role of apoptosis in the development and function of T lymphocytes. Cell Res. 2005;15:749–769. doi: 10.1038/sj.cr.7290345. [DOI] [PubMed] [Google Scholar]
- 8.Sturm A, de Souza HS, Fiocchi C. Mucosal T cell proliferation and apoptosis in inflammatory bowel disease. Curr Drug Targets. 2008;9:381–387. doi: 10.2174/138945008784221198. [DOI] [PubMed] [Google Scholar]
- 9.Tait SW, Ichim G, Green DR. Die another way: Non-apoptotic mechanisms of cell death. J Cell Sci. 2014;127:2135–2144. doi: 10.1242/jcs.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall DM, McCormick BA. Bacterial secreted effectors and caspase-3 interactions. Cell Microbiol. 2014;16:1746–1756. doi: 10.1111/cmi.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dörmer P, Spitzer E, Frankenberger M, Kremmer E. Erythroid differentiation regulator (EDR), a novel, highly conserved factor I. Induction of haemoglobin synthesis in erythroleukaemic cells. Cytokine. 2004;26:231–242. doi: 10.1016/j.cyto.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Jung MK, et al. Erythroid differentiation regulator 1, an interleukin 18-regulated gene, acts as a metastasis suppressor in melanoma. J Invest Dermatol. 2011;131:2096–2104. doi: 10.1038/jid.2011.170. [DOI] [PubMed] [Google Scholar]
- 13.Kim HJ, et al. Erythroid differentiation regulator 1 (Erdr1) is a proapototic factor in human keratinocytes. Exp Dermatol. 2011;20:920–925. doi: 10.1111/j.1600-0625.2011.01354.x. [DOI] [PubMed] [Google Scholar]
- 14.Jung MK, et al. Recombinant Erdr1 suppresses the migration and invasion ability of human gastric cancer cells, SNU-216, through the JNK pathway. Immunol Lett. 2013;150:145–51. doi: 10.1016/j.imlet.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Dörmer P, Spitzer E, Möller W. EDR is a stress-related survival factor from stroma and other tissues acting on early haematopoietic progenitors (E-Mix) Cytokine. 2004;27:47–57. doi: 10.1016/j.cyto.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schenten D, et al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity. 2014;40:78–90. doi: 10.1016/j.immuni.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlinson V, et al. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010;1:e29. doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 26.Izawa T, et al. Crosstalk between RANKL and Fas signaling in dendritic cells controls immune tolerance. Blood. 2007;110:242–250. doi: 10.1182/blood-2006-11-059980. [DOI] [PubMed] [Google Scholar]
- 27.Theill LE, Boyle WJ, Penninger JM. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol. 2002;20:795–823. doi: 10.1146/annurev.immunol.20.100301.064753. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, et al. RANKL regulates Fas expression and Fas-mediated apoptosis in osteoclasts. J Bone Miner Res. 2005;20:107–116. doi: 10.1359/JBMR.041022. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZX, Yang L, Young KJ, DuTemple B, Zhang L. Identification of a previously unknown antigen-specific regulatory T cell and its mechanism of suppression. Nat Med. 2000;6:782–789. doi: 10.1038/77513. [DOI] [PubMed] [Google Scholar]
- 30.Taylor RC, Cullen SP, Martin SJ. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 31.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimonkevitz R, Colon S, Kappler JW, Marrack P, Grey HM. Antigen recognition by H-2-restricted T cells. II. A tryptic ovalbumin peptide that substitutes for processed antigen. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- 33.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 34.Zanin-Zhorov A, et al. Cutting edge: T cells respond to lipopolysaccharide innately via TLR4 signaling. J Immunol. 2007;179:41–44. doi: 10.4049/jimmunol.179.1.41. [DOI] [PubMed] [Google Scholar]
- 35.Datta De D, Datta A, Bhattacharjya S, Roychoudhury S. NF-kappaB mediated transcriptional repression of acid modifying hormone gastrin. PLoS One. 2013;8:e73409. doi: 10.1371/journal.pone.0073409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Wardwell LH, Huttenhower C, Garrett WS. Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr Infect Dis Rep. 2011;13:28–34. doi: 10.1007/s11908-010-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]