Tzul et al. (1) report different unfolding rates and similar folding rates for a number of thioredoxins. The authors interpret this result as evidence of the principle of minimal frustration. Their study includes several resurrected Precambrian thioredoxins that we have previously prepared and characterized (2–5).
We agree that the principle of minimal frustration is essential to understand protein evolution. However, approximate folding-rate invariance is easily explained without invoking this principle. Thioredoxin kinetic stability relies on a transition state that is substantially unstructured (5, 6). Therefore, mutations that changed unfolding rates to tune kinetic stability during evolution likely had much less effect on folding rates, as implied by the well-known principles of ϕ-value analysis (7).
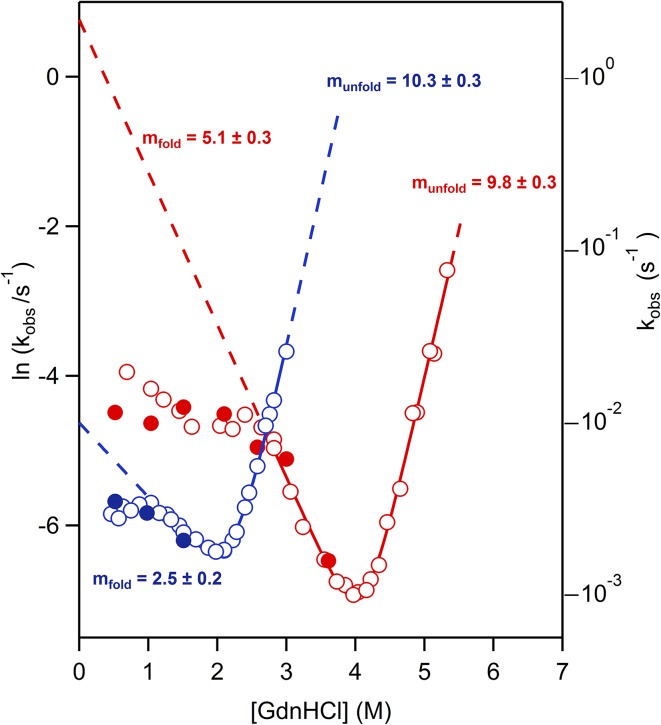
Moreover, our experimental results are not consistent with folding-rate invariance being a general feature of thioredoxins. Fig. 1 shows folding–unfolding rates for the modern Escherichia coli thioredoxin and a resurrected Precambrian thioredoxin. The unfolding of the ancestral protein is ∼three orders-of-magnitude slower than the unfolding of the modern protein, indicating enhanced kinetic stability. However, in clear disagreement with the results reported in Tzul et al. (1) for the very same proteins, we find the following two features. (i) There are deviations from linearity (rollovers) in the folding branches at low denaturant concentrations. “Rollovers” are commonly attributed to proline isomerization and the presence of significantly populated intermediate states (8). Such kinetic complexities reflect ruggedness of the folding landscape and have been previously reported to occur for thioredoxins (9). (ii) There is a faster folding rate for the ancestral protein. The ancestral versus modern folding-rate enhancement extrapolates to ∼two orders-of-magnitude at low denaturant concentrations and it is linked to a higher slope of the folding branch (the folding m value). This finding suggests a role for the residual structure of the unfolded state, because the unfolding slope is unchanged. Substantial folding-rate enhancement is also observed in the rollover region. The intriguing possibility arises that the higher folding rate for the ancestral protein is an adaptation to inefficient Precambrian folding chaperones.
Fig. 1.
Folding–unfolding rates for the modern E. coli thioredoxin (blue) and the resurrected thioredoxin corresponding to the last common ancestor of the cyanobacterial, Deinococcus and Thermus groups (LPBCA thioredoxin; red). The proteins were purified, as we have previously described (2–5). Rate-constant values were determined at pH 7 (Hepes buffer 50 mM) from the time-dependence of the protein fluorescence (open datapoints), following protocols we have previously described and used for thioredoxins (5, 6). We also followed kinetics using interrupted refolding with double-jump unfolding assays (closed datapoints), a methodology that we have previously described (10) and that provides a direct determination of amount of native protein. The protein concentrations used in interrupted refolding experiments are ∼10-fold higher than those used in fluorescence kinetic experiments. The agreement between the results obtained with the two methodologies therefore supports that our data are not distorted by protein association. Multiexponential kinetic profiles were observed at denaturant concentrations corresponding to the rollover regions and only data corresponding to the slower phase (i.e., the phase leading to the native state) are shown in these cases. Monoexponential kinetic profiles were observed at denaturant concentrations outside the rollover regions. Rate constants derived from these monoexponential profiles were fitted using a two-state kinetic model. These fits are shown with continuous lines, whereas dashed lines represent their extrapolation outside the experimental range of monoexponential kinetics. Despite the uncertainties involved in such extrapolations, it is clear that the ancestral versus modern rate enhancement for the folding from the unfolded state approaches ∼two orders-of-magnitude under native conditions. Values of the kinetic denaturant m values derived from the fits are given in kJ·mol−1·M−1 alongside the corresponding folding and unfolding branches. GdnHCl, guanidinium hydrochloride.
Why are these two features not apparent in the data of Tzul et al. (1)? We obtained our kinetic data at pH 7 using guanidinium hydrochloride, whereas pH 2 was used in Tzul et al. Such an acidic pH is destabilizing, thus allowing a weaker denaturant (urea) to be used and bringing the unfolding rates to the stopped-flow (milliseconds) time scale. However, destabilizing conditions may buffer the experimental consequences of ruggedness in folding landscapes. In fact, it is well known that kinetic intermediates are more readily observed under strongly stabilizing conditions (8). Furthermore, extensive protonation of residues at acidic pH may distort the folding landscapes in a manner that is not physiologically and evolutionary relevant. Note that comparatively few organisms are acidophiles and that many acidophiles actually pump out protons to keep a neutral intracellular medium.
We conclude that the experiments at pH 2 reported by Tzul et al. (1) miss important features of ancestral thioredoxin folding and do not provide any clear evidence for the principle of minimal frustration in the evolution of folding landscapes.
Acknowledgments
Work in the authors’ laboratory is supported by European Fund of Local Development Funds and Grants BIO2015-66426-R from Ministry of Economy and Competitiveness/European Fund of Local Development (to J.M.S.-R.) and P09-CVI-5073 from the “Junta de Andalucía” (to B.I.-M.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Tzul FO, Vasilchuck D, Makhatadze GI. Evidence for the principle of minimal frustration in the evolution of protein folding landscapes. Proc Natl Acad Sci USA. 2017;114:E1627–E1632. doi: 10.1073/pnas.1613892114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Jimenez R, et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 2011;18:592–596. doi: 10.1038/nsmb.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingles-Prieto A, et al. Conservation of protein structure over four billion years. Structure. 2013;21:1690–1697. doi: 10.1016/j.str.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risso VA, et al. Mutational studies on resurrected ancestral proteins reveal conservation of site-specific amino acid preferences throughout evolutionary history. Mol Biol Evol. 2015;32:440–455. doi: 10.1093/molbev/msu312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero-Romero ML, et al. Selection for protein kinetic stability connects denaturation temperatures to organismal temperatures and provides clues to Archaean life. PLoS One. 2016;11:e0156657. doi: 10.1371/journal.pone.0156657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godoy-Ruiz R, et al. Natural selection for kinetic stability is a likely origin of correlations between mutational effects on protein energetics and frequencies of amino acid occurrences in sequence alignments. J Mol Biol. 2006;362:966–978. doi: 10.1016/j.jmb.2006.07.065. [DOI] [PubMed] [Google Scholar]
- 7.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 8.Matouschek A, Kellis JT, Jr, Serrano L, Bycroft M, Fersht AR. Transient folding intermediates characterized by protein engineering. Nature. 1990;346:440–445. doi: 10.1038/346440a0. [DOI] [PubMed] [Google Scholar]
- 9.Georgescu RE, Li JH, Goldberg ME, Tasayco ML, Chaffotte AF. Proline isomerization-independent accumulation of an early intermediate and heterogeneity of the folding pathways of a mixed α/β protein, Escherichia coli thioredoxin. Biochemistry. 1998;37:10286–10297. doi: 10.1021/bi9805083. [DOI] [PubMed] [Google Scholar]
- 10.Ibarra-Molero B, Sanchez-Ruiz JM. Are there equilibrium intermediate states in the urea-induced unfolding of hen egg-white lysozyme? Biochemistry. 1997;36:9616–9624. doi: 10.1021/bi9703305. [DOI] [PubMed] [Google Scholar]