Significance
Macrophages are immune cells equipped with multiple double-stranded RNA (dsRNA) sensors designed to detect viral infection and amplify innate antiviral immunity. However, many coronaviruses can infect and propagate in macrophages without activating dsRNA sensors. Here we present a function of murine coronavirus nonstructural protein 15 in preventing detection of viral dsRNA by host sensors. We show that coronaviruses expressing a mutant form of nonstructural protein 15 allow for activation of dsRNA sensors, resulting in an early induction of interferon, rapid apoptosis of macrophages, and a protective immune response in mice. Identifying the strategies used by viruses to evade detection provides us with new approaches for generating vaccines that elicit robust innate immune responses and protective immunity.
Keywords: coronavirus, nsp15, endoribonuclease, dsRNA, interferon
Abstract
Coronaviruses are positive-sense RNA viruses that generate double-stranded RNA (dsRNA) intermediates during replication, yet evade detection by host innate immune sensors. Here we report that coronavirus nonstructural protein 15 (nsp15), an endoribonuclease, is required for evasion of dsRNA sensors. We evaluated two independent nsp15 mutant mouse coronaviruses, designated N15m1 and N15m3, and found that these viruses replicated poorly and induced rapid cell death in mouse bone marrow-derived macrophages. Infection of macrophages with N15m1, which expresses an unstable nsp15, or N15m3, which expresses a catalysis-deficient nsp15, activated MDA5, PKR, and the OAS/RNase L system, resulting in an early, robust induction of type I IFN, PKR-mediated apoptosis, and RNA degradation. Immunofluorescence imaging of nsp15 mutant virus-infected macrophages revealed significant dispersal of dsRNA early during infection, whereas in WT virus-infected cells, the majority of the dsRNA was associated with replication complexes. The loss of nsp15 activity also resulted in greatly attenuated disease in mice and stimulated a protective immune response. Taken together, our findings demonstrate that coronavirus nsp15 is critical for evasion of host dsRNA sensors in macrophages and reveal that modulating nsp15 stability and activity is a strategy for generating live-attenuated vaccines.
Coronaviruses are a family of positive-sense RNA viruses that infect humans and animals and cause respiratory, gastrointestinal, or neurologic disease. Coronaviruses can emerge from animal reservoirs to cause significant epidemics in humans, as exemplified by the Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) outbreak in 2002–2003 and Middle East Respiratory Syndrome coronavirus (MERS-CoV), which was recognized as an emerging virus in 2012 (1, 2). A remarkable feature of many coronaviruses is their ability to infect macrophages and delay triggering of antiviral sensors that would otherwise activate production of type I IFN (IFN-α/β) (3–5). Coronaviruses encode multiple antagonists that likely impede or delay the activation of type I IFN and IFN-stimulated genes (ISGs), thereby contributing to pathogenesis (6). A recent report using SARS-CoV infection of mice documented that delayed IFN signaling contributes to disease (7). The goal of this study was to identify viral factors that contribute to the delay in the activation of IFN in response to coronavirus infection.
To investigate coronavirus antagonism of the early IFN response, we use mouse hepatitis virus strain A59 (MHV-A59). This model coronavirus replicates in multiple murine cell types, including macrophages, and can cause acute hepatitis or lethal encephalitis, depending on the site of injection. The viral genomic RNA comprises 32 kb. Two-thirds of the genome encodes a large replicase polyprotein, whereas the remainder of the genome codes for structural proteins and strain-specific accessory proteins (Fig. 1A). The replicase polyprotein is processed by viral proteases into 16 nonstructural proteins (nsps) that assemble in the host endoplasmic reticulum to generate convoluted membranes and double-membrane vesicles (DMVs), which are the sites of viral RNA synthesis (8, 9). Coronavirus RNA replication proceeds via the generation of a nested set of negative-strand RNAs that serve as templates for synthesis of mRNAs and new positive-strand genomes (10). Double-stranded RNA (dsRNA) intermediates, which are potent stimulators of cytoplasmic innate sensors, are produced during this process and associate with the DMVs (9). A potential function for coronavirus DMVs may be to sequester viral dsRNA away from host dsRNA sensors; however, it is unclear if DMVs alone are sufficient to prevent activation of the host innate immune response. Here we report that coronavirus nonstructural protein 15 (nsp15), a highly conserved nidovirus component with endoribonuclease activity, acts in conjunction with the viral replication complex to limit the exposure of viral dsRNA to host dsRNA sensors.
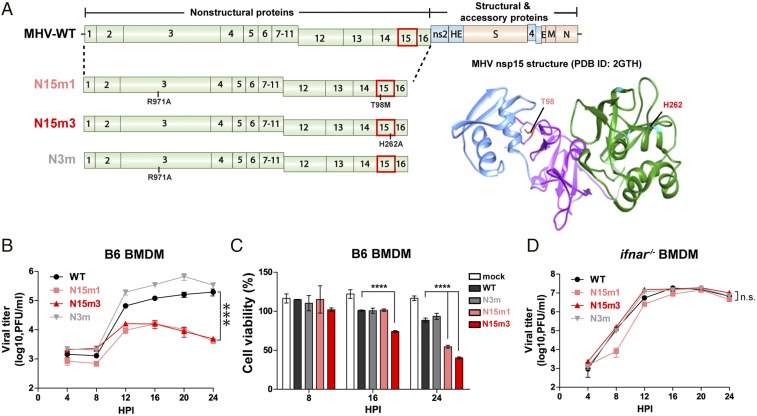
Fig. 1.
MHV nsp15 mutant viruses are attenuated for replication and induce rapid cell death in macrophages. (A) Schematic diagram of the MHV-A59 genome and the location of mutations in mutant viruses. (Right) Crystal structure of MHV nsp15. N domain (blue), M domain (purple), C domain (green), and residues (T98 and H262) are indicated (PDB ID code 2GTH). (B–D) Growth kinetics of viruses in (B) B6 or in (D) ifnar−/− BMDMs, and (C) the cell viability of infected B6 BMDMs. BMDMs were infected with WT or mutant MHV at an MOI of 0.1. Cell supernatants were collected for plaque assay, and the cells were lysed for CellTiter Glo assay at the indicated time points. Values were analyzed using a nonlinear regression test (B and D) or a two-way ANOVA test by time (C). ***P < 0.001; ****P < 0.0001; n.s., not significant. Data are representative of two to three independent experiments and presented as the mean ± SD.
Bioinformatic analysis revealed that nsp15 contains a domain with distant homology to cellular endoribonucleases. The nsp15 endoribonuclease, termed NendoU, is highly conserved among vertebrate nidoviruses (coronaviruses and arteriviruses) (11). Structural and biochemical studies revealed that the SARS-CoV and MHV nsp15, and the arterivirus ortholog nsp11, form oligomers to cleave RNA molecules with a preference for the 3′-ends of uridylates (12–16). The role of endoribonuclease activity in nidovirus replication and pathogenesis, however, is not well understood. In one study, researchers were unable to recover human coronavirus 229E encoding an endoribonuclease catalytic site mutant, and therefore concluded that nsp15 was essential for coronavirus replication (17). On the other hand, MHV encoding nsp15 catalytic site mutations replicated to reduced titers (∼1 log) in a fibroblast cell line (18), suggesting that nsp15 activity is important but not required for MHV replication. Similarly, mutagenesis studies of arterivirus nsp11 yielded a range of results: some mutations could not be recovered in viable virus, whereas others allowed recovery with a range of phenotypes (19, 20). Overexpression studies of coronavirus nsp15 (21) and arterivirus nsp11 (20, 22, 23) demonstrated antagonism of innate immune responses, but there was no direct evidence to suggest that these proteins counteract innate immunity in the context of viral infection. Here we report that nsp15 is not required for viral RNA synthesis per se, but acts to mediate evasion of host dsRNA sensors. Viruses that contain a mutation in nsp15, which either renders nsp15 unstable or disables endoribonuclease activity, stimulate MDA5-dependent IFN production and activate host dsRNA sensors. The nsp15 mutant viruses induce apoptotic cell death and exhibit reduced replication in macrophages. Importantly, we show that nsp15 is a key component of coronavirus pathogenesis, highlighted by loss of dissemination of nsp15 mutant viruses to target organs and an absence of pathology in mice. We show that nsp15 mutant viruses can elicit a protective immune response against subsequent challenge with WT virus.
Results
Coronaviruses Encoding Mutant Forms of nsp15 Exhibit Impaired Replication in Macrophages.
Here we characterize two independent nsp15 mutant viruses of MHV-A59, termed N15m1 and N15m3, and report that these viruses exhibited impaired viral replication and induced rapid cell death during infection of mouse bone marrow-derived macrophages (BMDMs) (Fig. 1). The parental WT MHV-A59 (termed MHV-WT) and all mutant viruses were generated using reverse genetics (24) and were subjected to deep sequencing to validate the genotypes. N15m3 contains an alanine substitution in the catalytic histidine (H262A) residue of nsp15, which was previously shown to inactivate endoribonuclease activity (17, 25). N15m1 contains a methionine substitution at a highly-conserved threonine residue (T98M) and an alanine substitution at arginine-971 (R971A) in nsp3 (Fig. 1A and Fig. S1A). We also generated a mutant virus with a single substitution in nsp3 (R971A), termed N3m, to distinguish the effect of the substitution in nsp3 from the substitution in nsp15 of N15m1 virus. To date, we have been unable to isolate a virus containing only the T98M substitution, which may indicate that additional substitutions are necessary to stabilize the T98M mutant virus.
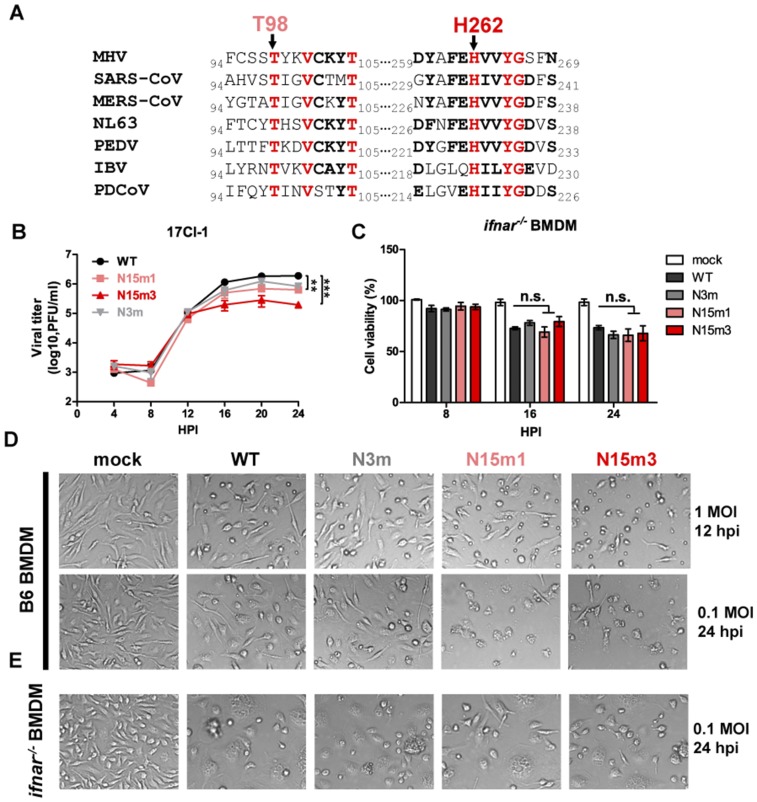
Fig. S1.
MHV nsp15 mutant viruses induce rapid cell death in macrophages. (A) Clustal W was used to align the complete amino acid sequence of nsp15 from the listed CoVs, with the region surrounding the highly conserved T98 and H262 depicted. (B) Growth kinetics in 17Cl-1 cells (MOI 0.1). Values were analyzed using a nonlinear regression test. **P < 0.01; ***P < 0.001. (C) ifnar−/− BMDMs were infected at an MOI of 0.1 and harvested at indicated times for CellTiter Glo assay; n.s., not significant. (D and E) Cytopathic effect was observed under bright-field microscopy (magnification, 100×), with rapid cell death in nsp15 mutant viruses infected B6 BMDM (D) and syncytia formation in all virus-infected ifnar−/− BMDMs (E). Data are representative of two to three independent experiments and error bars represent the mean ± SD.
MHV-WT and the three mutant viruses replicate efficiently in a murine fibroblast cell line, 17Cl-1 cells, indicating that WT nsp15 is not required for viral replication in fibroblasts (Fig. S1B), which is in agreement with a previous report describing nsp15 catalytic mutants (18). We report that infection of BMDMs generated from C57BL/6 mice (B6) with nsp15 mutant viruses resulted in limited production of progeny virus and more rapid cell death compared with WT virus infection (Fig. 1 B and C and Fig. S1D). This result is in contrast to the robust replication of MHV-WT and N3m, which replicate to high titers in BMDMs with minimal cell death as late as 24 h postinfection (hpi) (Fig. 1 B and C and Fig. S1D). Efficient replication of the nsp15 mutant viruses was restored in BMDMs generated from mice with a knockout of the type I IFN receptor (ifnar−/−) (Fig. 1D). In the absence of IFN signaling, all tested viruses induced similar cell death kinetics (Fig. S1C and Fig. S1E). These results implicate type I IFN signaling as a major contributor to the replication deficiency of nsp15 mutants.
MHV nsp15 Mutant Viruses Stimulate an Early and Robust Induction of Type I IFN in Macrophages.
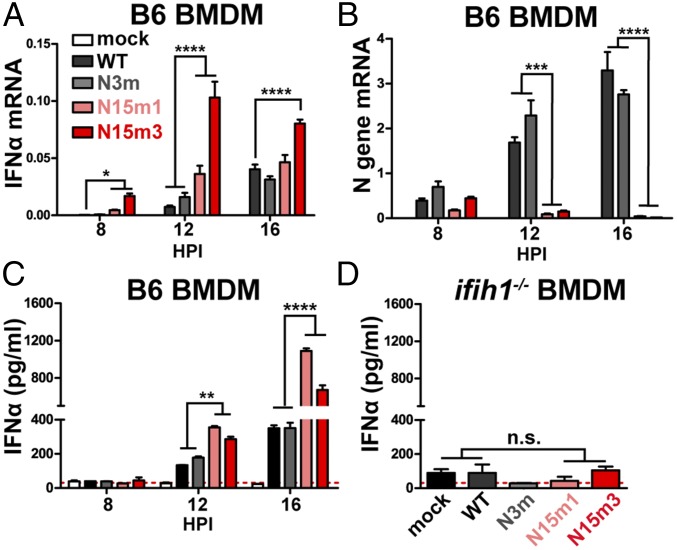
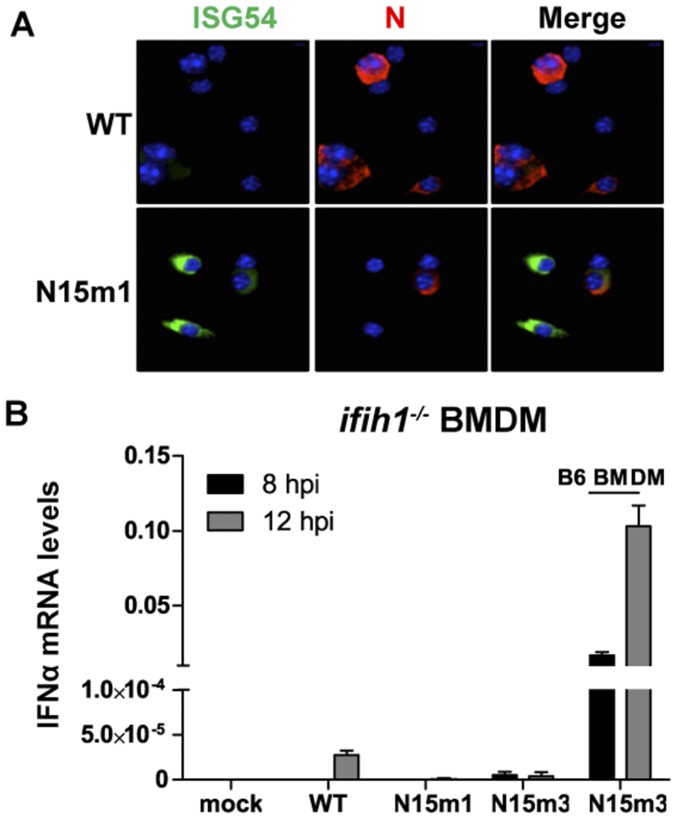
Given the importance of IFN signaling in controlling nsp15 mutant virus infection of macrophages, we hypothesize that WT nsp15 antagonizes the type I IFN system. To address this question, B6 BMDMs were infected with MHV-WT or mutant viruses and harvested at 8, 12, and 16 hpi to evaluate the level of IFN-αmRNA and protein. We found that the nsp15 mutant viruses stimulated an early and more robust induction of IFN-α compared with MHV-WT or N3m virus (Fig. 2). IFN-α mRNA was significantly elevated at 8 hpi in nsp15 mutant-infected cells, with WT or N3m virus requiring 12 hpi to reach similar levels (Fig. 2A). The detection of IFN-α mRNA in nsp15 mutant virus-infected cells peaked at 12 hpi and was slightly reduced at 16 hpi (Fig. 2A), corresponding to a reduction in the number of viable cells (Fig. 1C). We noted a significant reduction in the level of nucleocapsid (N) gene mRNA in nsp15 mutant virus-infected cells at 12 and 16 hpi (Fig. 2B), time points when IFN-α expression was significantly elevated (Fig. 2A). The concentration of IFN-α protein was significantly higher in cell supernatants obtained from nsp15 mutant-infected cells, compared with WT- or N3m-infected cells (Fig. 2C). Additionally, increased detection of ISG54 in N15m1-infected cells was observed using immunofluorescence (Fig. S2A). Consistent with a previous report (26), activation of IFN during MHV infection was dependent on production of MDA5 (encoded by the ifih1 gene), as BMDMs derived from ifih1−/− mice did not express IFN when infected by either WT or mutant viruses (Fig. 2D and Fig. S2B). Overall, these results demonstrate that nsp15 mutant viruses stimulate an early and robust type I IFN response in B6 BMDMs, indicating that nsp15 functions as a type I IFN antagonist during MHV infection.
Fig. 2.
Nsp15 mutant viruses trigger an early induction of type I IFN in macrophages. (A–C) B6 BMDMs were infected with WT or mutant MHV at an MOI of 0.1. At indicated time points, total RNA was extracted and analyzed for (A) the mRNA levels of IFN-α11 or (B) N gene by qPCR. The levels of mRNA relative to β-actin mRNA were expressed as 2-ΔCT [ΔCT = CT(gene of interest) − CT(β-actin)]. (C) The cell supernatant was collected for the detection of secreted IFN-α by quantitative ELISA. (D) ifih1−/− BMDMs were infected with WT or mutant MHV at an MOI of 0.1. At 16 hpi, the cell supernatant was collected for ELISA. Values were analyzed using a two-way ANOVA test by time in A–C or an unpaired t test in D. *P < 0.05; n.s., not significant. **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are representative of two to three independent experiments and presented as the mean ± SD. Red dashed line is limit of detection.
Fig. S2.
MHV nsp15 mutant viruses induce type I IFN in macrophages. (A) B6 BMDMs were infected with WT or N15m1 MHV at an MOI of 0.1. At 12 hpi, infected cells were fixed with 4% formaldehyde and stained with Hoechst 33342 (nucleus; blue), antinucleocapsid (N; red), and anti-ISG54 (green). Immunofluorescence was detected by confocal microscopy. (Magnification, 200×.) (B) ifih1−/− BMDMs were infected at an MOI of 0.1. IFN-α11 mRNA levels were determined by quantitative PCR. Values were relative to β-actin. Data are representative of two to three independent experiments and error bars represent the mean ± SD.
MHV nsp15 Mutant Viruses Trigger Rapid Apoptotic Cell Death in Macrophages.
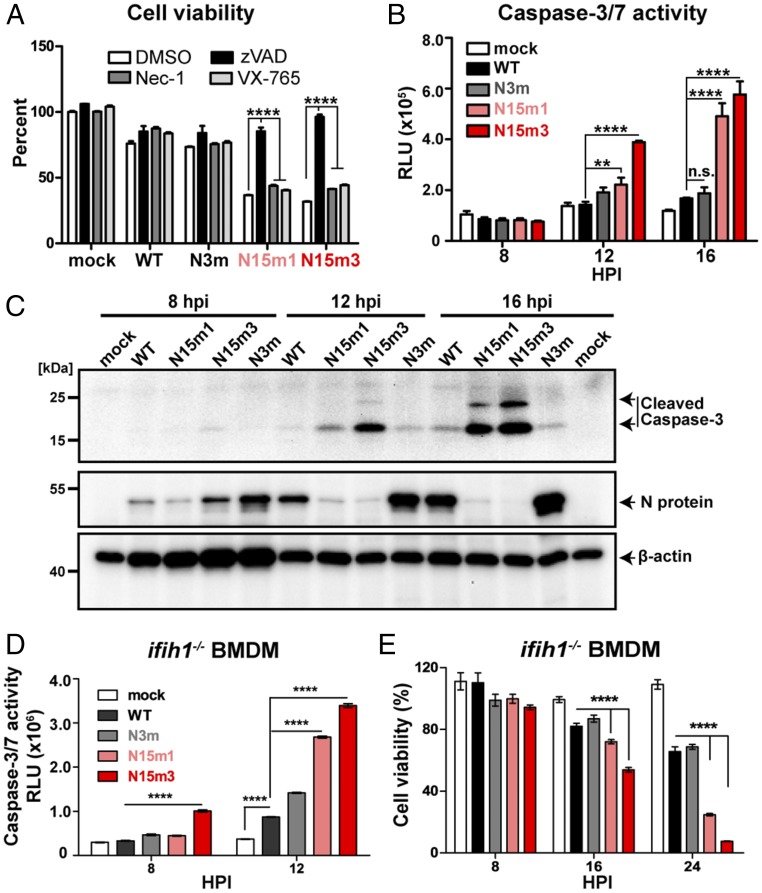
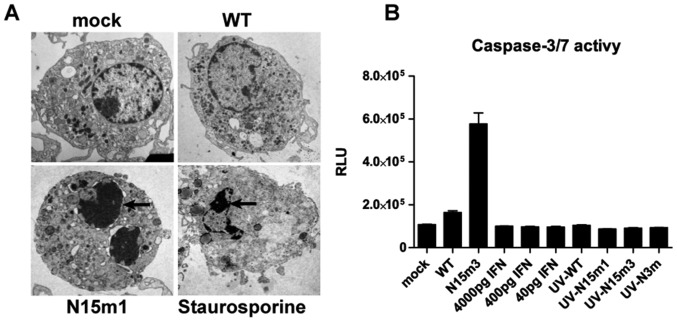
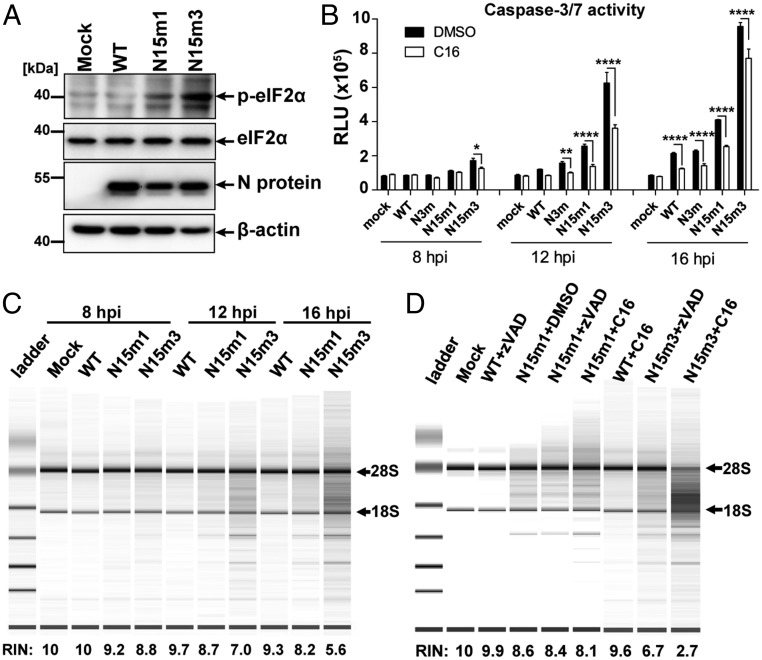
Because we observed rapid cell death in nsp15 mutant-infected macrophages (Fig. 1C and Fig. S1D), we investigated the cell death pathways triggered during viral replication. We first evaluated the effects of various inhibitors on virus-infected cells. The pan-caspase inhibitor zVAD (27) prevented virus-induced cell death (Fig. 3A). In contrast, neither Necrostatin-1 (Nec-1), a receptor interacting serine/threonine kinase 1 (RIPK1) inhibitor (28), nor caspase-1 inhibitor VX-765 (29), prevented virus-induced cell death (Fig. 3A). These data suggest that infection of BMDMs by nsp15 mutant viruses activates apoptotic cell death rather than RIPK1/RIPK3-dependent necroptosis or caspase-1–mediated pyroptosis (30, 31). However, we note that zVAD—a cysteine protease inhibitor—may also affect viral replication (32); therefore, we investigated other hallmarks of apoptotic cell death, including the activation of caspase-3/7. We observed enhanced caspase-3/7 activity in nsp15 mutant virus-infected BMDMs (Fig. 3B). Furthermore, increased levels of cleaved caspase-3 products were detected by Western blotting (Fig. 3C). Finally, condensed chromatin and nuclear fragmentation were observed by electron microscopy in nsp15 mutant-infected cells, but not in BMDMs infected by MHV-WT (Fig. S3A). Taken together, these data demonstrate that nsp15 mutant viruses trigger early apoptosis in macrophages.
Fig. 3.
MHV nsp15 mutant viruses induce early apoptosis in macrophages. (A) B6 BMDMs were infected with WT or mutant MHV at an MOI of 0.1 and subsequently treated with either DMSO, zVAD (20 μM), Nec-1 (25 μM), or VX-765 (20 μM). Cell viability was measured at 24 hpi by a CellTiter Glo assay. Results are reported relative to DMSO-treated mock cells and were analyzed using a two-way ANOVA test by virus. (B) B6 BMDMs were infected with WT or mutant MHV at an MOI of 0.1. At indicated time points, caspase-3/7 activity was determined by a Caspase-Glo 3/7 assay. Values are displayed in relative light units (RLU) and were analyzed using a two-way ANOVA test by time. (C) B6 BMDMs were infected with WT or mutant MHV at an MOI of 0.1. At indicated time points, cell lysates were collected for the detection of cleaved–caspase-3, N protein, and β-actin by Western blotting. (D and E) ifih1−/− BMDMs were infected at an MOI of 0.1. (D) Cell viability and (E) caspase-3/7 activity were evaluated. Values were analyzed using two-way ANOVA test by time. Data are representative of two to three independent experiments and presented as the mean ± SD in A and B. **P < 0.01; ****P < 0.0001; n.s., not significant.
Fig. S3.
N15m1 induces a rapid apoptotic cell death. (A) B6 BMDMs were treated with staurosporine (1 μM) or infected with WT or N15m1. Condensation of nuclear chromatin (black arrows) indicative of apoptosis was determined by electron microscopy at 16 hpi. (B) B6 BMDMs were infected or treated with IFN-α or UV-inactivated cell supernatant of infected-cell at 16 hpi. (Magnification, 4,000–6,000×.) Caspase-3/7 activity was measured at 16 h postinfection or posttreatment. Values were displayed in RLU. Data are representative of two to three independent experiments and presented as the mean ± SD.
To evaluate the possibility that the observed apoptotic cell death was induced by IFN-α production alone, we treated B6 BMDMs with various doses of IFN-α and evaluated caspase-3/7 activity. We found that IFN-α treatment did not induce caspase-3/7 activity (Fig. S3B). Furthermore, no elevated caspase-3/7 activity was observed in BMDMs treated with UV-inactivated supernatant, which contained replication-defective virus and IFN-α (Fig. S3B). These results indicate that active virus replication, and not IFN-α alone, is required for induction of apoptosis. Using a complementary approach, we evaluated virus-induced apoptosis in ifih1−/− BMDMs. These cells lack MDA5 expression and, as shown in Fig. 2D, do not induce IFN during viral infection. However, other dsRNA-sensing pathways remain intact in these cells. We found that infection of ifih1−/− BMDMs with nsp15 mutant viruses induced elevated levels of caspase-3/7, which promoted rapid cell death (Fig. 3 D and E). These results implicate a non-MDA5–dependent pathway as an additional mechanism in the induction of apoptosis. Taken together, these data indicate that nsp15 mutant virus infection can activate multiple independent dsRNA-sensing pathways to induce apoptotic cell death.
MHV nsp15 Mutant Viruses Activate Host dsRNA Sensors.
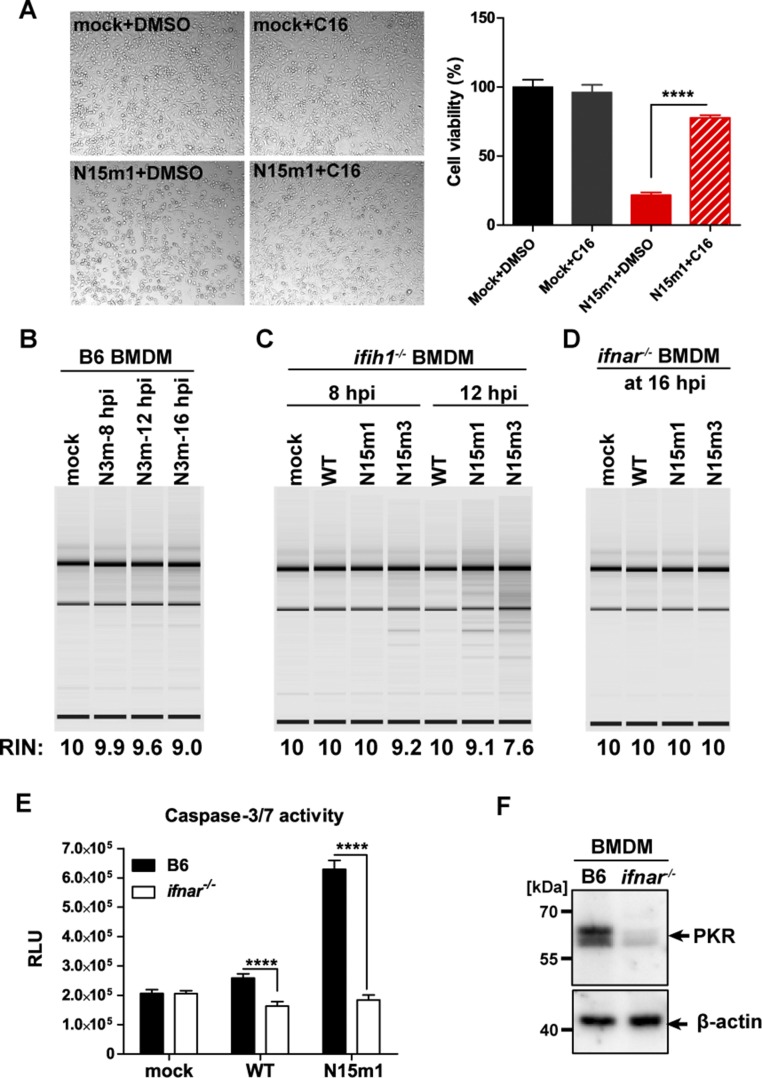
Because our data suggest that additional dsRNA-sensing pathways are activated in response to nsp15 mutant virus replication, we investigated other dsRNA-sensing pathways that have been linked to apoptotic cell death. To this end, we examined the protein kinase R (PKR) and the 2′-5′ oligoadenylate synthetase (OAS)/RNase L systems (33–37). First, we evaluated the level of phosphorylated eIF2α, an indicator of PKR activation (35, 37), during infection. Increased phosphorylation of eIF2α was observed in BMDMs infected with nsp15 mutant viruses compared with infection with WT virus (Fig. 4A). Addition of C16, a specific kinase inhibitor of PKR (38), significantly reduced the levels of caspase-3/7 activity (Fig. 4B) and prevented cell death (Fig. S4A). These data suggest that the PKR pathway is activated during MHV infection of BMDMs. We also assessed the degradation of RNA, which is an indicator of active 2′-5′ OAS/RNase L system signaling (39). Degradation of RNA was observed as early as 8 hpi in cells infected with the nsp15 mutant viruses, but not in cells infected by MHV-WT or N3m (Fig. 4C and Fig. S4B). Degradation of RNA was not inhibited by blocking apoptosis with either zVAD or C16 (Fig. 4D). This result indicated that activation of OAS/RNase L was independent of PKR activation. In addition, robust RNA degradation was observed in nsp15 mutant virus-infected ifih1−/− BMDMs (Fig. S4C), suggesting that the OAS/RNase L system was activated in the absence of IFN. Finally, neither RNA degradation (Fig. S4D) nor elevated caspase-3/7 activity (Fig. S4E) was observed in ifnar−/− BMDMs despite the fact that robust viral replication was present (Fig. 1D). This could be because of low basal expression of PKR (Fig. S4F) and OAS in ifnar−/− macrophages, the latter of which was previously reported (40). Taken together, these data indicate that nsp15 mutant viruses activate multiple independent host dsRNA-sensing systems in macrophages, including MDA5, PKR, and OAS/RNase L. In the absence of functional nsp15, the collective stimulation of these pathways by viral dsRNA results in robust type I IFN induction, apoptotic cell death, and severely reduced viral titer.
Fig. 4.
Infection of MHV nsp15 mutant viruses activates host dsRNA sensors. (A) B6 BMDMs were infected with WT or mutant MHV at an MOI of 1. At 8 hpi, cells were lysed and 20 μg cell lysate was evaluated for phospho-eIF2α, eIF2α, viral N protein, and β-actin by Western blotting. (B) PKR inhibitor C16 blocks virus-induced apoptosis in macrophages. B6 BMDMs were infected with WT or mutant MHV (MOI of 0.1) and subsequently treated with the PKR inhibitor C16 (1 μM). Cells were collected and evaluated for caspase-3/7 activity at indicated time points. Values are displayed in RLU and presented as the mean ± SD, *P < 0.05; **P < 0.01; ****P < 0.0001, unpaired t test. (C) RNA degradation pattern of 200 ng total RNA extracted from mock or infected B6 BMDMs (MOI 0.1) evaluated using a bioanalyzer. (D) RNA degradation pattern of RNAs from mock or infected B6 BMDMs (MOI 0.1) treated with C16 inhibitor (1 μM) or zVAD (20 μM) at 16 hpi. The RNA integrity numbers (RIN) and the positions of 28S and 18S ribosomal RNAs are shown to the bottom and the right of the image, respectively. Data are representative of two to three independent experiments.
Fig. S4.
MHV nsp15 mutant viruses activate dsRNA sensors in macrophages. (A) B6 BMDMs were infected with WT or N15m1 at an MOI of 0.1. Cytopathic effect (Left) (magnification, 100×) and cell viability was evaluated at 18 hpi. ****P < 0.0001, unpaired t test. (B–D) BMDM were infected at an MOI of 0.1. RNA degradation was evaluated in virus-infected (B) B6 BMDMs, (C) ifih1−/− BMDMs, and (D) ifnar−/− BMDMs. (E) B6 or ifnar−/− BMDMs were infected with WT or N15m1 at an MOI of 0.1. Caspase-3/7 activity was measured at 18 hpi by a Caspase-Glo 3/7 assay. ****P < 0.0001, unpaired t test. (F) Protein level of PKR in uninfected BMDMs by Western blotting. Data are representative of two to three independent experiments and presented as the mean ± SD.
T98M Mutation Destabilizes nsp15 Protein and Impairs Endoribonuclease Activity.
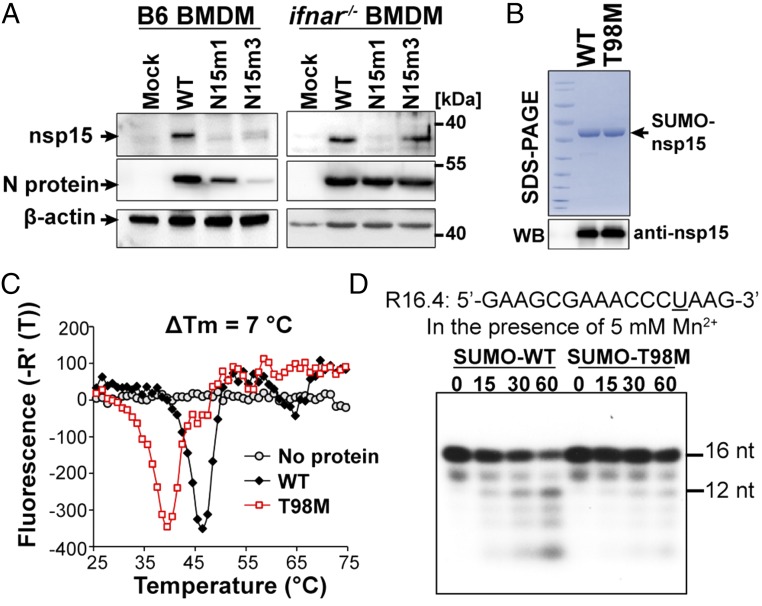
As shown above, the N15m1 and N15m3 viruses repeatedly exhibited similar phenotypes, despite encoding different mutations in nsp15. Because the N15m1 mutant virus is unique to this study, we investigated the effect of the T98M mutation on the nsp15 protein. The T98M mutation resides at the interface between the N-terminal domain and middle domain of nsp15 (Fig. 1A, Right). The endoribonuclease activity of nsp15 requires the formation of stable oligomers, which may be destabilized by the T98M mutation (25). Western blot analysis revealed that levels of nsp15 were significantly reduced in both N15m1- and N15m3-infected cells compared with WT-infected cells (Fig. 5A, Left). It is possible that this phenotype could be because of the limited viral replication of nsp15 mutant viruses observed in B6 BMDMs or reduced detection of nsp15 mutant proteins by antiserum. However, compared with N15m3, the level of N15m1 nsp15 protein remained low during infection of ifnar−/− BMDMs, despite similar N protein levels (Fig. 5A, Right) and growth kinetics for all viruses tested (Fig. 1D). We also note that detection of nsp15 protein by antiserum was not affected by the T98M mutation, as both recombinant WT and T98M nsp15 were equally detectable (Fig. 5B, Lower). These results suggest that the T98M mutation destabilizes nsp15, resulting in a decrease in the steady-state level of the protein in N15m1-infected cells.
Fig. 5.
T98M mutation causes nsp15 protein instability. (A) BMDMs infected with WT or N15m1 virus at an MOI of 0.1 were lysed at 16 hpi and viral N protein, nsp15, and β-actin were detected by Western blotting. (B) WT and the T98M mutant of nsp15 were expressed and purified from E. coli. Coomassie blue staining shows the purified SUMO-tagged WT and T98M nsp15, which were detected by anti-nsp15 antibody using Western blotting (Lower). (C) DSF thermal shift analysis of nsp15 WT (black) and nsp15-T98M mutant (red) proteins. (D) A radiolabeled RNA molecule R16.4 was treated over time with WT nsp15 or T98M in the presence of 5 mM Mn2+. At the indicated time points, an aliquot of the reaction was analyzed on a denaturing 20% polyacrylamide gel. The sequence of RNA R16.4 is shown above the gel image. The only uridylate, at position 13, is underlined. Data are representative of two to three independent experiments.
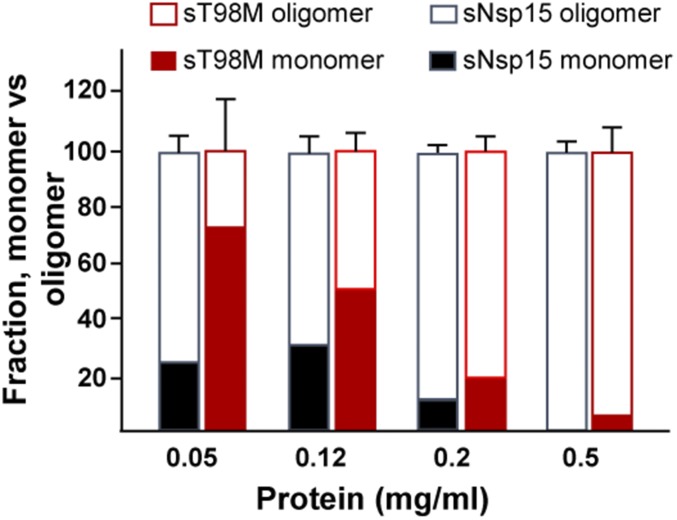
To further evaluate the effect of the T98M mutation on the nsp15 protein, we cloned and expressed codon-optimized versions of the WT and T98M nsp15 as SUMO-fusion proteins in Escherichia coli (Fig. 5B, Upper). Based on the structure of nsp15, the SUMO tag will not affect the assembly of oligomers, including hexamers, as previously reported (12–14). To determine if the T98M mutation destabilizes the protein, differential scanning fluorimetry (DSF) was used to measure the stability of nsp15 in response to heat (41). DSF assesses the denaturation of proteins in the presence of a hydrophobic dye that will fluoresce upon binding to hydrophobic protein sequences. This technique has been used to monitor interactions between viral protein subunits (42). WT nsp15 undergoes a major transition to the denatured state at 47 °C (Fig. 5C). In contrast, the T98M mutant denatured at 40 °C, 7 °C lower than the WT nsp15, which is consistent with our hypothesis that the T98M mutation renders nsp15 less stable. Using dynamic light-scatter spectrometry, we report that the majority of WT nsp15 assembles to form oligomers at a protein concentration of 0.05 mg/mL. In contrast, the majority of the T98M nsp15 protein is detected in the monomeric form under these conditions, suggesting that the T98M mutation impairs nsp15 oligomerization (Fig. S5). Thus, our in vitro characterization data for WT and T98M nsp15 proteins strongly support our prediction that the T98M mutation decreases protein stability. We reasoned that the instability of nsp15 imposed by the T98M mutation might impact endoribonuclease activity, which may in turn contribute to the N15m1 phenotype during infection. Indeed, the T98M mutant exhibited significantly reduced RNA cleavage activity in vitro compared with WT protein (Fig. 5D). Altogether, these results demonstrate that the T98M mutation, like H262A, impairs nsp15 endoribonuclease function, albeit by different means.
Fig. S5.
T98M mutation alters the oligomerization of nsp15. Dynamic light scattering evaluating the percent monomer and hexamer present at increasing concentrations of WT and T98M nsp15 (see detailed description in SI Materials and Methods). Data are representative of two to three independent experiments and presented as the mean ± SD.
Loss of nsp15 Endoribonuclease Activity Affects the Distribution of dsRNA in Virus-Infected BMDMs.
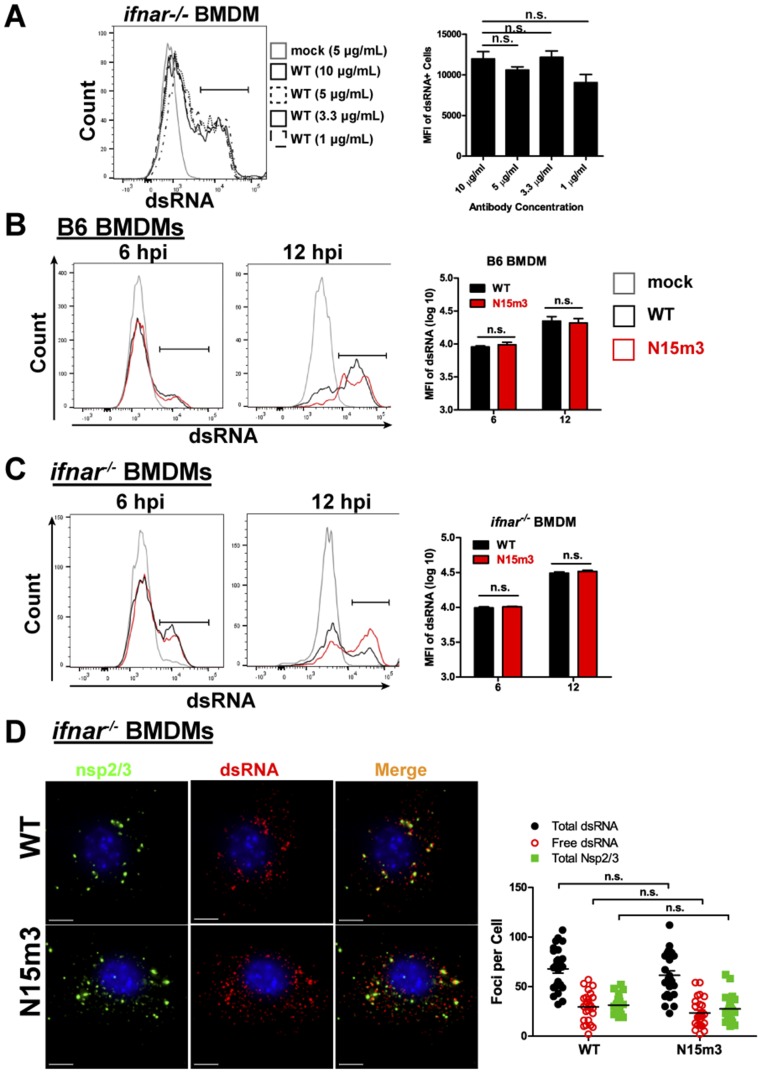
Based on our findings that highlight the importance of nsp15 endoribonuclease activity, we initially hypothesized that nsp15 may prevent accumulation of viral dsRNA in the cytoplasm, thereby reducing the likelihood of triggering host dsRNA sensors. To address this possibility, we measured the abundance of dsRNA during infection of BMDMs by flow cytometry. We predicted that the loss of nsp15 endoribonuclease activity in the mutant viruses would correspond to an increase in immunofluorescence intensity of dsRNA. However, we did not observe a significant difference between WT- and N15m3-infected cells in either B6 or ifnar−/− BMDMs at early or late phases of infection (Fig. S6 A–C), implying that nsp15 does not reduce the amount of dsRNA in infected cells.
Fig. S6.
Evaluation of dsRNA levels in infected-macrophages. (A) ifnar−/− BMDMs were infected with WT virus at an MOI of 0.1. At 6 hpi, cells were stained with anti-dsRNA at stated concentrations and analyzed by flow cytometry. (B) B6 or (C) ifnar−/− BMDMs were infected with WT or N15m3 virus at an MOI of 0.1. At 6 and 12 hpi, cells were stained for dsRNA and analyzed by flow cytometry. BMDMs were sorted by removing debris in FSC-A/SSC-A plot, selecting single cells by FSC-H/FSC-A plot, and gating on dsRNA+ cells. (D) ifnar−/− BMDMs were infected with WT or N15m3 at MOI of 0.1, fixed at 12 hpi, and stained with anti-dsRNA, anti-nsp2/3, and Hoescht 33342. The foci from 25 images were counted using IMARIS software program. Values were analyzed by a unpaired t test and n.s. stands for not significant. Data are representative of two to three independent experiments. (Scale bars, 5 μM.)
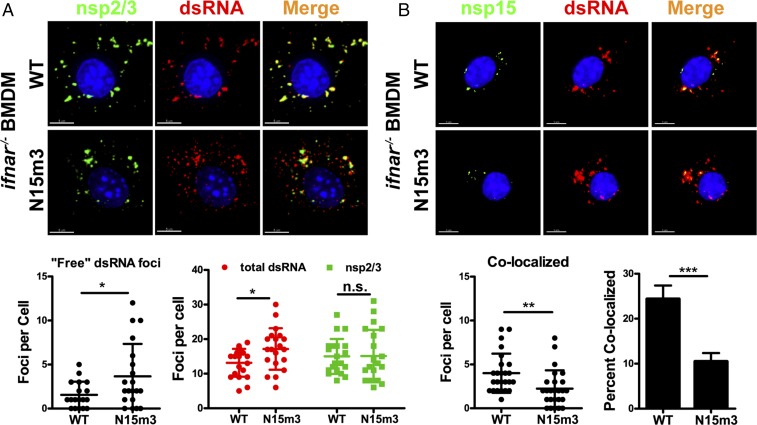
Previous studies reported that coronavirus dsRNA associates with replication complexes early during infection and is buried in DMVs, which are thought to protect viral RNA from host sensors (8, 9, 43). Additionally, our group and others have shown that nsp15 also associates with replication complexes (44, 45) and interacts with single-strand RNA and dsRNA (13). Therefore, we hypothesized that that nsp15 may function to maintain the association of dsRNA with replication complexes. We predicted that loss of nsp15 endoribonuclease activity may allow for accumulation of dsRNA that is not associated with replication complexes (“free” dsRNA). Such free dsRNA might be vulnerable to detection by cytoplasmic dsRNA sensors. To test this hypothesis, we used ifnar−/− BMDMs because we showed that replication of both WT and nsp15 mutant viruses is equivalent in these cells. The subcellular localization of dsRNA and nsp2/3, which are hallmark components of viral replication complexes (8), was detected using confocal microscopy with specific antibodies. Interestingly, N15m3 infection yielded more dsRNA foci that did not colocalize with nsp2/3 foci (Fig. 6A). To quantify these free dsRNA foci, we used an automated immunofluorescence analysis program (IMARIS) to identify and count the number of dsRNA and nsp2/3 foci. We found that there were similar levels of nsp2/3+ replication complexes in N15m3- and WT-infected cells at 6 hpi; however, more free-dsRNA foci were detected in N15m3-infected cells than in WT-infected cells (Fig. 6A). Consistent with previous reports (43, 46), dsRNA was dispersed late in infection, leading to a similar increase in the number of free-dsRNA foci in both WT- and N15m3-infected cells (Fig. S6D). We note that the early dispersal pattern of dsRNA in N15m3-infected cells coincides with the early activation of dsRNA sensors and the IFN response (Figs. 2–4). Taken together, these data suggest that nsp15 may not affect the abundance of cytosolic dsRNA, but rather functions to maintain the association of dsRNA with replication complexes, likely by sequestering dsRNA within DMVs (8).
Fig. 6.
Mutation of nsp15 affects dsRNA distribution in virus-infected BMDMs. ifnar−/− BMDMs were infected with WT or N15m3 at an MOI of 0.1. Cells were fixed at 6 hpi and stained with (A) anti-nsp2/3, anti-dsRNA, and Hoescht 33342 or (B) anti-nsp15, anti-dsRNA, and Hoescht 33342. Surfaces for puncta were created based on dsRNA and nsp2/3 fluorescence, and fluorescence was measured within each surface. The foci from 25 images were counted using IMARIS software program. (A) Images of subcellular localization of dsRNA and nsp2/3 (Upper) and quantification of foci (Lower). (B) Images of subcellular localization of dsRNA and nsp15 (Upper) and quantification of foci (Lower). Percent colocalization of nsp15 with dsRNA was calculated by dividing dsRNA+ nsp15+ foci by total dsRNA foci. Values were analyzed by an unpaired t test and error bars represent the mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant. Data are representative of two to three independent experiments. (Scale bars, 5 μm.)
To further elucidate the relationship between nsp15 and dsRNA during viral infection, we visualized nsp15 and dsRNA localization by immunofluorescence using specific antibodies. As expected, we found that the number of dsRNA foci that colocalized with nsp15 foci was significantly reduced in N15m3-infected cells compared with WT-infected cells (Fig. 6B). These results demonstrate that nsp15 localization with dsRNA—and therefore with replication complexes—is dependent on endoribonuclease activity, the lack of which may result in more free cytoplasmic dsRNA available for detection by host sensors.
nsp15 Mutations Profoundly Attenuate Murine Coronavirus Infection in Vivo.
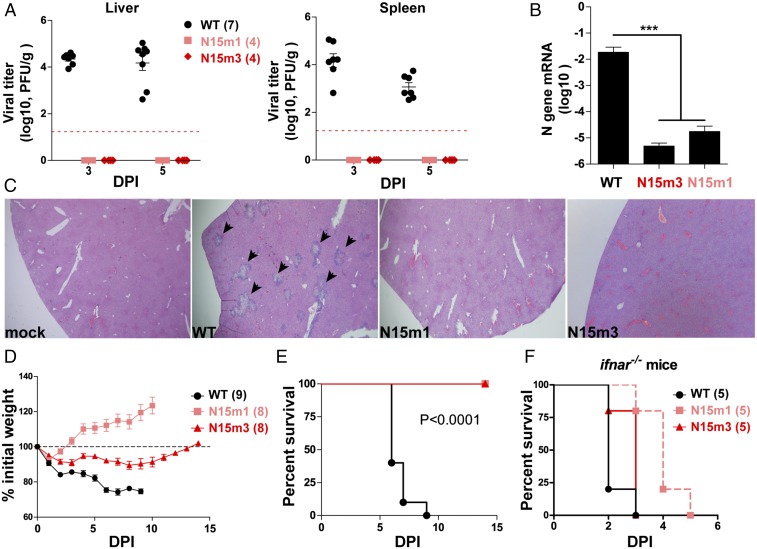
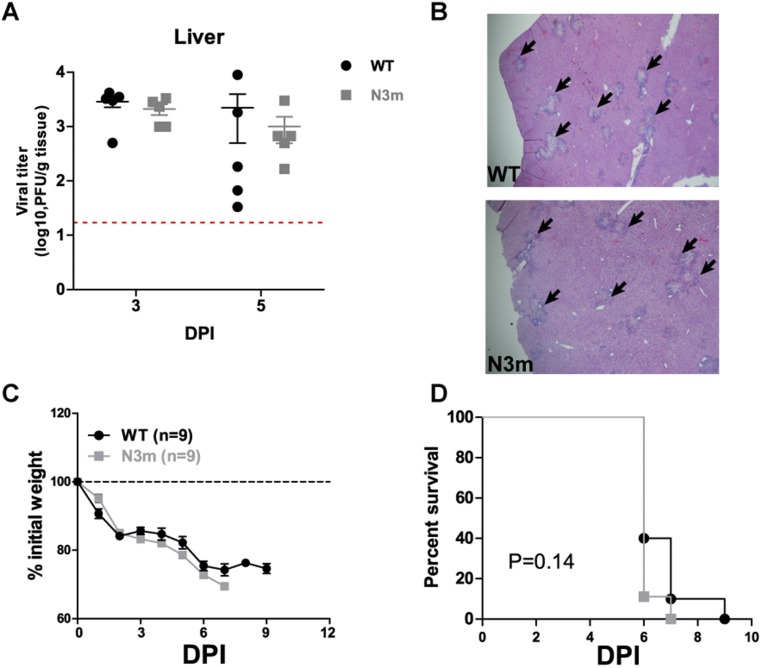
Because nsp15 mutant viruses induce a robust IFN response and activate host dsRNA sensors, we wanted to determine if the loss of nsp15-mediated antagonism of innate immune responses alters the pathogenesis of murine coronavirus. We first infected C57BL/6 mice intraperitoneally with 6 × 104 plaque-forming units (pfu) of WT or mutant virus and evaluated viral burden in target organs and pathology in the liver. Strikingly, standard plaque assays that enumerate infectious particles failed to detect virus in the livers and spleens of mice inoculated with nsp15 mutant viruses at 3 and 5 d postinfection (dpi) (Fig. 7A). However, high viral titers were detected in the same target organs of mice infected with WT or N3m virus (Fig. 7A and Fig. S7A). N gene transcripts, albeit low levels, were only detectable by qPCR in the mesenteric lymph node (MLN) of mice infected with nsp15 mutant virus at 1 dpi (Fig. 7B). Histological examination of the livers revealed typical lesions associated with infection by MHV-WT and N3m, but not in nsp15 mutant virus-infected mice (Fig. 7C and Fig. S7B).
Fig. 7.
MHV nsp15 mutant viruses are highly attenuated in mice. Six-week-old C57BL/6 mice were intraperitoneally inoculated with 6.0 × 104 pfu of WT or mutant viruses. (A) Liver and spleen were harvested at 3 and 5 dpi (DPI) and tested for viral titer by plaque assay. Red dashed line indicates the limit of detection. (B) At 24 hpi, MLN were harvested for RNA extraction. The N gene mRNA levels were measured by quantitative PCR and relative to β-actin. ***P < 0.001, unpaired t test. (C) At 5 dpi, mouse livers from A were harvested for pathology evaluation by H&E staining. Typical lesions of MHV infection in liver were indicated by arrowheads. (Magnification, 40×.) Data are representative of four mice per group. (D and E) Mice were inoculated by intracranial injection with 600 pfu of WT or mutant viruses. Viral pathogenicity was evaluated by (D) body weight loss and (E) survival rate. Data are a pool of two independent experiments. The P values of survival rate were calculated using a log-rank test. (F) Twelve- to 14-wk-old ifnar−/− mice were intraperitoneally inoculated with 50 pfu of virus and monitored for mortality. The P values of survival rate were calculated using a log-rank test. WT vs. N15m1, P = 0.0047; WT vs. N15m3, P = 0.0719; N15m1 vs. N15m3, P = 0.0145. Mouse numbers (n) are indicated in parentheses. Data are a pool of two independent experiments. Error bars in A, B, and D represent the mean ± SEM.
Fig. S7.
N3m virus phenocopies WT virus in mouse infection. (A and B) Six-week-old C57BL/6 mice (five mice per group) were intraperitoneally inoculated with 6.0 × 104 pfu of virus. (A) Viral titer in liver at 3 and 5 dpi was determined and (B) liver pathology at 5 dpi was evaluated by H&E staining. (Magnification, 40×.) Black arrowheads indicate the liver lesions caused by MHV infection. Red dash line indicates the limit of detection. (C and D) Six-week-old C57BL/6 mice were intracranially injected with 600 pfu of WT or mutant viruses. Viral pathogenicity was evaluated by (D) body weight loss and (E) survival rate. Data are a pool of two independent experiments. Error bars in A and C represent the mean ± SEM P value of survival rate was calculated using a log-rank test.
A lethal challenge model of MHV-A59 was used to determine if nsp15 mutant viruses are attenuated. Injection of 600 pfu of WT virus into the mouse cranium is sufficient to induce lethal encephalitis (47, 48). As expected, all WT- or N3m-infected mice lost body weight (Fig. 7D and Fig. S7C) and succumbed to infection by 7–9 dpi (Fig. 7E and Fig. S7D). In contrast, all nsp15 mutant virus-infected mice survived the infection (Fig. 7E) and exhibited only mild or transient weight loss, from which they recovered over time (Fig. 7D). These data reveal that nsp15 mutant viruses are profoundly attenuated and exhibit minimal pathogenicity, even in this sensitive model. To evaluate the replication of these viruses in the absence of IFN signaling, ifnar−/− mice were intraperitoneally infected with 50 pfu of WT or nsp15 mutant virus. All mice succumbed to infection, indicating that all tested viruses could replicate and induce lethal disease in the absence of IFN signaling (Fig. 7F). Interestingly, N15m1-infected mice exhibited delayed mortality compared with WT-infected mice (P = 0.0047), suggesting that either the instability of nsp15 or the addition of the mutation in nsp3 may contribute to viral attenuation.
MHV nsp15 Mutant Viruses Confer Protective Immunity against WT Virus Infection.
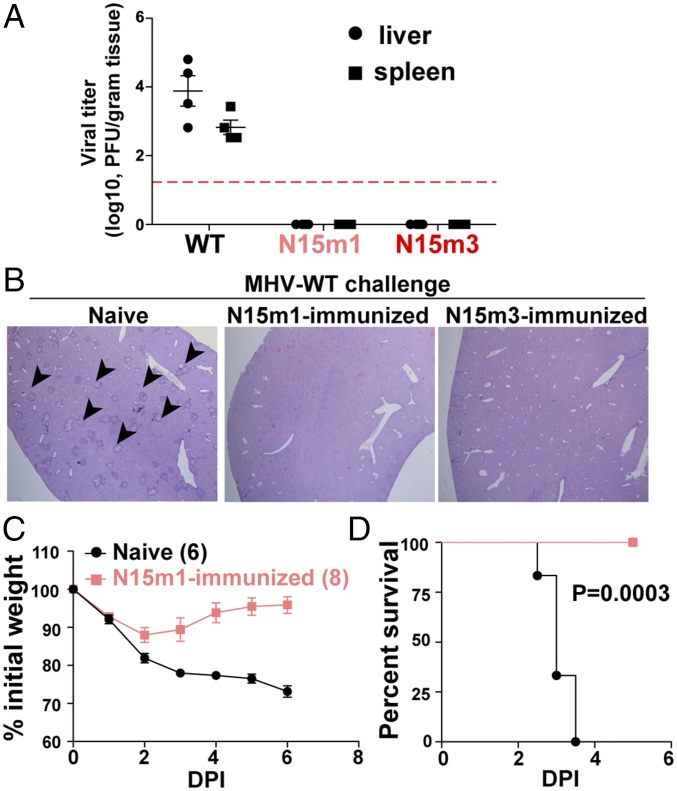
Because the nsp15 mutant viruses are highly attenuated for virulence in vivo, we wanted to determine if these viruses could elicit protective immunity against subsequent challenge with WT virus. B6 mice were infected intraperitoneally with 6 × 104 pfu of nsp15 mutant viruses. Four weeks later, the same mice, as well as naïve, age-matched mice, were challenged with 6 × 104 pfu of WT virus and viral burden and liver pathology were assessed at 5 dpi. In contrast to naïve mice, mice that had been inoculated with nsp15 mutant viruses before challenge with MHV-WT produced undetectable viral titers in tested organs (Fig. 8A) and no liver pathology observed (Fig. 8B). Even more striking were the results from intracranial infection. Here we challenged mice that had been previously inoculated 7 wk prior with N15m1 or naïve mice with a 10-fold lethal dose of MHV-WT. We found that the immunized mice experienced only minor weight loss and fully recovered from infection, whereas the age-matched naïve mice succumbed to infection (Fig. 8 C and D). This finding demonstrates that immunization with nsp15 mutant virus protects mice from a subsequent lethal challenge, suggesting that nsp15 mutant viruses can elicit strong, protective immune memory in mice, which highlights their potential as vaccine candidates.
Fig. 8.
MHV nsp15 mutant viruses confers protective immunity against WT virus infection. Ten-week-old naïve C57BL/6 mice or mice immunized with mutant virus 4 wk prior (from Fig. 7A) were intraperitoneally inoculated with 6.0 × 104 pfu of WT virus. At 5 dpi, organs were harvested for (A) viral titration and (B) liver pathology. Red dashed line in A indicates limit of detection. Images of liver sections in B are representative of four mice per group. (Magnification, 40×.) Black arrowheads indicate the liver lesions caused by MHV infection. (C and D) Thirteen-week-old naïve mice and mice immunized seven weeks prior with N15m1 (from Fig. 7E) were challenged with 6.0 × 103 pfu WT virus by intracranial inoculation. Viral pathogenicity was evaluated by (C) body weight loss and (D) percent survival. Mouse numbers (n) are indicated in parentheses. The P values of survival rate were calculated using a log-rank test. Data are a pool of two independent experiments. Error bars in A and C represent the mean ± SEM.
Discussion
RNA viruses that replicate via dsRNA intermediates can be detected as “nonself” by host dsRNA sensors. These sensors include several cytoplasmic RIG-I–like receptors (RLRs), such as RIG-I and MDA5. Activation of RLRs stimulates the production of type I IFN, which up-regulates additional dsRNA sensors, including PKR and the OAS/RNase L system. Collectively, activation of these systems promotes the expression of ISGs and the subsequent execution of an antiviral state within the cell. Importantly, expression of these dsRNA sensors is maintained at a relatively high basal level in macrophages and microglia, which allows for an immediate response against invading viral pathogens (49). To counteract these systems, many viruses have evolved strategies to prevent the early activation of dsRNA sensors. Here we describe a previously unrecognized role for coronavirus nsp15 in derailing the activation of dsRNA sensors in macrophages.
Previous studies using overexpression of SARS-CoV nsp15 (21, 50) and arterivirus nsp11 (20, 22, 23) provided an initial indication that these proteins can act as IFN antagonists. However, as shown here, only by studying nsp15 in the context of viral replication in macrophages and in mice was the nature of the IFN antagonism revealed. We generated murine coronaviruses that encode mutations in nsp15 and compared the replication of WT and mutant viruses in multiple cell types. We found that replication of the mutant viruses was impaired in B6 BMDMs and activated MDA5, PKR, and the OAS/RNase L system. Activation of these dsRNA sensors stimulated IFN and promoted early apoptosis of macrophages, which limited the production of progeny virus in cell culture. In mice, we found that nsp15 mutant viruses were highly attenuated and able to elicit a protective immune response against subsequent challenge with WT virus. Overall, these results reveal a critical function of nsp15 in blocking the activation of host dsRNA sensors, such that disabling nsp15 stability or activity remarkably attenuates coronavirus pathogenesis.
Previous studies have shown that coronaviruses encode multiple proteins that antagonize recognition of viral dsRNA and prevent activation of the host innate immune response. One such antagonist, coronavirus nsp16, is a 2′O-methyltransferase (2′O-MTase) that provides a cap structure at the 5′-end of viral mRNAs to prevent their detection by host sensor MDA5 (51). SARS-CoV and MHV mutant viruses that lack 2′O-MTase activity activate the type I IFN response and are attenuated in macrophages and mice (51, 52). Many additional viral antagonists have been described, and an emerging theme is that these viral proteins may have tissue- or cell type-specific roles. For example, the strain of murine coronavirus used in this study also encodes accessary protein ns2. This protein is a 2′,5′-phosphodiesterase (PDE) that cleaves 2′,5′-oligoadenylate, the product of OAS. Cleavage of 2′,5′-oligoadenylate prevents RNase L-mediated degradation of RNA (39). Loss of ns2 PDE activity attenuates viral pathogenicity in the liver but not in the brain, suggesting a liver-specific effect of ns2 activity (53, 54). In contrast, our results demonstrate that mutation of nsp15 is sufficient to attenuate viral pathogenesis in both the liver and the brain. Furthermore, we detected activation of the OAS/RNase L pathway in nsp15 mutant virus-infected cells despite the presence of functional ns2 (Fig. 4C). We speculate, therefore, that there may be a hierarchy of coronaviral proteins that modulate host innate immune responses. Evidence for such a hierarchy was recently reported in the context of MERS-CoV (55–58). These studies revealed that MERS-CoV NS4a and NS4b inhibit dsRNA sensors. Specifically, MERS-CoV NS4a encodes a dsRNA-binding protein that limits the activation of PKR, whereas NS4b encodes a PDE, which inhibits RNase L activity similar to MHV ns2. However, Rabouw et al. reported that deletion of NS4a alone did not activate PKR, suggesting that MERS-CoV encodes redundant mechanisms to suppress recognition by, and activation of, dsRNA sensors (57). To that end, we posit that nsp15 may play a dominant role over ns2/NS4a/Ns4b activity in antagonizing host dsRNA sensors, such that loss of ns2, NS4a, or NS4b alone is not sufficient to activate these sensors in all cell types. Future studies are needed to elucidate the hierarchy of coronavirus proteins, including nsp15, ns2, NS4a, NS4b, the recently described antagonism of nsp14 (43), and others (6), that collectively antagonize innate immunity and contribute to tissue-specific viral pathogenesis.
The mechanism by which nsp15 endoribonuclease activity suppresses the activation of dsRNA sensors is unknown. Previously, pestivirus and Lassa virus were shown to encode viral ribonucleases that prevent activation of host sensors by degrading viral dsRNA (59, 60). Given these examples, it seemed logical to evaluate the relative abundance of viral dsRNA in WT- and nsp15 mutant-infected cells (Fig. 6 and Fig. S6). Indeed, while this manuscript was under review, Kindler et al. reported increased accumulation of viral dsRNA in nsp15 mutant virus-infected cells and suggested that nsp15 may be part of a viral RNA decay pathway (61). However, our analysis indicates no difference in the accumulation of dsRNA in cells infected with viruses that express either functional or catalytically-inactive nsp15. We note that the nsp15 catalytic mutant virus reported here (N15m3) encodes a different mutation (H262A) than the virus reported by Kindler et al. (H277A) (61), which may account for the difference in results. Another possibility is that nsp15 may recognize and cleave specific dsRNA targets and that mutations in nsp15 may influence target selection. Bhardwaj et al. found that SARS-CoV nsp15 can bind to a highly conserved, hairpin-structured RNA molecule derived from the 3′ untranslated region of the viral genome (13). This dsRNA-like molecule contains a right-angle turn and a loop with multiple potential cleavage sites. The authors reported that only the site in the right-angle turn could be cleaved, indicating that nsp15 cleavage can be influenced by RNA structure, although it is important to note that these studies were performed in vitro using nonphysiologically relevant manganese concentrations. Future studies are therefore needed to evaluate nsp15-mediated dsRNA cleavage in the context of virus infection.
Another feature of nsp15 is that it colocalizes with membrane-associated viral replication complexes (44, 45). Previous studies suggest that viral dsRNA may be sequestered within membrane-associated replication complexes as a means of protecting it from detection by host sensors (9). It is possible that the functional activity of nsp15 may occur in association with such viral structures. Indeed, the majority of dsRNA was colocalized with replication complex proteins in WT-infected cells. In contrast, in nsp15 mutant-infected cells, we observed early dispersal of dsRNA foci away from replication complexes (Fig. 6), which coincided with early activation of dsRNA sensors. We speculate, therefore, that nsp15 may function as a “gatekeeper” to sequester viral dsRNA within replication complexes and away from host dsRNA sensors. Further studies are needed to fully elucidate the mechanisms used by nsp15 to potentially hide or degrade viral RNA and ultimately prevent activation of host dsRNA sensors.
In summary, this study provides insights into the role of nsp15 as an antagonist of host dsRNA sensors during coronavirus infection in macrophages and in mice and provides new directions for developing live-attenuated vaccines.
Materials and Methods
Cells, Viruses, Antibodies, and Chemicals.
Baby hamster kidney cells expressing the MHV receptor (BHK-R) were kindly provided by Mark Denison, Vanderbilt University Medical Center, Nashville, TN. The murine fibroblast 17Cl-1 cell line was maintained in DMEM containing 5% FCS DMEM. The L929 cell line was a gift of Francis Alonzo, Loyola University Chicago, Maywood, IL. Differentiated BMDMs were maintained in bone marrow macrophage media containing DMEM (#10-017-CV, Corning) supplemented with 30% L929 cell supernatant, 20% FCS, 1% l-glutamine, 1% sodium pyruvate, and 1% penicillin/streptomycin. WT MHV strain A59 (GenBank accession no. AY910861) was generated by reverse genetics and full-genome–sequenced. Rabbit anti-nsp2/3 serum (anti-D3) and anti-nsp15 serum (anti-D23) were previously reported by our laboratory (8, 44). Mouse anti-N (J3.3) was a gift from John Fleming (University of Wisconsin–Madison, WI). The following antibodies were purchased commercially (catalog number and company indicated): mouse anti–β-actin (#A00702, Genscript), donkey anti-rabbit-HRP (#711-035–152, Jackson ImmunoResearch), goat anti-mouse-HRP (#1010-05, SouthernBiotech), anticleaved–caspase-3 (#Asp175, Cell Signaling Technology), anti-dsRNA (#K1, Scicons), anti-ISG54 (#PA3845, ThermoFisher), anti-eIF2α (#sc-133132, Santa Cruz), and anti–p-eIF2α (#sc-12412, Santa Cruz). Chemical inhibitors were obtained from the following sources: pan-caspase inhibitor zVAD (#627610, Millipore), Nec-1 (#480065, Millipore), PKR inhibitor C16 (#527450, Millipore), and VX-765 (#F7120, UBPbio), staurosporine (#ALX-380–014, Enzo Life Sciences).
Mutant Viruses and Deep Sequencing.
To generate MHV mutant viruses, nucleotide changes were incorporated into the MHV-A59 genome through PCR mutagenesis (primers available upon request) of cDNA fragments. Subsequent generation of virus by reverse genetics was performed as previously described by Yount et al. (24). Viral genomic RNA from in vitro transcription (mMESSAGE mMACHINE T7 Transcription Kit; AM1344, Invitrogen Ambion) of ligated cDNA fragments was electroporated into BHK-R cells. Cell supernatant was collected as viral stock following observation of cytopathic effects. Infectious clones were plaque-purified, propagated on BHK-R cells, and titrated on 17Cl-1 cells. Mutant viruses were maintained exclusively in BHK-R cells, which do not produce or respond to IFN. All virus stock preparations and plaque-purified isolates used in this study were full-genome deep-sequenced (Kansas State University diagnostic laboratory). Briefly, viral RNA was extracted from virus stock using QIAamp MinElute Virus Spin Kit (57704, Qiagen), used to generate a cDNA Library and sequenced by Miseq or Ion Torrent technology. Mutant MHV sequences were aligned to the WT MHV-A59 synthetic construct (GenBank accession no. AY910861).
Infection and Mouse Experiments.
BMDMs in 12- or 24-well plates were infected with indicated viral strains at a multiplicity of infection (MOI) of 0.1 or 1 in serum-free media. For growth kinetics analysis, cell-culture supernatants were collected at indicated time points and titrated by plaque assay on 17Cl-1 cells. For mouse infection, all experiments were performed using protocols reviewed and approved by the Loyola University Chicago Institutional Animal Care and Use Committee (IACUC). For intracranial infections, 6-wk-old C57BL/6 female mice (Jackson Laboratory) were inoculated with 600 pfu virus and monitored for body weight daily and killed when weight loss was over 25% according to the IACUC protocol. For intraperitoneal infection, 6-wk-old mice were injected with 60,000 pfu and organs were collected at indicated time points. Evidence of liver pathology was determined by H&E staining.
Cell Death Assays.
Cell viability and caspase-3/7 activity were measured using CellTiter Glo (G7571, Promega) and Caspase-Glo 3/7 (G8091, Promega), respectively, according to the manufacturer’s protocols.
DSF Assay.
Recombinant MHV nsp15 or T98M proteins were diluted in storage buffer (10% glycerol, 20 mM Tris⋅Cl, pH 7.5, 300 mM NaCl, and 5 mM β-ME) at different concentrations. Size measurement was carried out by Zetasizer Nano-S dynamic light scattering (Malvern Instruments) at room temperature. Each sample was measured at least three times. The average intensity and size distributions are shown in Fig. S5.
RNA Cleavage Assay.
The standard RNA cleavage assay used 1 × 104 cpm of 5′-end radiolabeled RNA substrate (1 μM final RNA concentration) and 0.026 μM nsp15 in 50 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 1 mM DTT, and 5 mM MnCl2 at 30 °C. The endoribonuclease reactions were terminated by the addition of a gel-loading buffer that contained 7.5 M urea. Products were separated by electrophoresis in 20% polyacrylamide gels containing 7.5 M urea. Gels were wrapped in plastic and exposed to a PhosphorImager screen for quantification using Molecular Dynamics software.
SI Materials and Methods
Cells, Viruses, Antibodies, and Other Reagents.
Baby hamster kidney cells expressing the MHV receptor (BHK-R) were kindly provided by Mark Denison, Vanderbilt University Medical Center, Nashville, TN, and cultured in DMEM (#12100-046, Gibco) supplemented with 10% heat-inactivated FCS and G418 (0.8 mg/mL) (SV30069, HyClone) to maintain selection for MHV receptor expression. The 17Cl-1 cell line was maintained in DMEM containing 5% FCS DMEM. The L929 cell line was a gift of Francis Alonzo, Loyola University Chicago, Maywood, IL, and maintained in DMEM (#10-017-CV, Corning) supplemented with 10% FCS, 1% l-glutamine, 1% sodium pyruvate, 1% nonessential amino acids, and 1% penicillin/streptomycin. Differentiated BMDMs were maintained in bone marrow macrophage (BMM) media containing DMEM (#10-017-CV, Corning) supplemented with 30% L929 cell supernatant, 20% FCS, 1% l-glutamine, 1% sodium pyruvate, and 1% penicillin/streptomycin. WT MHV strain A59 (GenBank accession no. AY910861) was generated by reverse genetics and full-genome–sequenced. Rabbit anti-nsp2/3 serum (anti-D3) and anti-nsp15 serum (anti-D23) were previously reported by our laboratory (8, 44). Mouse anti-N (J3.3) was a gift from John Fleming, University of Wisconsin–Madison, WI. The following antibodies were purchased commercially (catalog number and company indicated): mouse anti–β-actin (#A00702, Genscript), donkey anti–rabbit-HRP (#711-035-152, Jackson ImmunoResearch), goat anti–mouse-HRP (#1010-05, SouthernBiotech), anticleaved–caspase-3 (#Asp175; Cell Signaling Technology), anti-dsRNA (#K1, Scicons), anti-ISG54 (#PA3845, ThermoFisher), anti-eIF2α (#sc-133132, Santa Cruz), and anti–p-eIF2α (#sc-12412, Santa Cruz). Chemical inhibitors were obtained from the following sources: pan-caspase inhibitor zVAD (#627610, Millipore), Nec-1 (#480065, Millipore), PKR inhibitor C16 (#527450, Millipore), VX-765 (#F7120, UBPbio), and staurosporine (#ALX-380-014, Enzo Life Sciences).
Generation of BMDM.
Bone marrow was collected from femurs of C57BL/6 (#000664, The Jackson Laboratory), ifih−/− (#015812, The Jackson Laboratory), or ifnar−/− mice (breeder pair provided by Deborah Lenschow, Washington University in St. Louis, St. Louis, MO). For generation, 5 × 106 bone marrow cells were plated in 100 × 26-mm Petri dishes (25387-030, VWR) with 15 mL BMM media with 50 µM β-mercaptoethanol. After 3 d of incubation at 37 °C with 5% CO2, 10 mL of BMM media was added. Following another 3 d of differentiation, BMDMs were washed in cold 1× PBS, incubated for 30 min at 4 °C in 10 mL 1×, PBS then gently rinsed from the plates by manual pipetting. Next, 1 × 107 cells/mL were suspended in BMM media with 10% DMSO and stored in liquid nitrogen until use. For preparation of L929 cell supernatant, 3.75 × 105 L929 cells were plated in 75 mL DMEM in a T150 flask (#10-126-34, Thermofisher). After 6 d of incubation at 37 °C/5% CO2, the supernatant was harvested, filtered, and stored at −20 °C until use. For viral infections, BMDMs were thawed and plated in 100 × 26-mm Petri dishes in BMM media without β-mercaptoethanol. After 3 d of incubation at 37 °C/5% CO2, cells were plated onto tissue culture dishes for subsequent infection experiments after 24-h incubation.
Mutant Viruses and Deep Sequencing.
To generate MHV mutant viruses, nucleotide changes were incorporated into the MHV-A59 genome through PCR mutagenesis (primers available upon request) of cDNA fragments. Subsequent generation of virus by reverse genetics was performed as previously described by Yount et al. (24). Viral genomic RNA from in vitro transcription (mMESSAGE mMACHINE T7 Transcription Kit; AM1344, Invitrogen Ambion) of ligated cDNA fragments was electroporated into BHK-R cells. Cell supernatant was collected as viral stock following observation of cytopathic effects. Infectious clones were plaque-purified, propagated on BHK-R cells, and titrated on 17Cl-1 cells. Mutant viruses were maintained exclusively in BHK-R cells, which do not produce or respond to IFN. All virus stock preparations and plaque-purified isolates used in this study were full-genome deep-sequenced (Kansas State University diagnostic laboratory). Briefly, viral RNA was extracted from virus stock using QIAamp MinElute Virus Spin Kit (57704, Qiagen), used to generate a cDNA Library and sequenced by Miseq or Ion Torrent technology. Mutant MHV sequences were aligned to the WT MHV-A59 synthetic construct (GenBank accession no. AY910861).
Viral Titration by Plaque Assay.
To determine virus growth kinetics, BMDMs in 24-wells plates were infected with indicated viral strains at an MOI of 0.1 in serum-free media. After 1-h incubation, inoculum was replaced with fresh, complete medium. Cell culture supernatants were collected at indicated time points and titrated by plaque assay on 17Cl-1 cells. To determine the viral titer in organs, a portion of tissue was homogenized with 1.0-mm diameter zirconia/silicon beads (11079110z, BioSpec Products) using an automated homogenizer (MP Biomedicals) at 6.0 m/s for 40 s for 50-mg tissue in serum-free DMEM. The homogenized organs were centrifuged at 450 × g for 5 min and the supernatants were titrated for viral plaques on 17Cl-1 cells. Titers were obtained from three independent assays for each sample. Graphs of virus kinetics were generated using Prism software (GraphPad Software).
Cell Death Assays.
Cell viability and caspase-3/7 activity were measured using CellTiter Glo (G7571, Promega) or Caspase-Glo 3/7 (G8091, Promega), respectively, according to the manufacturer’s protocol. Briefly, 3.0 × 105 BMDMs per well were plated in 24-well plates and infected at an MOI of 0.1 for 1 h in serum-free media. Media was replaced with complete media, or for addition of chemical inhibitors or IFN-α, viral inoculum was replaced with completed BMM media containing indicated concentration of inhibitors or IFN-α. Infected BMDMs were incubated for 24 h at 37 °C/5% CO2. BMDMs were washed twice with 1× PBS and lysed with 150 µL Glo reagent/medium mixture (1:1); 50 µL of cell lysate was used for measuring the luminescence signal.
Immunofluorescence.
BMDMs (1.5 × 105 cells per well) were plated on a glass coverslip in a 24-well plate for 24 h and subsequently infected with WT or mutant virus at an MOI of 0.1 in serum-free media. At 6 or 12 hpi, the infected cells were fixed with 4% formaldehyde in 0.095 M Pipes buffer (P1851, Sigma), permeabilized with 0.1% Triton X-100 (T8787, Sigma) in 1× PBS, and blocked with 5% NGS. Primary and secondary antibodies were used as follows: anti-NSP2/3 (1:1,500), anti-dsRNA (1:500), anti-ISG54 (1:500), anti-nucleocapsid (1:500), donkey anti-rabbit IgG alexafluor 488 (1:1,000; A-21441, Invitrogen), and goat anti-mouse IgG Alexa Fluor 568 (1:1,000; A11004, Thermofisher). Nuclei were visualized with Hoescht 33342 (1:2,500; H1339, Life Technologies). Cells were imaged by collecting z-stack images with a Deltavision wide-field fluorescent microscope (Applied Precision, GE) equipped with a digital camera (CoolSNAP HQ, Photometrics). Images were taken with a 20× or 100× lens. Samples were excited with light generated by an Insight SSI solid-state illumination module (Applied Precision, GE) and deconvolved with SoftWoRx deconvolution software (Applied Precision, GE). All images were collected under identical acquisition conditions and processed using Imaris 7.6.4 (Bitplane).
FACS Analysis.
Triplicate wells of BMDMs (6.0 × 105) were plated in a 12-well plate (Corning) for 6 hpi experiments or (3.0 × 105) in a six-well plate (Corning) for 12 hpi experiments to lessen syncytia formation. After 24 h, BMDMs were subsequently infected with WT or mutant virus at an MOI of 0.1. At 6 or 12 hpi, cells were collected and fixed with 4% formaldehyde in 1× PBS, permeabilized with 0.1% Triton X-100 in 1× PBS, and blocked with 1× PBS containing 5% NGS and 0.5% sodium azide. Cells were labeled with fixable viability dye (65-0865-14, eBioscience), anti-dsRNA (5 µg/mL), and goat anti-mouse (1:1,000) secondary antibody labeled with Alexa Fluor 568. Cells were analyzed using a LSR Fortessa cell analyzer (BD Bioscience). Flow cytometry data were analyzed using FlowJo software (TreeStar).
Quantification of IFN-α Production by RT-qPCR and ELISA.
BMDMs in a 12-well plate were mock-infected or infected with virus at an MOI of 0.1. At indicated time points, monolayer cells were used for RNA extraction using an RNeasy Mini Kit (74104, Qiagen), and cell culture supernatants were collected for ELISA. To determine IFN-α11, β-actin, or MHV-A59 N gene mRNA production, total RNAs were extracted and an equal amount of RNA (500 ng) was used for cDNA synthesis using Rt2 HT First Strand Kit (330401, Qiagen). qPCR was performed with specific primers for mouse IFN-α11 (PPM03050B-200, Qiagen), mouse β-actin (PPM02945B-200, Qiagen) or MHV-A59 N gene using RT2 SYBR Green qPCR Mastermix (330502, Qiagen) in the Bio-Rad CFX96 system. The thermocycler was set as follows: one step at 95 °C (10 min), 40 cycles of 95 °C (15 s), 60 °C (1 min) and plate read, one step at 95 °C (10 s), and a melt curve from 65 °C to 95 °C at increments of 0.5 °C/0.05 s. Samples were evaluated in triplicate and data are representative of three independent experiments. The levels of mRNA were relative to β-actin mRNA and expressed as 2−ΔCT [ΔCT = CT(gene of interest) − CT(β-actin)]. To measure secreted IFN-α, 50 µL cell culture supernatant was used for assay using a mouse IFN-α ELISA kit (BMS6027, eBioscience) following the manufacturer’s instructions.
Electron Microscopy.
B6 BMDMs (6.0 × 105 per well) in a 12-well plate were infected with virus at an MOI of 0.1 in serum-free DMEM. Control cells were treated with BMM media. Following a 1-h incubation at 37 °C/5% CO2, media was replaced with BMM media. Uninfected controls were treated with 1 µM staurosporine prepared in BMM media to induce apoptosis. Cells were further incubated at 37 °C/5% CO2 for 16 h then prepared for EM. Cells were washed in 1× PBS then incubated for 30 min at 4 °C in 1× PBS. Cells were gently collected from plates using a pipette, pelleted at 1,200 rpm for 5 min at 4 °C, and fixed (4% glutaraldehyde in 0.1 M cacodylate buffer). Cell sections were imaged by the Electron Microscopy Core Facility at Loyola University Chicago.
Western Blotting.
B6 BMDMs (12.0 × 105 per well) were media-treated or infected with virus at an MOI of 0.1 in serum-free DMEM for 1 h at 37 °C/5% CO2. At indicated time postinfection, cells were lysed with cold lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium ortho-vanadate, 1 µg/mL leupeptin, 1 mM PSMF). Cells in lysis buffer were incubated on ice for 10 min then pelleted at 14,000 rpm for 10 min at 4 °C. Cell-free supernatant was diluted in 2× sample buffer (10% glycerol, 5% β-ME, 3% SDS, 7.5 mg/mL Trizma-base, bromophenol blue). Samples were separated by electrophoresis through 10% or 4∼15% gradient acrylamide PAGE-gel and transferred to PVDF Immuno-Blot membranes (162-0177, Bio-Rad). Membranes were suspended in blocking buffer (5% BSA in TBST) for 2 h at room temperature. Primary and secondary antibodies were used as follows: anticleaved–caspase-3 (1:1,000), anti–β-actin (1:5,000), anti-N protein (1:200), anti–p-eIF2α (1:600), anti-eIF2α (1:600), donkey anti-rabbit-HRP (1:5,000), goat anti–mouse-HRP (1:5,000), mouse anti–goat-HRP (1:5,000). Membranes were washed three times with 1× TBST and chemiluminescence was visualized using Western Lightning Plus-ECL reagent (50-904-9326, PerkinElmer) in a FluorChem E machine (ProteinSimple).
Purification of the Recombinant MHV nsp15 Protein from Escherichia coli.
The cDNA sequences of WT MHV nsp15 and T98M were amplified using the appropriate primers and subcloned into the pET-His-SUMO vector using fusion PCR. The constructs harboring the WT nsp15 or T98M mutant gene were transformed into Rosetta (DE3) pLys cells. A single colony was inoculated in 100 mL of Terrific Broth (TB) in the presence of 50 μg/mL of ampicillin and 17 μg/mL chloramphenicol and incubated at 37 °C overnight. The culture was then transferred into 3 L of TB media and cultured until the OD600 reached 0.8. The temperature was decreased to 16 °C and isfopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM for 20 h. The cells were collected by centrifugation at 8,000 × g for 10 min and cell pellets were suspended in lysis buffer containing 10% glycerol, 50 mM Hepes (pH 7.0), 400 mM NaCl, 5 mM β-ME, and 10 mM imidazole. The lysate was centrifuged at 15,000 × g for 30 min and the supernatant was loaded onto a Ni-NTA column. The Ni-NTA column was subsequently washed three times with lysis buffer containing 20 mM imidazole. The protein was eluted with elution buffer containing 500 mM imidazole and then the buffer was changed to 10% glycerol, 20 mM Tris⋅Cl (pH 7.5), and 5 mM β-ME. The sample was further purified by Mono Q-Sepharose with a gradient of Tris buffer containing 0–1 M NaCl. nsp15 was quantified by SDS/PAGE compared with a known concentration of BSA, and stored at −80 °C in a buffer containing 10% glycerol, 20 mM Tris-Cl, pH 7.5, 300 mM NaCl, and 10 mM β-ME.
Dynamic Light Scattering.
Recombinant MHV nsp15 or T98M proteins were diluted in storage buffer (10% glycerol, 20 mM Tris-Cl, pH 7.5, 300 mM NaCl, and 5 mM β-ME) at different concentrations. Size measurement was carried out by Zetasizer Nano-S dynamic light scattering (Malvern Instruments) at room temperature. Each sample was measured at least three times. The average intensity and size distributions are shown in Fig. S5.
DSF Assay.
DSF was carried out in the Stratagene MX3005P real-time PCR machine. The samples contain 1× SYPRO Orange, 10 μM of the recombinant protein. All samples were heated at a rate of 0.5 °C/min, and the fluorescence intensity and Tm (melting temperature) were determined.
Bioanalyzer RNA Analysis.
Equal amounts of total RNA purified from BMDMs were analyzed on an Agilent 2100 Bioanalyzer using RNA Nano LabChips.
Mouse Experiments.
All experiments were performed using protocols reviewed and approved by the Loyola University Chicago IACUC. C57BL/6 mice were purchased from the Jackson Laboratory. For intracranial infections, 6-wk-old mice were inoculated with 600 pfu in 20 µL PBS. Infected mice were monitored for body weight daily and euthanized when weight loss was over 25% according to the IACUC protocol. Statistical analysis of survival rate was evaluated using the log-rank test. For intraperitoneal infection, 6-wk-old mice were injected with 60,000 pfu in 100 µL PBS. Organs were collected at indicated time points and evaluated for viral replication. Liver pathology was evaluated using H&E staining by the Tissue Processing Core Facility at Loyola University Chicago.
Sequence Alignment.
Nsp15 T98 region from representative strains of coronavirus subgroups using Clustal W. α-Coronavirus: NL63 (Amsterdam I strain, AY567487), PEDV (CV777 strain, NC_003436); β-coronavirus: MHV, (A59 strain, AY910861); SARS-CoV (MA15 strain, FJ882957); MERS-CoV (EMC strain, JX869059); γ-coronavirus: IBV (Beaudette strain, NC_001451); δ-coronavirus: PDCoV (KJ567050).
Acknowledgments
We thank our colleagues at Loyola University Chicago: Dr. Francis Alonzo for assistance with generating bone marrow-derived macrophages, Dr. Timothy O’Brien for assistance with the statistical analysis, and Aaron Volk for assistance with editing the manuscript; and Ying-Ching Chuang (Indiana University) for protein purification. This work was supported by the National Institutes of Health Grant R01 AI085089 (to S.C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.S.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618310114/-/DCSupplemental.
References
- 1.Perlman S, Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: Recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu J, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: Implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung CY, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: Possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindler E, Thiel V, Weber F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enjuanes L, Almazán F, Sola I, Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 11.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricagno S, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, et al. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nedialkova DD, et al. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J Virol. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, et al. A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J Virol. 2016;90:4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov KA, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H, et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posthuma CC, et al. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, et al. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus suppresses both MAVS and RIG-I expression as one of the mechanisms to antagonize type I interferon production. PLoS One. 2016;11:e0168314. doi: 10.1371/journal.pone.0168314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, et al. The nonstructural protein 11 of porcine reproductive and respiratory syndrome virus inhibits NF-κB signaling by means of its deubiquitinating activity. Mol Immunol. 2015;68:357–366. doi: 10.1016/j.molimm.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, et al. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-β induction. Mol Immunol. 2011;48:1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yount B, Denison MR, Weiss SR, Ralph S, Baric RS. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarino LA, et al. Mutational analysis of the SARS virus Nsp15 endoribonuclease: Identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth-Cross JK, Bender SJ, Weiss SR. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pronk GJ, Ramer K, Amiri P, Williams LT. Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by REAPER. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- 28.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 29.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D-W, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 32.Martin U, et al. Antiviral effects of pan-caspase inhibitors on the replication of coxsackievirus B3. Apoptosis. 2007;12:525–533. doi: 10.1007/s10495-006-0015-y. [DOI] [PubMed] [Google Scholar]
- 33.Castelli JC, et al. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 34.Barber GN. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 2005;12:563–570. doi: 10.1038/sj.cdd.4401643. [DOI] [PubMed] [Google Scholar]
- 35.Hsu L-C, et al. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428:341–345. doi: 10.1038/nature02405. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman RJ. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: A new role for an old actor. Proc Natl Acad Sci USA. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saelens X, Kalai M, Vandenabeele P. Translation inhibition in apoptosis: Caspase-dependent PKR activation and eIF2-alpha phosphorylation. J Biol Chem. 2001;276:41620–41628. doi: 10.1074/jbc.M103674200. [DOI] [PubMed] [Google Scholar]
- 38.Zhu PJ, et al. Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell. 2011;147:1384–1396. doi: 10.1016/j.cell.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, et al. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birdwell LD, et al. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J Virol. 2016;90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 42.Hoover HS, et al. Phosphorylation of the brome mosaic virus capsid regulates the timing of viral infection. J Virol. 2016;90:7748–7760. doi: 10.1128/JVI.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becares M, et al. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol. 2016;90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi ST, et al. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Athmer J, et al. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. MBio. 2017;8:e02320-e16. doi: 10.1128/mBio.02320-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagemeijer MC, Vonk AM, Monastyrska I, Rottier PJM, de Haan CAM. Visualizing coronavirus RNA synthesis in time by using click chemistry. J Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mielech AM, et al. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J Virol. 2015;89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng X, et al. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L, et al. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol. 2013;87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei Y, et al. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menachery VD, et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roth-Cross JK, et al. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J Virol. 2009;83:3743–3753. doi: 10.1128/JVI.02203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol. 2011;85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thornbrough JM, et al. Middle East respiratory Syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio. 2016;7:e00258-e16–e16. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabouw HH, et al. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 2016;12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Niemeyer D, et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zürcher C, Sauter K-S, Mathys V, Wyss F, Schweizer M. Prolonged activity of the pestiviral RNase Erns as an interferon antagonist after uptake by clathrin-mediated endocytosis. J Virol. 2014;88:7235–7243. doi: 10.1128/JVI.00672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kindler E, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]