Significance
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression posttranscriptionally. The evolutionarily conserved miR-183/96/182 cluster, consisting of three related miRNAs, is highly expressed in maturing sensory receptor cells. However, its role in the functional maturation of sensory receptors has not been adequately addressed due to the lack of appropriate in vivo models. We show that deletion of miR-183/96/182 in mice leads to severe deficits in vision, hearing, balance, and smell. These deficits arise from defects in the timing and completion of terminal differentiation in sensory receptor cells associated with dysregulation of networks of genes involved in key processes, such as chromatin remolding and ciliogenesis. Thus, the miR-183/96/182 cluster has an essential role for the maturation of sensory receptors.
Keywords: miR-183/96/182 cluster, photoreceptors, hair cells, olfactory epithelium, terminal differentiation
Abstract
The polycistronic miR-183/96/182 cluster is preferentially and abundantly expressed in terminally differentiating sensory epithelia. To clarify its roles in the terminal differentiation of sensory receptors in vivo, we deleted the entire gene cluster in mouse germline through homologous recombination. The miR-183/96/182 null mice display impairment of the visual, auditory, vestibular, and olfactory systems, attributable to profound defects in sensory receptor terminal differentiation. Maturation of sensory receptor precursors is delayed, and they never attain a fully differentiated state. In the retina, delay in up-regulation of key photoreceptor genes underlies delayed outer segment elongation and possibly mispositioning of cone nuclei in the retina. Incomplete maturation of photoreceptors is followed shortly afterward by early-onset degeneration. Cell biologic and transcriptome analyses implicate dysregulation of ciliogenesis, nuclear translocation, and an epigenetic mechanism that may control timing of terminal differentiation in developing photoreceptors. In both the organ of Corti and the vestibular organ, impaired terminal differentiation manifests as immature stereocilia and kinocilia on the apical surface of hair cells. Our study thus establishes a dedicated role of the miR-183/96/182 cluster in driving the terminal differentiation of multiple sensory receptor cells.
Mammalian sensory epithelia, such as those underlying vison, hearing, smell, and balance, consist of ciliated sensory receptor cells. Although highly specialized, similarities in embryonic origin underlie common features in their development (1). Following specification, lineage-restricted postmitotic precursors become structurally and functionally mature through a series of cellular differentiation events, collectively described as terminal differentiation (2). During maturation, sensory receptor cells typically develop microtubule-based primary cilia and in some cases actin-based membrane protrusions on apical membranes, which are sensory organelles, while maintaining apical basal polarity within sensory epithelia. In photoreceptors, for example, the extension of outer segments (OSs), specialized sensory cilia, is central to postmitotic differentiation and necessary for light sensitivity (3). In auditory and vestibular hair cells, the proper formation of hair bundles is indispensable for detecting sound and head positions (4). Similarly, olfactory sensory neurons (OSNs), the odorant receptor cells in the olfactory epithelium, project multiple dendritic cilia into the mucous membrane of the nasal epithelium where olfactory signaling is initiated (5). Mature sensory epithelia are highly structured with specific spatial organization that is tied to their functions. Synchronized planar cell polarity (PCP) of hair cells in the inner ear, for example, provides their directional sensitivity (6), whereas the development of laminar architecture in the retina restricts photoreceptors to proper compartments for efficient wiring with secondary neurons (7).
MicroRNAs (miRNAs), a class of small, noncoding RNAs, have emerged as key regulators of the timing of sequential events in development, often by acting as modulators of molecular circuitries (8). A single polycistronic miRNA gene, the miR-183/96/182 cluster (miR-183 cluster) (9), is highly expressed across diverse types of sensory receptors (10). Phylogenetically, its orthologs are found throughout deuterostomes and protostomes and exhibit remarkably conserved expression in ciliated sensory neurons (11). In mouse retina, the three miRNAs of the miR-183 cluster, found mostly in photoreceptors (9), rapidly and coordinately increase immediately after birth and peak during the second week of life (9), suggesting a role in terminal differentiation of photoreceptors.
A gene trap allele of the miR-183 cluster, designated as miR183GT (12), was recently described. The phenotype of the miR183GT mice, however, was unexpectedly mild. Electroretinography (ERG) a-wave, which indicates terminal differentiation of photoreceptors, such as OS biogenesis, was essentially normal during early postnatal development, although there may be evidence suggesting synaptic transmission defects. Late-onset retina degeneration, from 6 mo of age, with susceptibility to acute light-induced damage of retina, suggested a function of the miR-183 cluster in homeostasis of adult retina. The lack of a major effect on early postnatal development of photoreceptors in miR183GT mice was surprising (13) and raised the possibility that the miR183GT allele could be hypomorphic. To address this issue, we created a null mutation of the miR-183 cluster through classic homologous recombination. This germline mutation causes severe deficits in vison, hearing, balance, and smell. Further analyses showed that these sensory deficits arise from profound defects in key terminal differentiation events. In the retina, failure in terminal differentiation is associated with dysregulation of gene networks involved in terminal differentiation timing control, ciliogenesis, and cone nuclear translocation. Thus, the miR-183 cluster is essential for the maturation of sensory receptors.
Results
Deletion of miR-183 Cluster Leads to Broad Loss of Sensory Functions in Adult Mice.
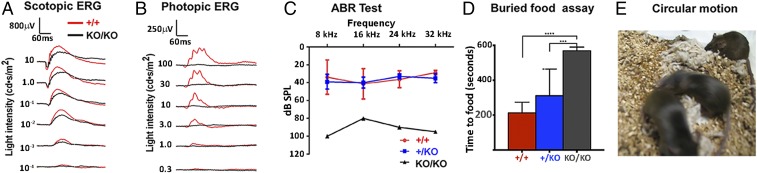
Northern blotting confirmed that the miR-183 cluster miRNAs were enriched in sensory organs (SI Appendix, Fig. S1A), and coordinately up-regulated in retina from birth [postnatal day 0 (P0)], reaching a maximum between P6 and P14 (SI Appendix, Fig. S1B), a period critical for photoreceptor maturation (SI Appendix, Fig. S1C). We deleted the entire genomic region of the miR-183 cluster by homologous recombination (SI Appendix, Fig. S2 A, B, and E) and generated a miR-183 cluster knockout allele, designated as miR-183 cluster-KO. Following germline transmission, we confirmed complete lack of expression of all miR-183 cluster members, including 5p (5′ arm) and 3p (3′ arm) miRNAs, by Northern blotting and qRT-PCR (SI Appendix, Fig. S2 C and D) in retinas homozygous for the miR-183 cluster-KO allele (miR-183 clusterKO/KO, or KO). Adult KO mice exhibited a wide range of sensory defects. Visual function measured by ERG showed a significant reduction of the responses in KO mice under dark-adapted conditions and almost complete elimination under light-adapted conditions, indicating compromised rod photoreceptor function and severe impact of the cone pathway (Fig. 1 A and B and SI Appendix, Fig. S3). Preliminary results of auditory brainstem response (ABR) tests also suggested severe hearing loss from KO mice (Fig. 1C). In addition, constant circling behavior (Fig. 1E), a sign of vestibular dysfunction (14), indicated a body balance defect. KO mice also showed compromised olfactory function in the buried food test (15) (Fig. 1D).
Fig. 1.
Sensory deficits of the visual, auditory, vestibular, and olfactory systems. (A and B) Superimposed ERG waveforms from representative miR-183 clusterKO/KO (KO/KO) and miR-183 cluster+/+ (+/+) littermates at 4 mo of age under dark-(scotopic; A) or light-adapted (photopic; B) condition. Quantitative measurements of ERG responses are shown in SI Appendix, Fig. S3. (C) ABR test was performed at 3 mo of age on four miR-183 cluster+/+, five miR-183 cluster+/KO, and two miR-183 clusterKO/KO littermates. The stimulus intensity began at 100 dB SPL (sound pressure level) and was decreased until the lowest level replicable response was obtained, considered to be threshold. Thresholds were compared using two-way ANOVA unweighted means analysis with within-frequency posttest comparisons to miR-183 cluster+/KO means corrected for multiple testing. Hearing loss was suggested in miR-183 clusterKO/KO group, with no loss in miR-183 clusterKO/+ mice. (D) Buried food assay revealed olfactory dysfunction in KO mice. Graph compares time required to retrieve a buried food pellet by miR-183 clusterKO/KO mice (569.9 ± 66.67, n = 10), miR-183 cluster+/KO (+/KO) (311.2 ± 153.4 s, n = 14) and miR-183 cluster+/+ (213.4 ± 60.99 s, n = 9) mice. ANOVA followed by post hoc Tukey test revealed significant differences between miR-183 clusterKO/KO and both miR-183 clusterKO/+ (***P < 0.001) and miR-183 cluster+/+ mice (F2, 30 = 16.20, ****P < 0.0001). But difference between miR-183 cluster+/KO and miR-183 cluster+/+ groups is not statistically significant. Mean time to food ± SEM is shown in seconds. (E) P30 litter showing circular motion of miR-183 clusterKO/KO littermates.
Maturation Deficiency of Cone and Rod Pathways as Measured by ERG.
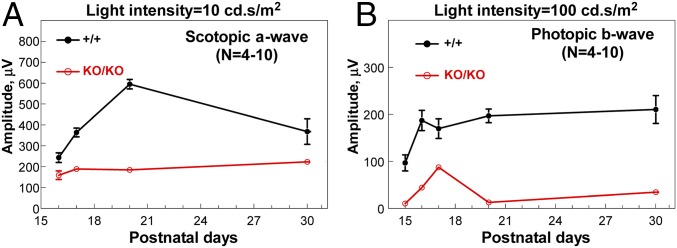
For wild-type mice, terminal differentiation of photoreceptors during P15–P20 was accompanied by dynamic changes in ERG responses (Fig. 2 A and B) (16). Rod-dependent scotopic a-wave increased almost three-fold, and cone-dependent photopic-b wave doubled during the period. Subsequently, scotopic a-wave showed a gradual decline over a period of 1 mo, whereas photopic b-wave maintained its peak level. This temporal change in wild-type ERG responses was largely absent in KO retinas. ERG responses from KO mice were consistently lower than wild type at each developmental time point, indicating deficiencies of rod and cone pathways, respectively, possibly due to failure in terminal differentiation (Fig. 2 A and B).
Fig. 2.
ERG responses over early postnatal days of photoreceptor maturation. Amplitudes of ERG scotopic a-wave (A) and photopic b-wave (B) are plotted over postnatal days. n = 4–10 at each time point involved. Error bars are SD (where not visible, values are too small to make bars visible).
Photoreceptors and ON-Bipolar Cells Are Affected by Deletion of miR-183 Cluster.
The loss of ERG b-wave in KO mice indicated abnormalities at either pre- or postsynaptic levels. Previous studies detected the three miRNAs of the cluster in bipolar and ganglion cells in addition to photoreceptors (9, 17). To resolve which cell populations are affected in the KO, retinas from adult wild-type and KO mice were compared morphologically by light microscopy. We observed a reduction in thickness in inner and outer segments initially, and in the photoreceptor outer nuclear layer (ONL) at later time points (SI Appendix, Fig. S4), indicating that photoreceptors were impacted by miR-183 cluster deletion. In wild-type mice, markers for inner retinal neurons gave the expected labeling patterns (SI Appendix, Fig. S5). Immunolabeling for CHX10, PAX6, GFAP, CALB (calbindin), and BRN3A were comparable between wild type and KO, suggesting no major defects in bipolar, amacrine, Müller glia, horizontal, and ganglion cell populations, respectively. PKCα, a marker for rod ON-bipolar cells, however, was labeled more intensely in the synaptic terminals of KO retina (SI Appendix, Fig. S5). PKCα (Prkca) is a predicted target for miR-183 cluster with a conserved binding site for miR-183 in its 5′-UTR, suggesting that loss of the miR-183 cluster may also have impacted the ON-bipolar cells. The major differences between the KO and wild type, however, were shown by S-opsin and RHO (rhodopsin) staining (SI Appendix, Fig. S5), revealing deficiencies in terminal differentiation of cone and rod photoreceptors.
Mispositioning of Nuclei, Delayed M-Opsin Expression, and OS Extension of Cone Photoreceptors Followed by Cone Cell Death.
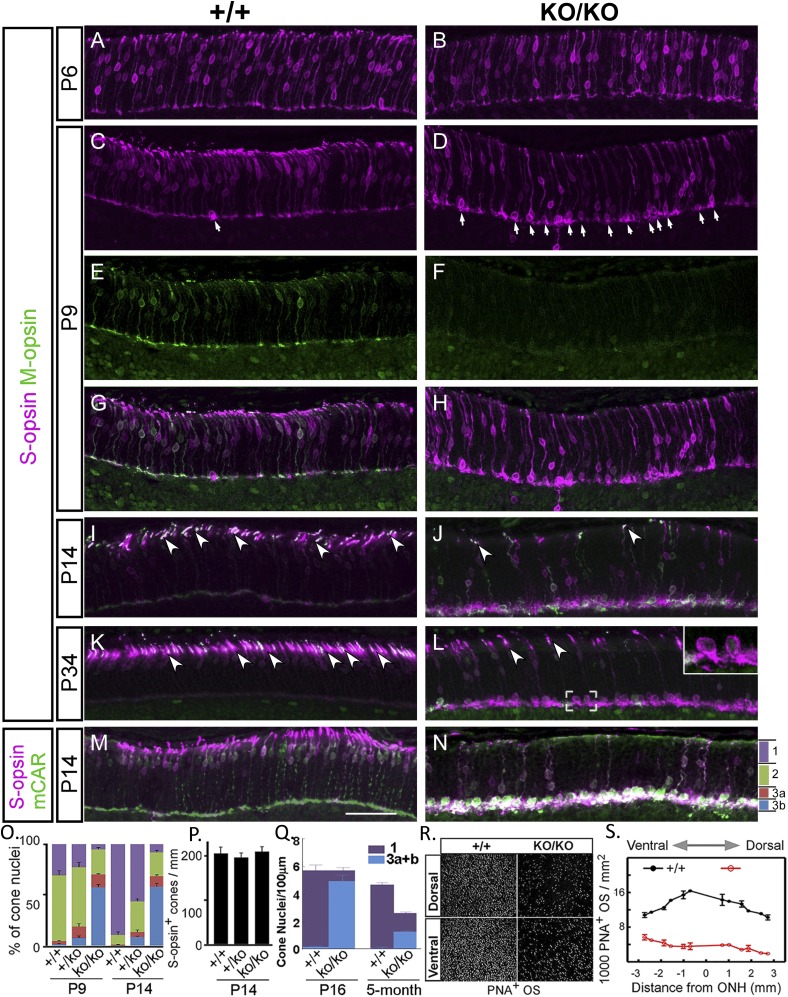
In early postnatal development, rod and cone precursors migrate to the outer retina to form the ONL, while exiting from mitotic division. Cone nuclei become confined apically within the ONL and their axons extend across the ONL to establish synaptic contact with horizontal and bipolar cells (18, 19). By P6, somas of cones, as defined by S-opsin staining, were scattered throughout ONL in both wild-type and KO retinas (Fig. 3 A and B). By P9, most cone nuclei in wild type appeared in the outer half of ONL. In miR-183 cluster-KO, this synchronized localization toward the outer edge was not detected; in contrast, some nuclei were aggregated in proximity to the outer plexiform layer (Fig. 3 C and D, arrows, and G and H). This trend became more obvious by P14 (Fig. 3 J and N) and P34 (Fig. 3L). The distribution of cone nuclei among four subdivisions (bins 1 through 3b) of the entire ONL was compared (Fig. 3O). Those that localized to the bottom of the ONL in the KO often did not have apical processes (Fig. 3L, Inset). Thus, it appeared that many cones in the KO lost their apical junctional connections at the outer limiting membrane and retracted to the bottom region of the ONL. Total cone counts at P14 remained similar among wild-type, heterozygous, and homozygous groups (Fig. 3P), indicating the size of cone population was normal initially. However, by 5 mo, cone nuclei in the inner one-third of the KO ONL were significantly reduced, bringing the total cone count down to about half of that in wild type (Fig. 3Q). Thus, cones that failed to position their nuclei properly were preferentially lost from the ONL.
Fig. 3.
Aberrant nuclear positioning and outer segment formation in cone photoreceptors. (A–N) Representative images of immunostaining for S-opsin, M-opsin, and cone arrestin (mCAR) in retina (inferior) at postnatal ages indicated. ONL is divided into four bins: 1 apical third (purple), 2 middle third (green), with the basal third equally divided into 3a (red) and 3b (blue) (N). (O) Distributions of S-opsin+ nuclei in the four bins at P9 and P14, respectively (percentage). (P) Number of cone nuclei (S-opsin+) per millimeter of ONL length at P14 (P > 0.05). (Q) Number of cone nuclei per micrometer of ONL length as determined by the unique cone nuclear morphology on thin plastic sections (0.5 µm) at 5 mo (P < 0.05) under high magnification. Bins are similarly defined as in N, except that blue indicates the bottom third combining the two basal subdivisions (bins 3a+b). (R) Representative images of PNA staining of flat mounts of dorsal and ventral retina at P14. (S) Distributions of PNA staining throughout retina at P14 over distances from optical nerve head (ONH). (Scale bar, 50 µm in all panels.) Error bar is SEM.
Whereas M-opsin staining was easily detectable as early as P9 in the ONL of wild-type mice (Fig. 3E), positive staining was not observed until P14 in the mutant (Fig. 3 F and J). The OS, as outlined by Opsin staining, was first detected as early as P6 in the wild type (Fig. 3C) and continued to extend until reaching adult length (Fig. 3 G, and I and K, arrowheads). In contrast, a short extension of OS from the cones first appeared at P9–P14 in the KO, and most cone outer segments failed to reach normal length even at P34 (Fig. 3 J and L, arrowheads, and N). Consequently, in the KO, few cones developed OS that extended enough to be visualized by PNA staining (Fig. 3 R and S). Thus, mispositioning of nuclei delayed M-opsin expression, and OS extension followed by cell death are the characteristics of cone photoreceptors in the early postnatal retina of KO mice, which collectively can account for ERG abnormalities of cone pathways in the mutant.
Delayed Rod Outer Segment Biogenesis Followed by Early-Onset Degeneration.
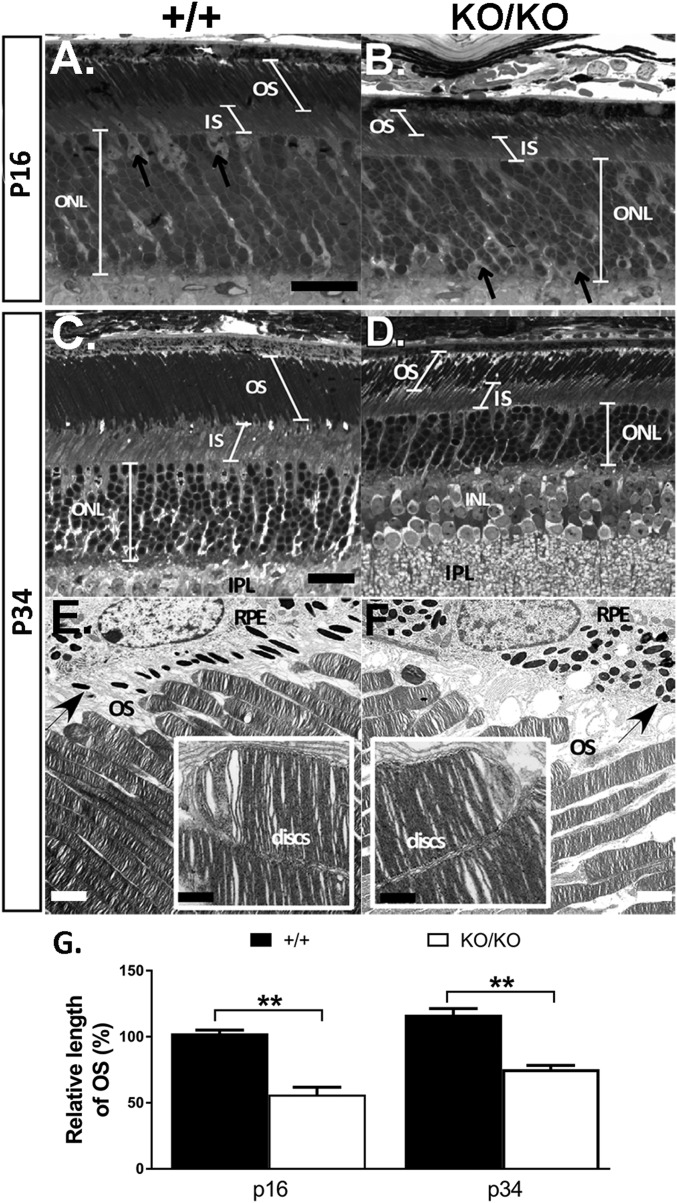
Rod maturation is marked by increasing RHO accumulation accompanied by OS biogenesis (SI Appendix, Fig. S1C) (7). By P16, OSs had developed substantially in wild type and further extended their length from P16 to 1 mo of age (Fig. 4 A, C, and G), reflecting normal OS biogenesis. In the KO retina, OSs appeared significantly shorter compared with wild type at P16 (Fig. 4 A and B). The OSs extended from P16 to P34, but never attained the wild-type length (Fig. 4 B, D, and G). Transmission electron microscopy confirmed that disk structure and organization of the KO was relatively normal at P34 (Fig. 4 E and F, Insets). The inner segment layer (IS) in the KO also appeared shorter at P16 and remained shorter than wild type at later ages (SI Appendix, Fig. S4).
Fig. 4.
Delayed OS biogenesis followed by early onset degeneration. Representative light (A–D) and electron (E and F) micrographs of retinas at P16 (A and B) and P34 (C–F) from miR-183 cluster+/+ (A, C, and E) and miR-183 clusterKO/KO (B, D, and F) mice. Arrows (E and F) point to melanin granules in RPE (retinal pigment epithelium), showing loss of polarized distribution in KO RPE. (G) Progression of OS elongation in miR-183 cluster+/+ and miR-183 clusterKO/KO mice from P16 to P34 shown as percentage of miR-183 cluster+/+ at P16. Measurements were taken from representative light micrographs of three mice at each age. **P < 0.01 (Student’s t test, P values = 0.004 and 0.003 for p16 and p34, respectively). [Scale bar: 16 µm (A–D), 2 µm (E and F), and 500 nm (Insets).] Quantitative measurements of OS, IS, and ONL throughout retina are shown in SI Appendix, Fig. S4.
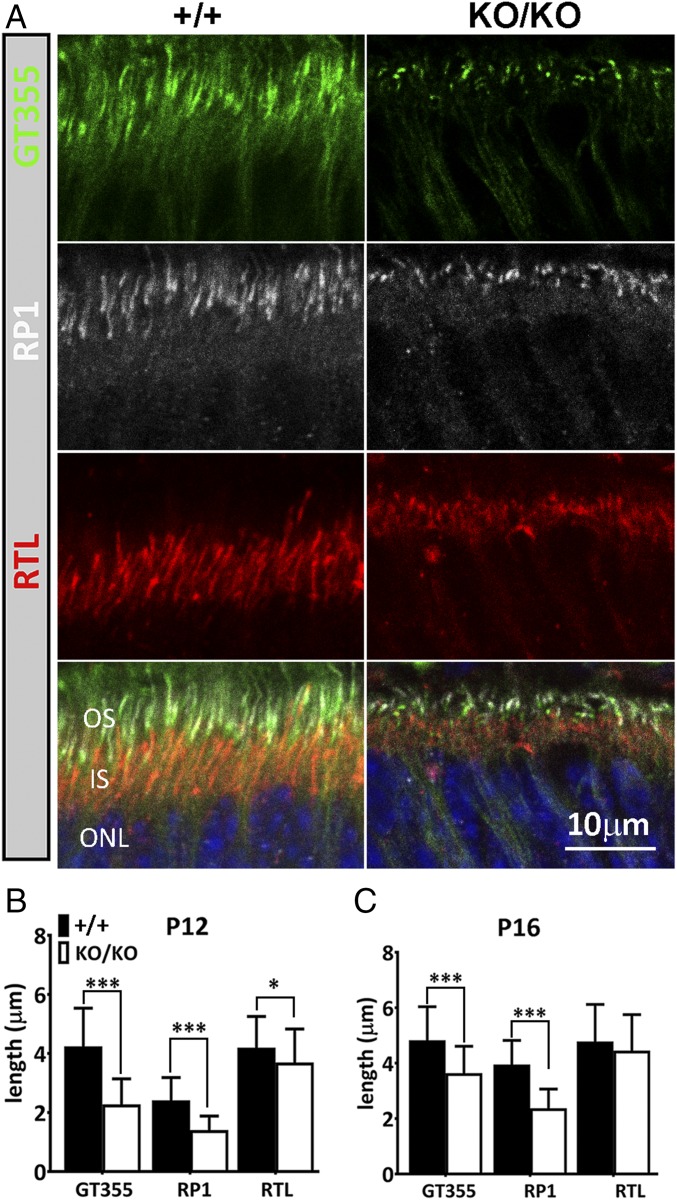
We further examined OS biogenesis with immunostaining of key ciliary components at P12 (Fig. 5 A and B), a period when OS extension of rod photoreceptors is active. Immunostaining of rootletin (RTL), a major component of the ciliary rootlet, which normally extends from the basal body downward toward the nucleus, revealed significant shortening of the rootlet domain in the KO photoreceptors compared with wild type at P12 (Fig. 5 A and B). Similarly, at P12, staining of glutamylated tubulin (GT355), a ciliary epitope normally localized to basal body and connecting cilium (20), revealed dot-like staining highlighting only basal bodies in the KO, whereas in the wild type, the GT355 domain became much more extended distally toward OSs (Fig. 5 A and B). RP1 is a photoreceptor-specific tubulin-binding protein confined to the ciliary axoneme of the OS (21). The immunostaining region for RP1 was significantly shortened in the KO at P12 compared with wild type (Fig. 5 A and B). By P16, however, all three ciliary domains in KO retina extended further (Fig. 5C). Again, this delayed extension of the RTL, GT355, and RP1 domains during early postnatal differentiation is consistent with delayed OS biogenesis in KO mice. In addition to delayed OS biogenesis in early postnatal days, ONL thinning was first seen at P31 (SI Appendix, Fig. S4) and continued to progress. By 5 mo of age more than half of the photoreceptors were lost in the KO retina (Fig. 4 C and D and SI Appendix, Fig. S4). Progressive loss of photoreceptors in the mutant, together with delayed OS extension, likely contributed to the reduction of scotopic a-waves. The morphological changes and ciliary marker staining in the KO retina are consistent with a delayed and defective ciliogenesis.
Fig. 5.
Delayed ciliogenesis of photoreceptors. (A) Frozen sections of retina from wild-type (+/+) and KO (KO/KO) mice at P12 were labeled with GT355 (green), anti-rootletin (RTL, red), and anti-RP1 (white) antibodies, respectively. Superimposed images with DAPI (blue) are shown at Bottom. (B and C) Quantitative measurements of lengths of the three ciliary domains at P12 (B) and P16 (C). Measurements were performed from representative images of three individual mice (n = 3) in Photoshop CC2015. Error bars are SD. *P < 0.05; ***P < 0.0001 (Student’s t test).
Delayed Up-Regulation of Key Photoreceptor Genes.
Whole-retina RNA-seq was performed comparing gene expression profiles between wild type and KO at P5, P11, and P27 (NCBI GEO repository, accession no. GSE95852). RNA-seq data of potential effector genes implicated in driving various phenotypic displays in the KO, which are either directly or indirectly regulated by the miR-183 cluster and discussed throughout this paper, are summarized in SI Appendix, Table S1 along with seed binding in 3′-UTR for direct targets, and relevant GO terms. Expressions of all predicted targets are summarized in Dataset S1.
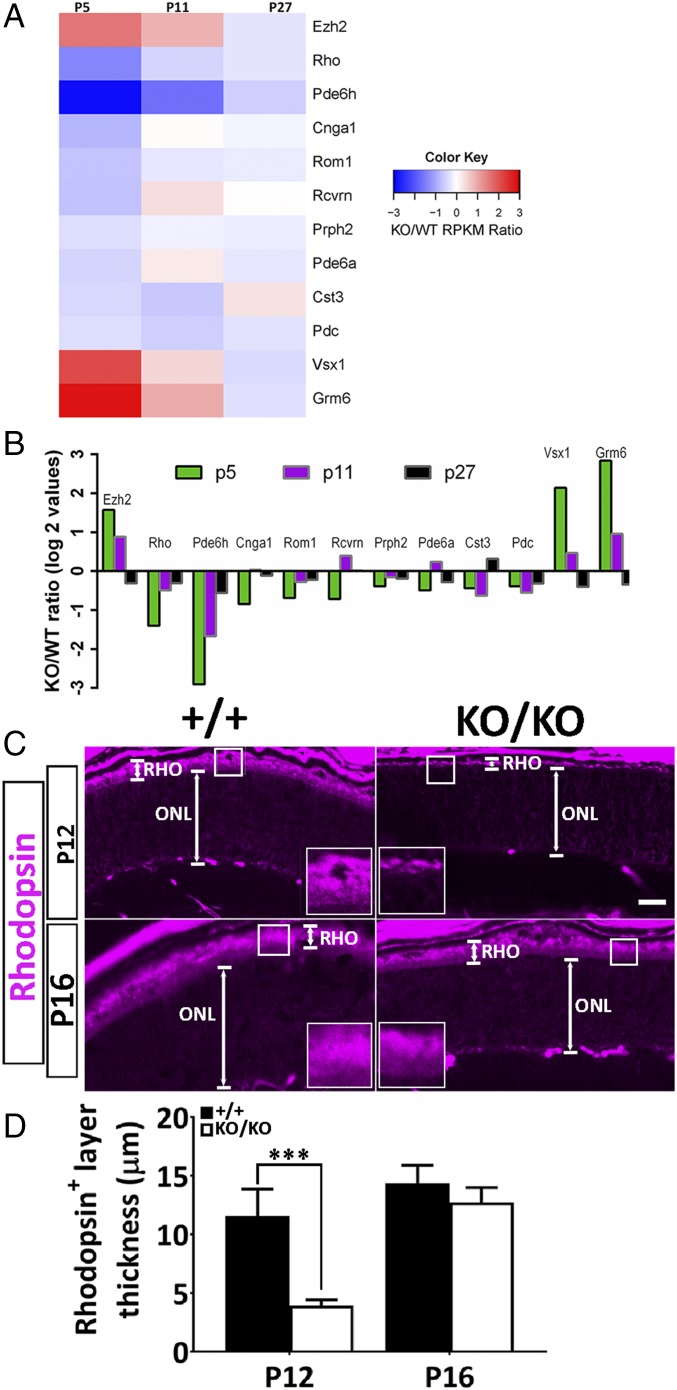
We first inspected temporal expressions of retinal genes potentially involved in driving terminal differentiation events (22) and identified a group of early-onset genes in the retina of KO mice that were initially repressed at P5 and gradually rose back to near wild-type levels by P27 (Fig. 6 A and B). Among those are genes critically involved in OS biogenesis, such as Rom1 and Rho [false discovery rate (FDR) < 0.05], and cone-specific cGMP phosphodiesterase genes (FDR < 0.05), which regulate intracellular cGMP levels (SI Appendix, Table S1). On the contrary, enhancer of zeste2 (Ezh2), the enzymatic component of the polycomb repressive complex 2 (PRC2), was transiently up-regulated threefold at P5 (FDR < 0.05; SI Appendix, Table S1) in KO retina, decreasing to near wild-type levels by P27 (Fig. 6 A and B). Ezh2, which is not a predicted target of the miR-183 cluster, catalyzes the trimethylation of lysine 27 on histone H3 to generate the repressive H3K27me3 marker at target genes (23). In mouse, there are specific temporal and spatial patterns of Ezh2 expression in retinal progenitors, accompanied by dynamic changes in histone modification on target genes (24). Interestingly, most of the early-onset retinal genes identified to be repressed at P5 in the KO (Fig. 6 A and B) are downstream targets of Ezh2 (22, 24, 25). Delayed up-regulation of Rho expression in the KO was also confirmed by immunostaining on P12 and P16 (Fig. 6 C and D) as well as P23 (SI Appendix, Fig. S5) retinas.
Fig. 6.
Delayed up-regulation of key photoreceptor genes. (A) Heat map of RNA-seq data from pooled retinas of miR-183 clusterKO/KO and age-matched wild-type mice at P5, P11, and P27, respectively, showing KO/WT RPKM (reads per kilobase of transcript per million mapped reads) ratio of the total of 12 genes that are key early-onset photoreceptor genes that were dysregulated in the KO retina at P5. KO/WT of Log2 values is also shown as bar graph in B. (C) Immunological staining of rhodopsin-positive domain (RHO, Insets) in the OS of wild-type (+/+) and KO (KO/KO) retinas at P12 and P16, respectively. (D) Quantitative measurements of RHO domains (in micrometers). N (biological replicates) = 3; error bars are SD. ***P < 0.0001 at P12 (Student’s t test).
Like Ezh2, Vsx1, a transcriptional factor regulating terminal differentiation of ON-bipolar cells (26), and Grm6, an ON-bipolar cell-specific glutamate metabotropic receptor (27), were also significantly up-regulated at P5 (FDR < 0.05; SI Appendix, Table S1) and gradually fell back to near wild-type levels by P27 (Fig. 6 A and B). Neither Vsx1 or Grm6 is predicted to be directly targeted by the miR-183 cluster. Conditional ablation of Ezh2 in mouse retina (Dkk3-Cre), however, led to the down-regulation of Vsx1 at P8 retina (25) and a reduced number of Vsx1-positive bipolar cells through mechanisms that remain elusive (24). Premature up-regulation of Vsx1 may subsequently contribute to the up-regulation of Grm6 and PKCα, although PKCα itself is a predicted target of the miR-183 cluster (SI Appendix, Table S1 and Fig. S5).
Resistance to Light-Induced Damage.
The miR-183 cluster has been shown to be regulated by light exposure (28). This observation led to the creation of a transgenic sponge allele (29), which reduced the levels of the miRNAs through sequestration, to test the role of the miR-183 cluster in protecting photoreceptors from light-induced damage. The adult miR-183 cluster sponge mice displayed a more severe retinal degeneration, relative to the controls following acute light exposure, suggesting a protective role. A similar finding was obtained in the hypomorphic miR183GT mutant (12). Our miR-183 cluster-KO mice, however, did not show increased susceptibility to light damage under similar exposure conditions (SI Appendix, Fig. S6 A and B). On the contrary, retinas of the congenic SV129 mice included as a control (30) underwent a much more severe degeneration. The apparent contradiction can be explained by a compromised OS biogenesis and delayed accumulation of rhodopsin in the OS of the KO retina, a phenotype not observed in either the sponge or gene trap model. Damage by acute light is dependent on capture of light energy and active phototransduction. Because shortened OS and lower level of rhodopsin reduce quantum catch, the increased resistance to light damage in the miR-183 cluster mutant would be an expected outcome (30).
Developmental Delay and Arrest in Cochlear and Vestibular Hair Cells.
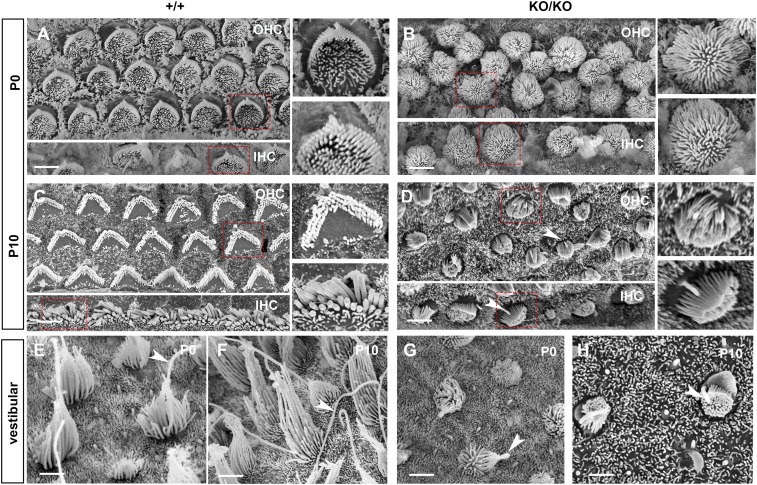
Scanning electron microscopy showed that at P0, hair cells of the organ of Corti in wild-type mice had hair bundles arranged with graded lengths, in a staircase-like arrangement, organized in the characteristic “V” shape, as a result of normal postdevelopmental remodeling (Fig. 7A). The kinocilium was seen clearly, adjacent to the tallest stereocilia at the point of the V. In contrast, in the KO at the same age, dense tufts of microvilli with no gradation in length were observed evenly distributed over the apical surface of hair cells, making the kinocilia difficult to identify, reminiscent of hair cell morphology during embryonic development (4) (Fig. 7B). From P0 onward, development of the cochlear sensory epithelium normally continues with excess stereocilia being resorbed, leaving only three rows of stereocilia and the kinocilium retracted. At P10, these changes were observed in wild-type mice (Fig. 7C). In the KO, however, kinocilia remained easily identifiable (Fig. 7D, arrows) and the hair bundle showed a slight gradation in length and appeared just starting to assume the V shape configuration (Fig. 7D). Overall, the KO cochlear hair cells manifest a highly immature morphology.
Fig. 7.
Developmental delay and arrest of cochlear and vestibular hair cells. Scanning electron micrographs showing the apical surface of sensory hair cells from miR-183 cluster+/+ (+/+, A and C with Insets, and E and F) and miR-183 clusterKO/KO mice (KO/KO, B and D with Insets, and G and H). Examination of hair cells from organ of Corti at P0 (A and B) revealed that stereocilia bundles in the mutant failed to develop the normal staircase arrangement, whereas kinocilium was not visible at the abneural pole (B) as in the wild-type littermate (A). Hair cells from the utricular macula (E and G) at P0 showed that both stereocilia and kinocilia (red arrows) appeared significantly shorter in the mutant (G). In organ of Corti, hair cell bundles at P10 comprise several rows of stereocilia in the mutant and do not form a typical staircase arrangement (D) as in the wild-type littermate control (C). Kinocilia in the mutant (arrows) were retained at P10 (D) and remained immature in appearance. (F and H) Showing hair cells from the utricular macula at P10. Similar to hair cells in the organ of Corti, stereocilia and kinocilia remained short and immature in the mutants (H). IHC, inner hair cell; OHC, outer hair cell.
Additionally, normal cochlear sensory cells are organized into three rows of outer and one row of inner hair cells (Fig. 7A and SI Appendix, Fig. S8C). These cells are strictly aligned with their neighboring cells in terms of planar orientation. This alignment is accomplished through PCP signaling and is critical for hearing. In the KO, hair cell alignment was disrupted, exhibiting no discernible rows at P0 (Fig. 7B), a phenotype that is similar to that seen in other PCP mutants (31) and can also be caused by poorly developed kinocilia.
The delay in hair bundle and kinocilium remodeling in the KO was also manifested in the vestibular organs. At P0, the kinocilia in KO bundles (Fig. 7G, arrowheads) resembled that of the wild type (Fig. 7E) but was much shorter. From P0 to P10 in the wild type, the kinocilia and stereocilia increased substantially in length, with stereocilia forming their characteristic staircase (Fig. 7 E and F). However, in the KO, both kinocilia and stereocilia remained short and the slope of the stereocilia staircase was attenuated (Fig. 7 G and H). Immunofluorescence of whole mount organ of Corti showed the finely structured microtubule arrangements protruding from the base of the kinocilium in wild-type cells (SI Appendix, Fig. S7 G and G′). The kinocilium in KO hair cells was not easily discernible, and acetylated microtubules in the subcuticular plate were increased and disorganized (SI Appendix, Fig. S7 H and H′). The results collectively point toward a delay/arrest in the postnatal maturation of auditory and vestibular cells.
Reduced Population of Mature Sensory Neurons in Olfactory Epithelium.
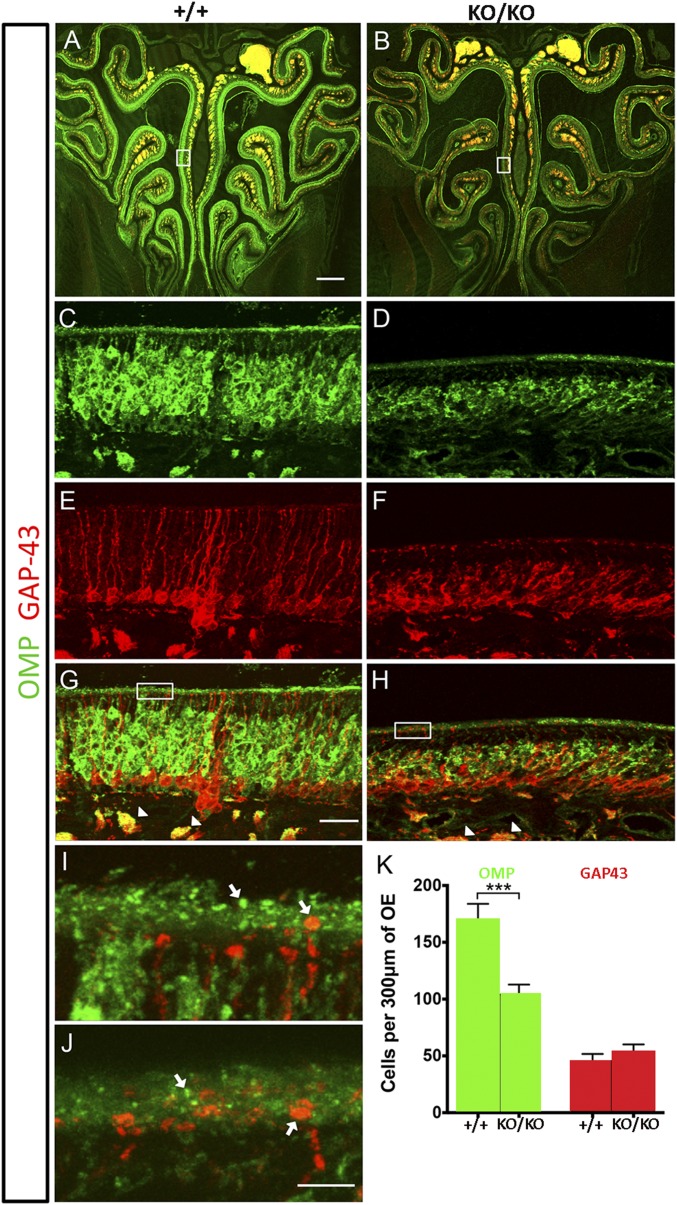
Whereas the number of immature OSNs was the same in KO and wild-type mice, the mature OSN population was reduced by ∼60% in the KO (Fig. 8 A–H and K). In addition, there were obvious changes in cell morphology for OSNs at different ages, with KO OSNs displaying a more compact and irregular shape than their wild-type counterparts (Fig. 8 C–H). The thickness of the olfactory epithelium was also notably reduced in KO mice, though the overall organization appeared comparable (Fig. 8 A–H). Closer inspection revealed differences in the dendritic knobs, which appeared to be less dense in KO mice (Fig. 8 I and J), supporting the idea that the reduced mature OSN population may be due to an olfactory ciliopathy.
Fig. 8.
Reduced population of mature sensory neurons in olfactory epithelium. (A–J) Immunofluorescence of the olfactory epithelium (OE) from 10-wk-old males showing both mature (OMP, green) and immature (GAP-43, red) olfactory sensory neurons (OSNs). (A and B) Low-magnification coronal sections of OE show reduced thickness in miR-183 clusterKO/KO mice compared with miR-183 cluster+/+ mice. (C–H) High magnification of the septal OE (boxed region from A and B) revealed abnormal OSN morphology in miR-183 clusterKO/KO mice compared with miR-183 cluster+/+ mice in both mature (C and D) and immature (E and F) neurons. Arrowheads indicate axon bundles (G and H). (I and J) High-magnification images of the OE ciliary region (boxed area from G and H) showed fewer dendritic knobs (indicated by arrows) in miR-183 clusterKO/KO mice, (J) compared with controls (I). (K) Graph showing OSN numbers in mutant and control mice revealed that mature OMP-positive cells were significantly reduced in miR-183 clusterKO/KO mice (103.9 ± 8.7, n = 4) compared with miR-183 cluster+/+ mice (170.1 ± 13.8, n = 4, *P < 0.001), whereas the number of Gap-43–positive immature neurons in miR-183 clusterKO/KO mice (53.88 ± 6.2, n = 4) and miR-183 cluster+/+ mice (45.2 ± 6.6, n = 4, P = 0.0.3432) was unchanged. Numbers shown as mean ± SEM and t test was used for comparison. (Scale bars, 200 μm in A and B, 25 μm in C–H, and 5 μm in I and J.)
Discussion
Targeted deletion of the miR-183 cluster led to dysfunctions of multiple sensory epithelia, including the retina, organ of Corti, vestibule organs, and olfactory epithelium in mice. All involve defects in key events of terminal differentiation. In photoreceptors, deletion of the miR-183 cluster led to delays in up-regulation of key photoreceptor genes, which in turn led to the delay in OS biogenesis. Similarly, postnatal developmental delay also manifested in auditory and vestibular hair cells as abnormal apical surface remodeling, a key postnatal developmental event. Our data thus support a role of miR-183 cluster in temporal regulation of terminal differentiation in photoreceptors and hair cells.
Many genes showed modified expression in the KO retina. Some are direct targets of the cluster, whereas others appear to be downstream targets. One of the latter category with potential for early effects on differentiation is EZh2, a methyl transferase mediating epigenetic repression of target genes (23). Ezh2 was up-regulated threefold in KO retina at P5 and then fell back to near normal by P27. Also, target genes of EZh2 in photoreceptors were repressed at P5. Rhodopsin, for example, is an essential structural building block of OS, comprising 90% of its mass, and dosage of rhodopsin is proportionally related to the length and volume of OSs in mice (32). Expression of Rho was reduced significantly in the KO at P5 gradually reaching wild-type levels by P27. Among other targets of Ezh2, Rom1, coding for an OS membrane protein required for morphogenesis and maintenance of OSs (33), was also reduced by 40% at P5, gradually approaching wild-type levels by P27. Delayed peaking of key photoreceptor genes in the mutant is consistent with shifted timing of terminal differentiation.
Indeed, Ezh2 is required in postnatal retina to maintain progenitor proliferation, and its decline coordinates the timing of terminal differentiation of retinal cell populations. Retinal-specific ablation of Ezh2 leads to acceleration of photoreceptor terminal differentiation with premature expression of key photoreceptor genes in the postnatal retina (22, 24). Dynamics of Ezh2 expression in the developing cochlear sensory epithelium also point to a regulatory role in terminal differentiation (34), but no similar data are available in the olfactory epithelium at present. Comparison of RNA-seq data among different sensory neuroepithelia of the KO mice can potentially provide insight into the functional relationship between gene networks modulated by the miR-183 cluster and the epigenetic mechanisms involving Ezh2 in regulating terminal differentiation.
In each of the sensory systems examined, miR-183 cluster-KO mice exhibit ciliary disruption, indicating that ciliogenesis, a key event in terminal differentiation of sensory receptor cells, is likely regulated by miR-183 cluster miRNAs. Ciliogenesis was delayed and incomplete in photoreceptors. Similar observations were made in hair cells of both organ of Corti and vestibule organs with defects of kinocilium extension and postnatal modification, whereas in the olfactory epithelium, there was a noticeable reduction in the density of dendritic knobs. Consistent with a regulatory role in ciliogenesis, several genes involved in ciliary length control, intraflagellar transport (IFT), and vesicle docking or apical membrane trafficking were dysregulated in the mutant retina at P11, a time point when photoreceptors are actively engaged in OS biogenesis (SI Appendix, Table S1).
Mak, the male germ cell-associated kinase, was up-regulated in the KO retina at P11 (SI Appendix, Fig. S8A and Table S1). Mak overexpression reduces the RP1 domain in mammalian cilia by phosphorylating RP1 (35), which is required for OS disk stacking (36). Mak is a predicted target of both miR-182 and miR-96 (SI Appendix, Table S1). Luciferase assays demonstrated that mimetics of the miR-183 cluster bound specifically to the mouse Mak 3′-UTR in HEK293 cells (SI Appendix, Fig. S8B). Loss of regulation of Mak by the miR-183 cluster during early OS biogenesis is consistent with ciliogenesis deficiency in the KO retina. Additional regulators of ciliogenesis that appeared dysregulated at P11 (SI Appendix, Table S1) included BBs10, Ttc21b, BBs12, cc2d2a, Lca5, Rp1, Rab8b, Cep250, and Mapre1, which have established roles in IFT, vesicle docking, or apical membrane trafficking in ciliogenesis (SI Appendix, Table S1). Among those, several are predicted targets of the miR-183 cluster, including Cep250, which was up-regulated at P5 (SI Appendix, Table S1), and Rab8b and Mapre1 (EB1), which were down-regulated. Whereas up-regulation of targets in the KO may be caused by loss of direct interactions with the miRNAs, down-regulation of others must result from downstream effects of miR-183 cluster mutation (SI Appendix, Table S1). Effects of miRNA regulation on individual targets, either direct or indirect, may not be large. Each of the ciliary pathway genes affected had relatively small changes in expression ranging from 1.8- to 2.8-fold only (SI Appendix, Table S1), arguing for a coordinated fine-tuning effect within the gene networks of ciliary biogenesis. This idea is consistent with ciliogenesis being tightly regulated at its beginning stage at P11. Expression of critical genes, either structural or regulatory, must be maintained within a narrow range. This task is uniquely suited for the biological roles of miRNAs, which serve to fine tune expressions of target genes and those networked with the target genes to keep them in relatively narrow ranges.
Nuclear positioning plays a role in development of a variety of cell types in vertebrates (37). In developing neuroepithelia, for example, nuclei of undifferentiated progenitors oscillate between the apical and the basal surfaces of polarized epithelia in synchrony with the cell cycle progression as a regulatory mechanism of cell division and differentiation, a process known as interkinetic nuclear migration (38). Another nuclear movement event, often referred to as nuclear translocation (NT), occurs following terminal mitosis and is considered as a terminal differentiation event to coordinate cellular migration (39) and synaptogenesis (19) within neuroepithelia. In vertebrate retina, NT occurs in precursors of ganglion (40), bipolar (41), and cone photoreceptor cells (19). In the wild-type retina, we observed that cone nuclei started moving apically from as early as P9 and were confined apically within the ONL by P14, confirming a previous report on a so-called late phase NT of cone nuclei in mouse retina (19). In the KO, however, the apical distribution of cone nuclei was largely abolished, indicating a regulatory role of the miR-183 cluster in late phase NT. Nuclear repositioning during development is mediated by interactions between SUN and KASH families, known as linkers of the nucleoskeleton to the cytoskeleton (LINC complexes) (42). Assemblies of LINC complexes connect the nuclear lamina to cytoplasmic cytoskeletal networks recruiting motors to mediate nuclear movement relative to cell bodies. Not surprisingly, cell autonomous cone nuclei mispositioning has been associated with mutations in members of the LINC complex such as KASH protein Syne-2/Nesprin-2, and SUN proteins SUN1/2 (43). Intriguingly, two of the components of the LINC complex, Syne2 and Kif2A, are predicted targets of the miR-183 cluster and showed modest increase at P5 by RNA-seq in the KO retina (1.5- and 1.3-fold, respectively, SI Appendix, Table S1). Syne2 is anchored on the inner leaflet of the nuclear envelope, whereas Kif2a is a microtubule-associated protein.
A similar phenotype of late NT deficiency has been described in the cone cyclic nucleotide-gated channel Cnga3 gene knockout mice (44), and a loss-of-function mutation of the gene coding for the catalytic subunit of cone-specific cGMP phosphodiesterase (Ped6c) in cpfl1 mice (45). Both Cnga3 and Ped6c genes are components of the vertebrate phototransduction cascade. The cGMP-gated Cnga3 regulates cationic influx in cones, and Pde6 cleaves intracellular cGMP in response to phototransduction activation. Loss-of-function mutation in either Cnga3 or Pde6c led to intracellular accumulation of cGMP, which was linked to photoreceptor death (45, 46). Whereas it is not known whether changes in intracellular cGMP levels in cones in either mutant played a role in the development of late-phase NT deficiency, it is conceivable that cGMP, an important second messenger, can impact many cellular homeostatic processes. In the KO retina, whereas expressions of genes coding for rod-specific PDE6 subunits α, β, and γ are near normal, cone-specific genes Pde6c (α′) and Pde6h (γ) are significantly reduced to 52% and 13% of wild-type levels, respectively, at P5 (SI Appendix, Table S1). Notably, Pde6h is a downstream target of Ezh2 (22) (Fig. 6 A and B). Thus, it appears that the miR-183 cluster regulates the late-phase NT directly and indirectly through parallel pathways, a notion supported by the observation that the miR-183 cluster-KO mutation led to a much more severe phenotype of deficient late NT than any of the above discussed mutations alone, possibly through additive or synergistic effects of disrupting parallel regulatory mechanisms. The importance of NT in cones is further highlighted by the preferential loss of cones with nuclei mislocalized to the bottom third of ONL after P14 in the KO mutant mice.
Our study uncovered striking postnatal developmental defects in auditory and vestibular hair cells where the miRNA-183 cluster is expressed during postnatal differentiation (47). In a previous study, defects in stereocilia remodeling of auditory hair cells were observed in an ENU-induced mouse mutant, Diminuendo, in which a point mutation disrupts the seed region of miR-96 (48), but the phenotype was much milder compared with KO mice. The mechanisms underlying this observation remain somewhat unclear as the seed change led to down-regulation of a set of genes with target sites complementary to the mutant seed, raising the possibility that Diminuendo is a dominant mutation. Our current study reveals that ablating the entire cluster results in severe developmental defects in both the auditory and vestibular hair cell stereocilia. It also indicates that miR-182 and -183 likely play previously unidentified, critical roles in both auditory and vestibular hair cell development and function.
Previous studies have found miR-183 cluster expression in the olfactory epithelium (9). Here we confirmed that all three miRNAs of the cluster were abundantly expressed in adult olfactory epithelium. We further show that loss of the miR-183 cluster not only significantly reduced the mature OSN population but also produced structural and functional changes to mature OSNs. Because OSNs undergo continuous turnover, the persistence of these olfactory phenotypes in adults demonstrates that the role of the miR-183 cluster in promoting maturation extends beyond olfactory epithelium development and is essential for OSN regeneration.
Taken together, the miR-183 cluster plays indispensable roles during sensory receptor maturation. Whereas there were typically ∼1,500 predicted target genes of the miR-183 cluster expressed in the retina at each stage examined, most had only small changes in expression in the KO (Dataset S1). This finding is consistent with a recent view that rather than acting as genetic switches of a few key individual genes in a binary fashion, miRNAs are fine tuners of large number of networked genes, through both direct interaction with their target genes and downstream network effects (49). The picture emerging from our current work is that the miR-183 cluster, with three component miRNAs that share redundancies (50), has evolved in ciliated sensory neurons to regulate key pathways of terminal differentiation. The miR-183 cluster-KO allele established in this study thus can serve as a valuable tool for further mechanistic investigation into the postmitotic developmental processes of sensory receptors.
Materials and Methods
The use of mice in this study has been approved by the National Eye Institute Animal Care and Use Committee, and handling of animals follows the NIH Animal Care and Use Guidelines (51). The miR-183 cluster deletion was created by homologous recombination. Functions of vison, hearing, and smell were evaluated by ERG, ABR test, and buried food assays, respectively. Expression of makers of the retina, cochlear, and vestibular sensory epithelia and olfactory epithelium was investigated by immunohistochemistry. Morphological changes in mutant retina and hair cells were examined by light and electron microscopy. Full descriptions of methodologies can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Sheldon Miller, Doris Wu, Fielding Hejtmancik, and Michael Redmond for critical reading of the manuscript; Megan Kopera for her assistance in mouse colony management of the miR-183 cluster-KO line; Hideko Takahashi for her assistance in in vitro fertilization during the study; Dr. Yingwei Chen for her help in genotyping and immunostaining experiments; and Matthew Brooks for his assistance in initial RNA-seq experiments. J.F., L.J, Y.L., P.L., J.L., H.Q., T.L., W.L., G.W., and L.D. were supported by NIH/National Eye Institute/Intramural Research Program (IRP); S.E., R.J.M., and B.K. were supported by NIH/National Institute on Deafness and Other Communication Disorders/IRP; and A.W. and L.B. were supported by NIH/National Institute of Neurological Disorders and Stroke/IRP, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Complete RNA-seq datasets from the miR-183 cluster knockout mice we generated in this study (C57BL/6 background) at postnatal 5-, 11-, and 27-d time points have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE95852).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619442114/-/DCSupplemental.
References
- 1.Streit A. The Cranial Sensory Nervous System: Specification of Sensory Progenitors and Placodes. StemBook; Cambridge, MA: 2008. [PubMed] [Google Scholar]
- 2.Falk N, Lösl M, Schröder N, Gießl A. Specialized cilia in mammalian sensory systems. Cells. 2015;4:500–519. doi: 10.3390/cells4030500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg AF, Moritz OL, Williams DS. Molecular basis for photoreceptor outer segment architecture. Prog Retin Eye Res. 2016;55:52–81. doi: 10.1016/j.preteyeres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- 5.Menco BP, Bruch RC, Dau B, Danho W. Ultrastructural localization of olfactory transduction components: The G protein subunit Golf alpha and type III adenylyl cyclase. Neuron. 1992;8:441–453. doi: 10.1016/0896-6273(92)90272-f. [DOI] [PubMed] [Google Scholar]
- 6.Ross AJ, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- 7.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decembrini S, et al. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci USA. 2009;106:21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 10.Weston MD, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce ML, et al. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci USA. 2013;110:E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busskamp V, et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron. 2014;83:586–600. doi: 10.1016/j.neuron.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Alagramam KN, Stahl JS, Jones SM, Pawlowski KS, Wright CG. Characterization of vestibular dysfunction in the mouse model for Usher syndrome 1F. J Assoc Res Otolaryngol. 2005;6:106–118. doi: 10.1007/s10162-004-5032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M, Crawley JN. 2009. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci Chapter 8:Unit 8.24.
- 16.Gresh J, Goletz PW, Crouch RK, Rohrer B. Structure-function analysis of rods and cones in juvenile, adult, and aged C57bl/6 and Balb/c mice. Vis Neurosci. 2003;20:211–220. doi: 10.1017/s0952523803202108. [DOI] [PubMed] [Google Scholar]
- 17.Karali M, et al. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- 19.Rich KA, Zhan Y, Blanks JC. Migration and synaptogenesis of cone photoreceptors in the developing mouse retina. J Comp Neurol. 1997;388:47–63. [PubMed] [Google Scholar]
- 20.Sun X, et al. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc Natl Acad Sci USA. 2016;113:E2925–E2934. doi: 10.1073/pnas.1523201113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Zuo J, Pierce EA. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J Neurosci. 2004;24:6427–6436. doi: 10.1523/JNEUROSCI.1335-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno K, et al. Transition of differential histone H3 methylation in photoreceptors and other retinal cells during retinal differentiation. Sci Rep. 2016;6:29264. doi: 10.1038/srep29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gall Trošelj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex’s evolutionary conserved function: The role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, et al. Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation. Dev Biol. 2015;403:128–138. doi: 10.1016/j.ydbio.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iida A, et al. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Dev Neurobiol. 2015;75:947–960. doi: 10.1002/dneu.22261. [DOI] [PubMed] [Google Scholar]
- 26.Shi Z, et al. Vsx1 regulates terminal differentiation of type 7 ON bipolar cells. J Neurosci. 2011;31:13118–13127. doi: 10.1523/JNEUROSCI.2331-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dryja TP, et al. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci USA. 2005;102:4884–4889. doi: 10.1073/pnas.0501233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krol J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Q, et al. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem. 2011;286:31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J Neurosci. 2001;21:53–58. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139:1806–1820. doi: 10.1242/dev.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price BA, et al. Rhodopsin gene expression determines rod outer segment size and rod cell resistance to a dominant-negative neurodegeneration mutant. PLoS One. 2012;7:e49889. doi: 10.1371/journal.pone.0049889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kevany BM, et al. Structural and functional analysis of the native peripherin-ROM1 complex isolated from photoreceptor cells. J Biol Chem. 2013;288:36272–36284. doi: 10.1074/jbc.M113.520700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Li W, Li W, Chai R, Li H. Spatiotemporal expression of Ezh2 in the developing mouse cochlear sensory epithelium. Front Med. 2016;10:330–335. doi: 10.1007/s11684-016-0459-6. [DOI] [PubMed] [Google Scholar]
- 35.Omori Y, et al. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci USA. 2010;107:22671–22676. doi: 10.1073/pnas.1009437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Lyubarsky A, Skalet JH, Pugh EN, Jr, Pierce EA. RP1 is required for the correct stacking of outer segment discs. Invest Ophthalmol Vis Sci. 2003;44:4171–4183. doi: 10.1167/iovs.03-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bone CR, Starr DA. Nuclear migration events throughout development. J Cell Sci. 2016;129:1951–1961. doi: 10.1242/jcs.179788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosodo Y. Interkinetic nuclear migration: Beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghashghaei HT, Lai C, Anton ES. Neuronal migration in the adult brain: Are we there yet? Nat Rev Neurosci. 2007;8:141–151. doi: 10.1038/nrn2074. [DOI] [PubMed] [Google Scholar]
- 40.Hinds JW, Hinds PL. Early ganglion cell differentiation in the mouse retina: An electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- 41.Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, et al. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michalakis S, et al. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci. 2005;46:1516–1524. doi: 10.1167/iovs.04-1503. [DOI] [PubMed] [Google Scholar]
- 45.Trifunović D, et al. cGMP-dependent cone photoreceptor degeneration in the cpfl1 mouse retina. J Comp Neurol. 2010;518:3604–3617. doi: 10.1002/cne.22416. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, et al. cGMP accumulation causes photoreceptor degeneration in CNG channel deficiency: evidence of cGMP cytotoxicity independently of enhanced CNG channel function. J Neurosci. 2013;33:14939–14948. doi: 10.1523/JNEUROSCI.0909-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sacheli R, et al. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9:364–370. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Lewis MA, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han YC, et al. An allelic series of miR-17 ∼ 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet. 2015;47:766–775. doi: 10.1038/ng.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin ZB, et al. Targeted deletion of miR-182, an abundant retinal microRNA. Mol Vis. 2009;15:523–533. [PMC free article] [PubMed] [Google Scholar]
- 51.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academics Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.