Significance
Integrins are cell-surface molecules that link extracellular ligands to the cytoskeleton. Force exerted by the cytoskeleton that is resisted by the ligand is thought to be important in activating the integrin by stabilizing an extended-open conformational state with high affinity for ligand. However, integrin αVβ8 does not interact with the cytoskeleton in the same way. Here, we show that, although the closely related integrins αVβ8 and αVβ6 bind the same ligand, pro-TGF-β1, their conformational responses to ligand binding and regulation by metal ions are quite different. These differences correlate with their distinct linkage to the cytoskeleton.
Keywords: integrins, allostery, pro-TGF-β1
Abstract
Integrins αVβ6 and αVβ8 are specialized for recognizing pro-TGF-β and activating its growth factor by releasing it from the latency imposed by its surrounding prodomain. The integrin αVβ8 is atypical among integrins in lacking sites in its cytoplasmic domain for binding to actin cytoskeleton adaptors. Here, we examine αVβ8 for atypical binding to pro-TGF-β1. In contrast to αVβ6, αVβ8 has a constitutive extended-closed conformation, and binding to pro-TGF-β1 does not stabilize the open conformation of its headpiece. Although Mn2+ potently activates other integrins and increases affinity of αVβ6 for pro-TGF-β1 25- to 55-fold, it increases αVβ8 affinity only 2- to 3-fold. This minimal effect correlates with the inability of Mn2+ and pro-TGF-β1 to stabilize the open conformation of the αVβ8 headpiece. Moreover, αVβ8 was inhibited by high concentrations of Mn2+ and was stimulated and inhibited at markedly different Ca2+ concentrations than αVβ6. These unusual characteristics are likely to be important in the still incompletely understood physiologic mechanisms that regulate αVβ8 binding to and activation of pro-TGF-β.
Integrins are cell-surface adhesion molecules that mediate cell–cell, cell–extracellular matrix, and cell–pathogen interactions. In mammals, 24 different integrins are formed by specific, noncovalent association of 18 α-subunits with 8 β-subunits (1–5). Almost all integrins link to the actin cytoskeleton through talin and kindlin-binding motifs in their β-subunit cytoplasmic domains and thus provide traction for cell migration. The only exceptions are integrin αVβ8, which binds through its β8-subunit to differentially express in adenocarcinoma of the lung (DAL-1, also known as Band 4.1B), and integrin α6β4, which links through its β4-subunit to intermediate filaments (6).
Here, we focus on the atypical integrin αVβ8 and compare it to integrin αVβ6. Integrins αVβ6 and αVβ8 are specialized for binding to and activation of pro-TGF-β1 and -β3. The pro-TGF-βs are biosynthesized and stored in tissues as latent forms. The dimeric TGF-β growth factor is kept latent by noncovalent association with its dimeric prodomain, which in turn is linked noncovalently and through disulfide bonds to a “milieu molecule” that stores the latent complex in the extracellular matrix or on the cell surface for subsequent, integrin-dependent activation. Integrins bind to an RGD motif that is present in the prodomains of pro-TGF-β1 and -β3. However, integrin binding alone is not sufficient for latent TGF-β activation by αVβ6; force is also required. Experiments suggest that tensile force is transmitted from the cytoskeleton to the integrin, is resisted by anchorage of TGF-β1 in the extracellular matrix or on cell surfaces, and results in distortion of prodomain straitjacket elements that loosely surround the growth factor followed by release of the growth factor (7, 8). In some systems, activation of TGF-β1 by αVβ8 is dependent on matrix metalloprotease (9) whereas in others activation requires linkage of pro-TGF-β1 to a surface, suggesting a role for force exertion (10).
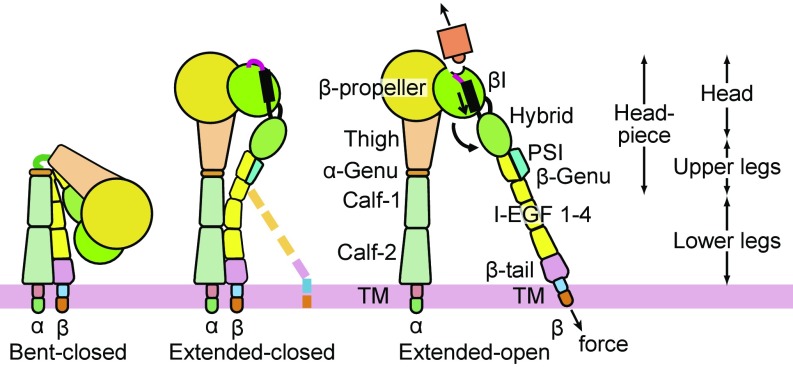
We examine here whether association of integrins αVβ6 and αVβ8 with different types of cytoskeletal adaptor proteins correlates with differences in regulation of the conformation and ligand-binding affinity of their ectodomains when they interact with pro-TGF-β1. Classical integrins that associate with talins and kindlins, including β1, β2, β3, and β7 integrins, as well as αVβ6, exhibit three conformational states, and conformational change is proposed to be regulated by the adaptor proteins and tensile forces that the cytoskeleton exerts when integrins bind to immobilized ligands (Fig. 1) (2, 4). Such integrins exhibit a bent-closed conformation in which the α- and β-leg domains are bent at their knees and the ligand-binding headpiece associates with the lower legs. Knee extension gives an extended-closed conformation in which the legs straighten and the head moves much farther from the ectodomain C-termini that connect to the transmembrane domains. Finally, in headpiece opening, conformational change occurs in the ligand-binding domain in the β-subunit, the βI domain. The βI domain is inserted into the hybrid domain. Rearrangements that increase affinity at the ligand-binding interface are relayed by βI domain α-helix pistoning to the hybrid domain interface, resulting in pivoting of the hybrid domain away from the α-subunit (Fig. 1). The extended-open integrin conformation has much higher ligand binding affinity than the bent-closed or extended-closed conformations (11, 12). Previously, it has been reported that integrin αVβ8 is constitutively extended and does not undergo headpiece opening in the presence of an RGD peptide (13). However, the affinity of the RGD peptide for αVβ8 was not known, and RGD peptides that lack an LXX(I/L) motif present in pro-TGF-β1 and -β3 have substantially lower affinity for αVβ6 than intact pro-TGF-β1 (14). RGD peptides are too small to be visualized in negative stain EM, and thus it was unclear whether RGD had remained bound, or had dissociated, before visualization of αVβ8 in EM. Furthermore, even if RGD had bound, it was unclear whether intact pro-TGF-β1 would be capable of stabilizing the open headpiece conformation of αVβ8. To stabilize the open conformation, ligands must bind with sufficiently higher affinity to the open than the closed conformation to drive conformational change to the higher energy open headpiece conformation. Because such stabilization has been seen with β1, β2, β3, and β6 integrins (2, 15), it was important to test whether αVβ8 was truly resistant to the ligand-induced headpiece opening with a high-affinity, biological ligand. Moreover, affinity measurements are of fundamental importance for understanding integrin interactions with their biological ligands. Therefore, we have measured the affinity for pro-TGF-β1 of αVβ8 and compared it to the affinity of αVβ6.
Fig. 1.
Schematics showing the three major integrin conformational states.
The metal ion dependence of the ligand-binding affinity of αVβ8 is also of interest. Integrin βI domains contain a Mg2+ ion and two Ca2+ ions. The Mg2+ ion directly coordinates ligand at the metal ion-dependent adhesion site (MIDAS). The two Ca2+ ions bind nearby and stabilize the conformation of ligand-binding loops. The adjacent-to-MIDAS (ADMIDAS) Ca2+ ion moves ∼6 Å between the open and closed conformations. Mn2+ is a commonly used activator of integrins and has been shown to work for integrin α4β7 by replacing Ca2+ at the ADMIDAS, which differs in coordination geometry in the closed and open conformations (16, 17). For several integrins, Mn2+ can induce headpiece opening even in the absence of ligand binding (2). Thus, it is interesting to compare effects of Mn2+ on αVβ6 and αVβ8 and explore correlations between headpiece opening and the ability of Mn2+ to increase ligand-binding affinity. Furthermore, because Ca2+, Mg2+, and Mn2+ may all contribute to regulating the integrin headpiece opening, we wondered if ligand-binding affinity would be regulated differently by metal ions in αVβ8 than in αVβ6. Here, we report that integrin αVβ8 is indeed atypical in conformation, in responsiveness to Mn2+ of its affinity for pro-TGF-β1, and in regulation of its ligand-binding affinity by Ca2+ and Mn2+.
Results
Negative Stain EM Comparisons of Integrin αVβ8 and αVβ6 Ectodomain and Headpiece Fragments and Their Complexes with pro-TGF-β1.
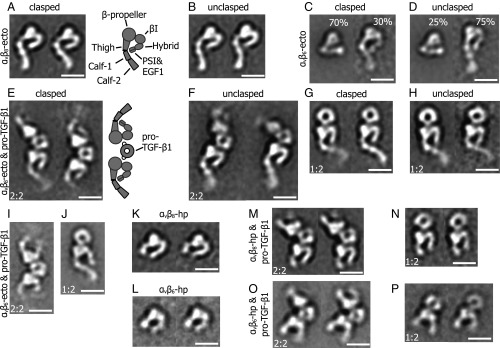
We carried out negative stain EM on six distinct preparations of αVβ8 including complexes with pro-TGF-β1. For comparison, we also carried out negative-stain EM on two αVβ6 ectodomain preparations. Integrin fragments and their complexes with pro-TGF-β1 were subjected to gel filtration (Fig. 2). Peak fractions were subjected to negative-stain EM, and >5,000 particles were subjected to multivariant grouping and averaging in 50 classes (Fig. 3 and Figs. S1–S8). The αVβ8 ectodomain was extended with its headpiece in the closed conformation, i.e., with the upper β-leg including the hybrid domain swung in, toward the αV subunit (Fig. 3 A and B). The αV- and β8-subunits are readily distinguished by the larger size of the β-propeller than the βI domain in the head. Discrete densities were clear in the αV-subunit for the β-propeller, thigh, calf-1, and calf-2 domains. Similar orientations between the thigh and calf-1 domains in class averages suggested that thigh and calf-1 domains adopted a uniform orientation after extension at the α-subunit knee. The β8-subunit densities were clear for the βI and the hybrid domain. The length of the hybrid domain density suggested that it might also include density for the PSI and I-EGF1 domains. No density was present for the β-subunit lower leg I-EGF2-4 and β-tail domains. Essentially, identical class averages were obtained for ectodomain preparations with and without a C-terminal coiled-coil clasp (Fig. 3 A and B). No density was evident for the lower β-leg in either preparation, suggesting that it is highly flexible. In contrast, αVβ6 ectodomain preparations adopted both bent-closed and extended-closed conformations (Fig. 3 C and D). The proportions of bent-closed and extended-closed particles were ∼3:1 in clasped and ∼1:3 in unclasped particles, suggesting that the clasp stabilizes the bent conformation.
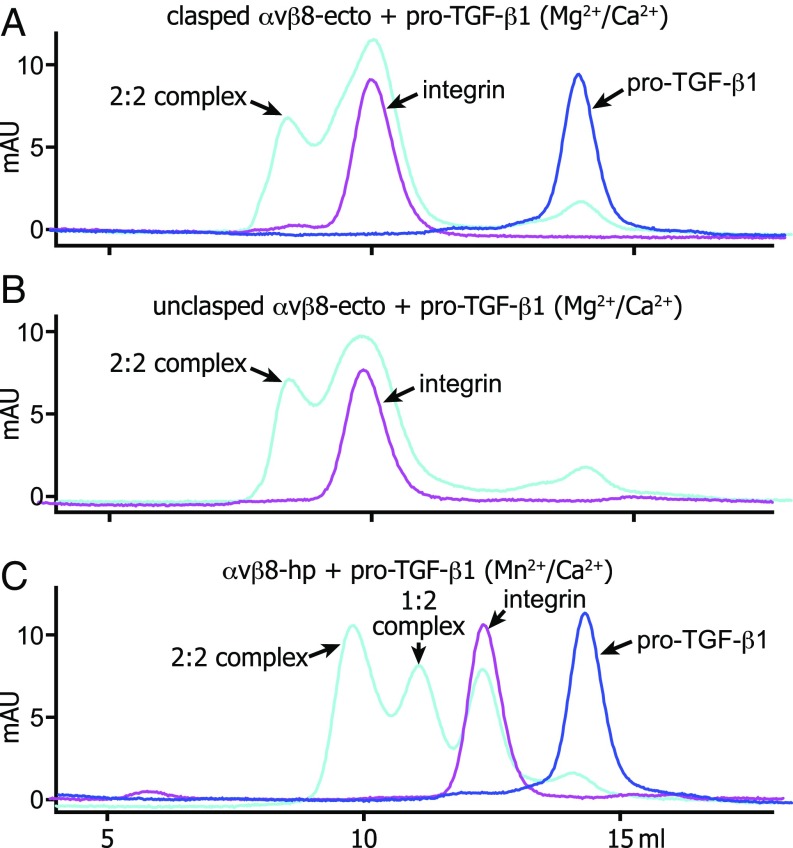
Fig. 2.
Gel filtration of integrin complexes with pro-TGF-β1. Superdex 200 gel-filtration profiles of clasped αvβ8 ectodomain (A), unclasped αvβ8 ectodomain (B), and αvβ8 headpiece (C) complex with pro-TGF-β1 in the presence of Mg2+ or Mn2+ in HBS buffer. To form a protein complex, 10 μg αvβ8 ectodomain or headpiece was mixed with 5 μg pro-TGF-β1 in 50 μL and incubated on ice for 30 min before injection.
Fig. 3.
Representative class averages of negatively stained αvβ8 and αvβ6 and their complexes with pro-TGF-β1. (A and B) The αvβ8 ectodomain (ecto) adopts an extended-closed conformation when either clasped (A) or unclasped (B). (C and D) The clasped and unclasped αvβ6 ectodomain exhibits both bent-closed and extended-closed conformations. The percentage of particles with each conformation is shown. (E–H) Clasped and unclasped αvβ8 ectodomain complexes with pro-TGF-β1 showed both 2:2 and 1:2 complexes. (I and J) The αvβ6 ectodomain 2:2 and 1:2 complexes with pro-TGF-β1. (K and L) The αvβ8 and αvβ6 headpieces adopt the closed conformation in the absence of bound ligand. (M and N) The 2:2 and 1:2 complexes of the αvβ8 headpiece and pro-TGF-β1. The headpiece remains closed. (O and P) The 2:2 and 1:2 complexes of the αvβ6 headpiece and pro-TGF-β1. Ligand binding induces headpiece opening. All samples were prepared in Mg2+/Ca2+, except the αvβ8 headpiece complex with pro-TGF-β1 was prepared in Mn2+/Ca2+. The αvβ6 class averages in I, J, L, O, and P were previously published (15, 18) and are shown for comparison. (Scale bar: 10 nm.)
Fig. S1.
Class averages of clasped αvβ8 ectodomain (5,445 particles) by multireference alignment and K-means classification.
Fig. S8.
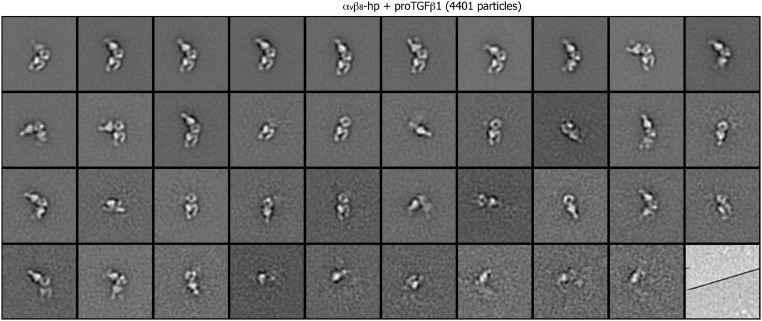
Class averages of αvβ8 headpiece complexed with pro-TGF-β1 (4,401 particles) by multireference alignment and K-means classification.
Fig. S3.
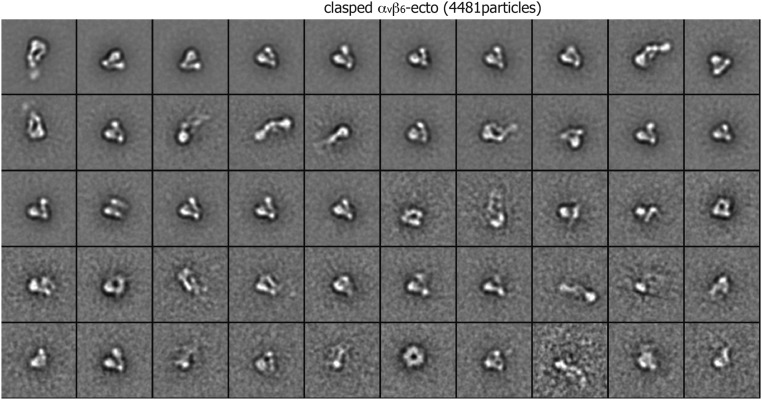
Class averages of clasped αvβ6 ectodomain (4,481 particles) by multireference alignment and K-means classification.
Fig. S4.
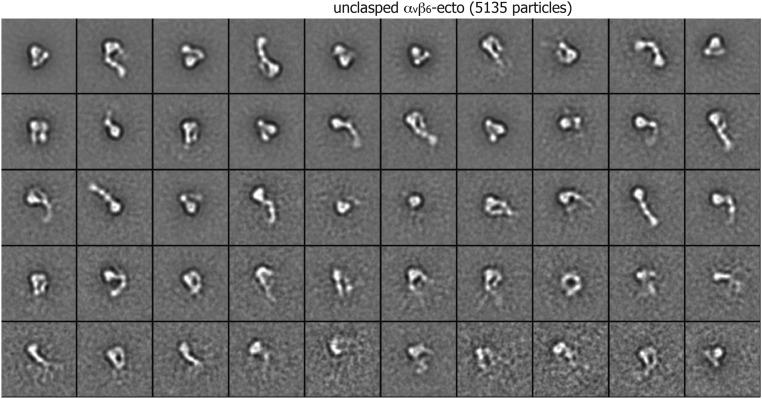
Class averages of unclasped αvβ6 ectodomain (5,135 particles) by multireference alignment and K-means classification.
Fig. S7.
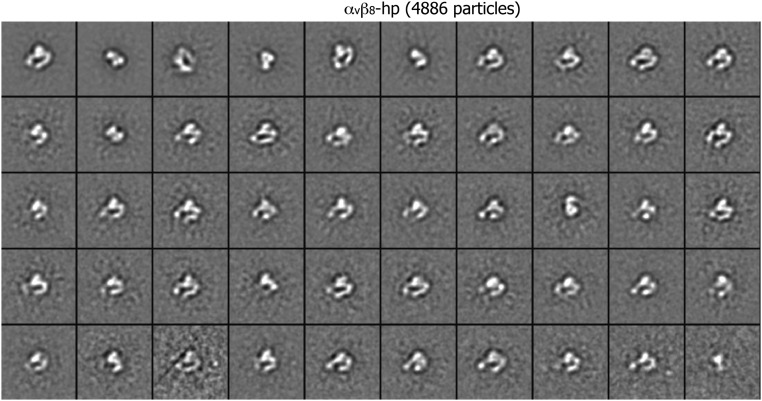
Class averages of αvβ8 headpiece (4,886 particles) by multireference alignment and K-means classification.
The αVβ8 ectodomain formed a complex with pro-TGF-β1 that was stable to gel filtration in buffer containing 1 mM Mg2+ and 1 mM Ca2+ (Fig. 2 A and B). Negative-stain EM on the complex peak showed class averages representing both 2:2 and 1:2 αVβ8:TGF-β1 complexes (Fig. 3 E–H). The integrins bound at the interface between their β-propeller and βI domains to the ring-shaped pro-TGF-β1. In both 2:2 and 1:2 αVβ8 complexes, the headpiece was closed, with the β-subunit hybrid domain pointing toward the interface between the αV thigh and calf-1 domains, as in the uncomplexed ectodomain. In contrast, in αVβ6:TGF-β1 complexes the headpiece was open with the hybrid domain swung away from the α-subunit (18) (Fig. 3 I and J).
Because Mn2+ potently activates integrins, we examined whether Mn2+ combined with pro-TGF-β1 could induce headpiece opening of αVβ8 and extended our studies to headpiece fragments. On its own, the αVβ8 headpiece was closed (Fig. 3K), as was the αVβ6 headpiece (Fig. 3L). We next examined particles from a 2:2 αVβ8:TGF-β1 complex peak from gel filtration in 1 mM Mn2+ and 0.2 mM Ca2+ (Fig. 2C). Most class averages showed 2:2 complexes, and a minority showed 1:2 complexes (Fig. 3 M and N). In 2:2 complexes, the better-resolved αVβ8 integrin headpiece, which was more coplanar with the grid, was clearly closed, whereas the hybrid domain was out of the plane for the other monomer (Fig. 3M). In 1:2 αVβ8:TGF-β1 headpiece complexes, the headpiece was clearly closed with an acute bend of the hybrid domain with respect to the head (Fig. 3N). In contrast, in αVβ6:TGF-β1 headpiece complexes, the headpiece was open, with an obtuse bend of the hybrid domain with respect to the head (Fig. 3 O and P).
Affinity of αVβ8 for Its Biological Ligand pro-TGF-β1 and Regulation by Metal Ions.
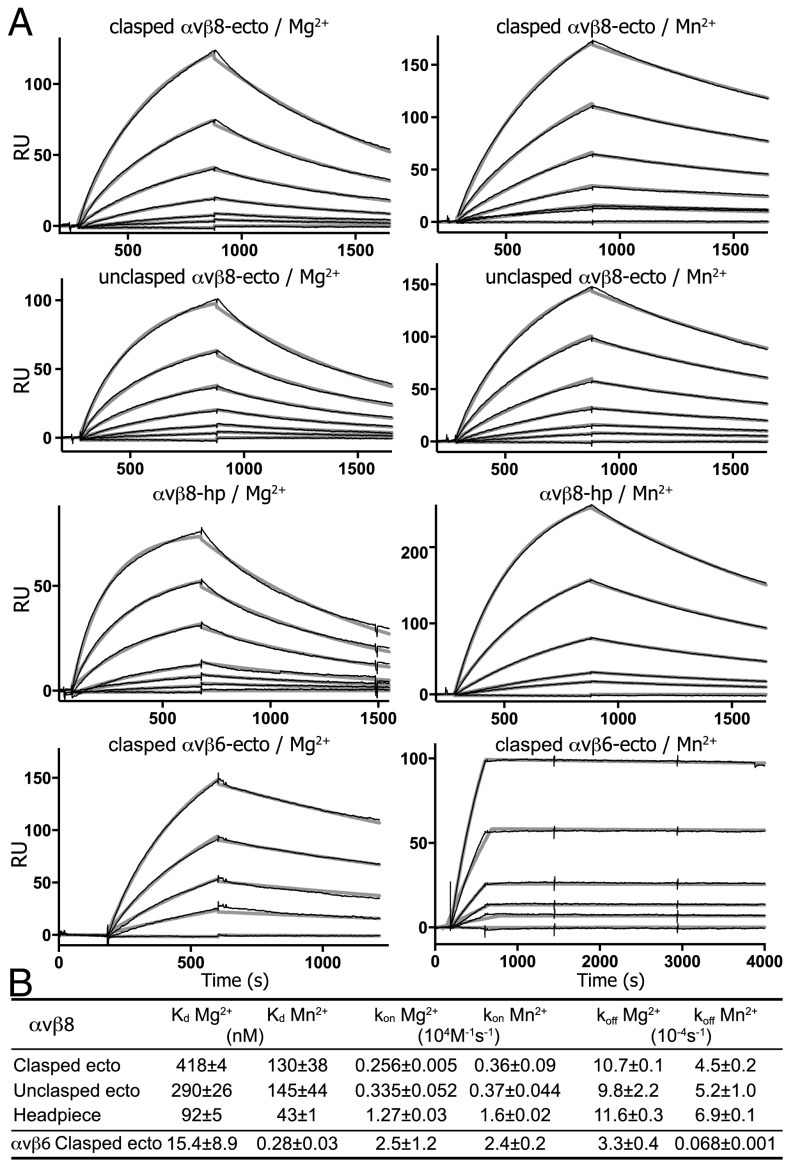
We measured affinities of our integrin preparations for pro-TGF-β1 using surface plasmon resonance (SPR). We used amine coupling to immobilize pro-TGF-β1 on the sensor chip and integrin preparations as analytes (Fig. 4A). Global fits to single on- and off-rates with a 1:1 Langmuir binding model were excellent, as shown by the black sensorgram and gray fit curves in Fig. 4A. Affinities (expressed here as Kd values) for pro-TGF-β1 of the αVβ8 clasped and unclasped ectodomains and headpiece ranged from 400 nM to 100 nM in 1 mM Mg2+ and 1 mM Ca2+ (Fig. 4 A and B). In contrast, the affinity for pro-TGF-β1 of the αVβ6 clasped ectodomain in 1 mM Mg2+ and 1 mM Ca2+ was much higher with a Kd of 15 nM (Fig. 4 A and B). In 1 mM Mn2+ and 0.2 mM Ca2+, affinity of the αVβ8 preparations increased only marginally by two- to threefold. In contrast, the affinity of αVβ6 increased much more, by 55-fold to 0.3 nM (Fig. 4 A and B). The on-rate in 1 mM Mg2+ of the αVβ8 clasped ectodomain was 10-fold slower than the on-rate of the αVβ6 clasped ectodomain (Fig. 4 A and B), whereas the off-rate for αVβ8 was faster than the off-rate for αVβ6. Mn2+ had little effect on the on-rate compared with Mg2+ for either integrin. In contrast, the off-rate was only marginally decreased in Mn2+ for αVβ8 whereas it was dramatically decreased by 50-fold for αVβ6. The effects on the off-rate are consistent with conformational change to the open headpiece after ligand binding that is stabilized by Mn2+ in αVβ6 and not in αVβ8.
Fig. 4.
Kinetics measurements of αvβ8 and αvβ6 binding to pro-TGF-β1 by SPR. (A) Sensorgrams show the overlay of fitting curve (thicker gray) and experimental curve (thinner black) of SPR measurement. Constructs and metal ions used are indicated. Concentrations were 800, 400, 200, 100, 50, 25, and 0 nM for the αvβ8 ectodomain; 400, 200, 100, 50, 25, 10, and 0 nM in Mg2+ and 200, 100, 50, 25, 10, and 0 nM in Mn2+ for the αvβ8 headpiece; and 200, 100, 50, 20, and 0 nM in Mg2+ and 100, 50, 20, 10, 5, and 0 nM in Mn2+ for the αvβ6 ectodomain. (B) Affinities and kinetic rates. Values are mean ± difference from mean of two independent experiments.
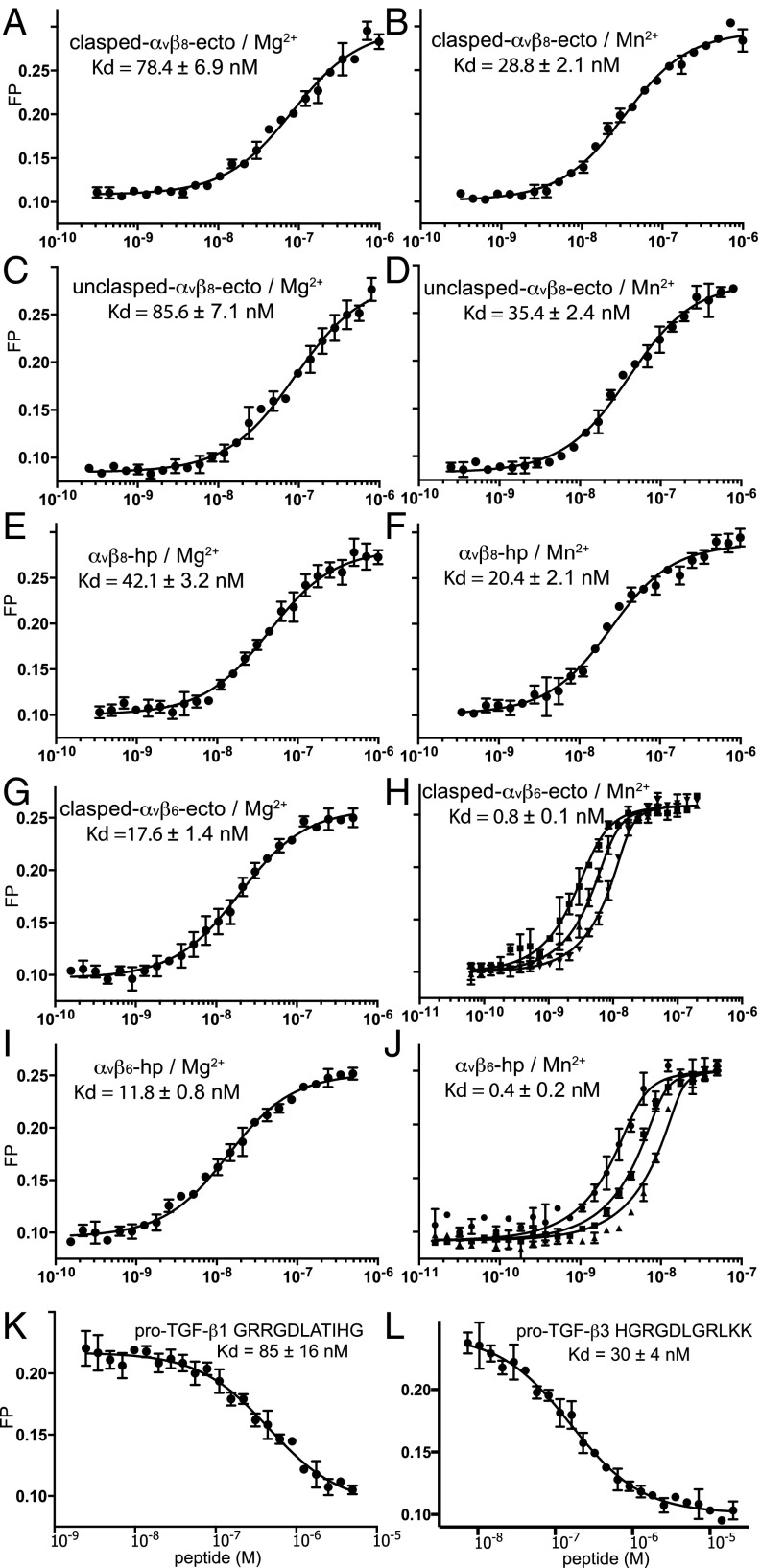
We used fluorescence polarization (FP) to extend measurements from the heterogeneous phase of SPR to the solution phase. A small fluorescent pro-TGF-β3 peptide tumbled rapidly; binding to the much larger integrin slowed tumbling and was measured as an increase in FP. We first measured affinity for fluorescent peptide using saturation binding with integrin. Affinities for the pro-TGF-β3 peptide of the αVβ8 preparations ranged from 42 nM to 86 nM, and affinity of the headpiece was roughly twofold higher than the ectodomain in the presence of 1 mM Mg2+ and 1 mM Ca2+ (Fig. 5 A, C, and E). In 1 mM Mn2+ and 0.2 mM Ca2+, affinity of the αVβ8 preparations increased about twofold (Fig. 5 B, D, and F). These results are consistent with SPR measurement in showing that the αVβ8 headpiece has slightly higher affinity to the ligand than the ectodomain and that Mn2+ only marginally enhances αVβ8 affinity. For comparison with αVβ8, we made similar measurements on αVβ6. In Mg2+, αVβ6 bound the TGF-β3 peptide with fourfold higher affinity than αVβ8 (Fig. 5 G and I). Mn2+ increased affinity of αVβ6 for the pro-TGF-β3 peptide by a further 22- to 30-fold (Fig. 5 H and J). Finally, inhibition of the FP assay with peptides from pro-TGF-β1 and -β3 that contained the RGDLXXI/L motifs showed threefold higher affinity to αVβ8 for pro-TGF-β3 than for pro-TGF-β1 (Fig. 5 K and L).
Fig. 5.
The αvβ6 and αvβ8 affinity for peptide ligands using fluorescence polarization. (A–J) Saturation binding of αvβ6 and αvβ8 ectodomain (ecto) and headpiece (hp) preparations to FITC-labeled pro-TGF-β3 RGD peptide (FITC-GRGDLGRLKK). Fluorescent FITC-labeled peptide was used at 10 nM and in H and J also at 5 nM and 20 nM. Data in H and J were fit globally to different probe concentrations to account for the effect of high-affinity binding on ligand depletion. (K and L) Affinity of the αvβ8 headpiece for pro-TGF-β1 and pro-TGF-β3 peptides measured by competition with FITC-labeled pro-TGF-β3 peptide in the presence of 1 mM Mg2+/Ca2+. The error bars in each plot represent the mean ± SD of triplicates. Errors in Kd values represent the fitting error.
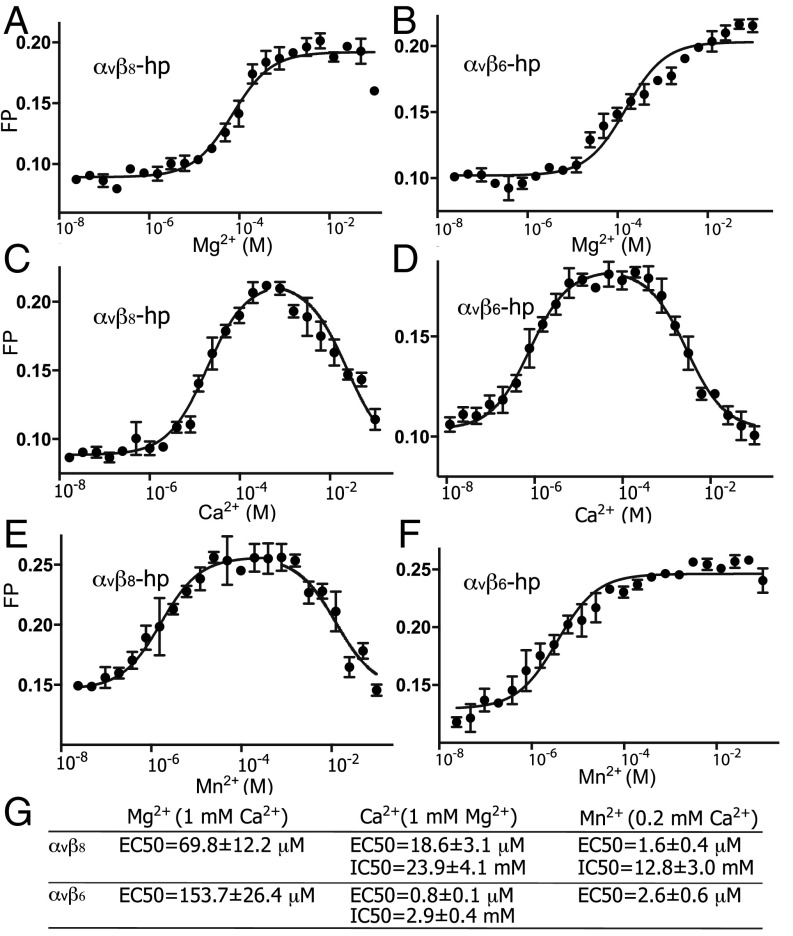
We next used FP to measure the influence of metal ions on TGF-β3 peptide ligand-binding affinity. Titration of Mg2+ in the presence of 1 mM Ca2+ showed that lower concentrations of Mg2+ were more effective in supporting ligand binding by αVβ8 than by αVβ6 (Fig. 6 A and B). Titration of Ca2+ in the presence of 1 mM Mg2+ showed that Ca2+ was required for ligand binding of αVβ8 with an EC50 of 19 μM, whereas a much lower concentration of Ca2+ was sufficient for ligand binding of αVβ6 with an EC50 of 0.8 μM. This value was lower than background Ca2+ levels present in laboratory solutions and required use of Ca2+–EGTA buffers for measurement. At higher concentrations, Ca2+ inhibited both integrins, with higher concentrations required to inhibit αVβ8 than αVβ6 (Fig. 6 C, D, and G). Mn2+ also showed distinctive effects on the two integrins. Mn2+ activated both integrins in the presence of 1 mM Ca2+ at lower effective concentrations than seen with activation by Mg2+. However, at higher concentrations Mn2+ inhibited ligand binding by αVβ8, an effect that was not seen with αVβ6 (Fig. 6 E and F). These results reveal several atypical features of αVβ8 in the regulation by metal ions of ligand binding.
Fig. 6.
Divalent cation dependence of αvβ8 and αvβ6 binding to ligand. (A–F) Fluorescence polarization was used to measure integrin headpiece binding to FITC-labeled pro-TGF-β3 peptide in varying concentrations of Mg2+ or Mn2+ with 1 mM Ca2+ and in varying concentrations of Ca2+ with 1 mM Mg2+. Ca2+/EGTA buffer was used for Ca2+ concentrations lower than 10 μM. Lines show nonlinear least square fitting of dose–response curves to the mean of triplicates; in C–E, two separate lines show fits to EC50 and IC50 values. Plotted points show mean ± SD of triplicates. All curves show fits to a 1:1 Langmuir model to [S3] or [S4] (Fig. S9); i.e., the Hill slope is fixed at 1. All plots fit this model well, except for Mg2+ and Mn2+ dependence of αvβ6, which may suggest two enhancing effects at different concentrations. (G) Summary of results showing mean ± fitting error.
Discussion
Here, we demonstrate that integrin αVβ8 is atypical in multiple respects. It is constitutively extended. Binding to its biological ligand, pro-TGF-β1, does not induce the open conformation of the αVβ8 headpiece, in agreement with previous findings using a low-affinity RGD peptide (13). The ability of pro-TGF-β1 to induce the open headpiece conformation of αVβ6 and not αVβ8 correlates with higher affinity binding with αVβ6 in Mg2+ and greater augmentation by Mn2+ of affinity with αVβ6 than αVβ8. Finally, the metal ion Mn2+ has a more complex effect on ligand binding by αVβ8 than by αVβ6.
We show that αVβ8 is constitutively extended and that its headpiece does not open when it complexes with pro-TGF-β1 either in the presence of Mn2+ or Mg2+. Previously, in the absence of ligand, αVβ8 was found to be largely extended, with ∼5% of unclasped but not clasped particles showing a bent-closed conformation, and Mn2+ was found to give little augmentation relative to Mg2+ of binding to the TGF-β1 prodomain (13). We did not find any unclasped or clasped class averages that clearly corresponded to bent-closed αVβ8, although some class averages appeared to correspond to the head region only, and we cannot rule out a small fraction of the bent-closed conformation (Figs. S1 and S2). Although complexes between pro-TGF-β1 and αVβ6 have previously been characterized in EM, the conformation in EM of the αVβ6 ectodomain on its own was not included (18). We examined here the clasped and unclasped αVβ6 ectodomain and found a ratio of extended to bent particles of about 1:3 and 3:1, respectively. This contrasts with the low frequency or lack of observation of bent αVβ8 reported here and previously (13). Our class averages for bent αVβ6 resemble the class average reported for bent αVβ8 (13). Based on these overall results it appears that αVβ8 can be present in a bent-closed conformation, but the bent conformation is less stable than the extended-closed conformation, whereas αVβ6 is similarly stable in these two conformations. Previously, RGD peptide and Mn2+ were found not to open the αVβ8 headpiece (13); however, we found it important to confirm these observations with pro-TGF-β1 for several reasons. First, the affinity of RGD for αVβ8 is unknown, and ELISAs that showed binding of αVβ8 to the TGF-β1 prodomain failed to show binding to the RGD-bearing ligand fibronectin (13). Second, it is common for ligands to dissociate from integrins during gel filtration or grid preparation as previously suggested for integrin αVβ3 and RGD peptide (19) and observed here for 2:2 integrin:pro-TGF-β1 complexes that yielded a number of class averages with 1:2 integrin complexes or integrins alone (Figs. S5 and S6). Visualization of the ligand itself, which is not possible with small peptides in EM, is the best confirmation that a ligand is bound.
Fig. S2.
Class averages of unclasped αvβ8 ectodomain (6,306 particles) by multireference alignment and K-means classification.
Fig. S5.
Class averages of clasped αvβ8 ectodomain complexed with pro-TGF-β1 (4,065 particles) by multireference alignment and K-means classification.
Fig. S6.
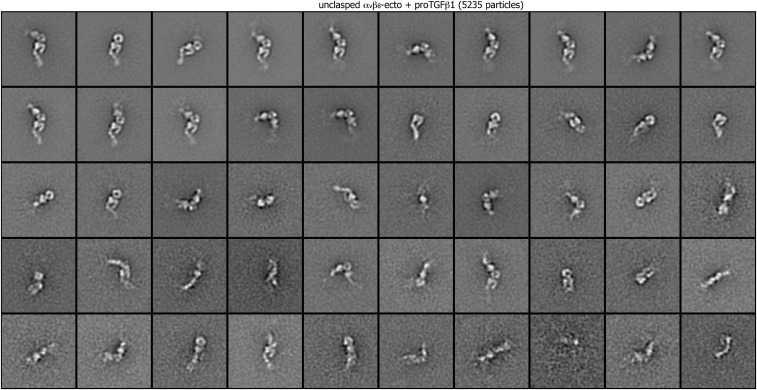
Class averages of unclasped αvβ8 ectodomain complexed with pro-TGF-β1 (5,235 particles) by multireference alignment and K-means classification.
By covisualizing αVβ8 and its biological ligand pro-TGF-β1 in complexes here in EM, we have definitively established that ligand binding does not stabilize the open headpiece conformation of αVβ8. Integrin αVβ6 served as a positive control in our study. The finding was verified with both integrin ectodomain and headpiece fragments in complexes of 2:2 and 1:2 stoichiometry and in Mg2+ and Mn2+. Previously, ligand binding has been shown to result in headpiece opening for integrins αVβ3, αIIbβ3, α5β1, αXβ2, and αVβ6 (2, 15). For integrin α4β7, binding to ligand in Mg2+ induced an intermediate headpiece conformation that is hypothesized to mediate transient, rolling adhesion, whereas binding to ligand in Mn2+ induced an open conformation that is hypothesized to mediate firm adhesion (20). Thus, maintenance of a ligand-bound integrin headpiece in the closed conformation in Mg2+, and lack of headpiece opening for a ligand-bound integrin in Mn2+, are unprecedented.
We have measured the affinity of αVβ8 for pro-TGF-β1, which is fundamental to understanding how αVβ8 binds and activates pro-TGF-β1 in vivo. Because other integrins exist in an ensemble of conformations, measurements of their affinities represent an average of the affinity of each conformation weighted by the population of that conformation in the ensemble. By using Fabs of the allostery conformation-specific antibodies to stabilize integrin α5β1 in open, closed, and extended conformations, we have been able to measure the affinity intrinsic to each integrin conformation (12). The affinity of fibronectin for the extended-open conformation of α5β1 is 1.4 nM and for the bent-closed and extended-closed conformations is ∼9,000 nM. Affinity measurements here of αVβ8 are unique for an integrin because, rather than measuring an average affinity for an ensemble of conformations, they measure the affinity of a single conformation, the extended-closed conformation.
Previous studies have suggested that αVβ8 has high affinity; however, rather than measuring affinity, the relative amount of binding in flow cytometry or ELISAs of αVβ8 was compared with another integrin such as αVβ3 (13, 21). The 100-nM affinity of αVβ8 for pro-TGF-β1 is indeed ∼100-fold higher than the affinity for biological ligand of the closed conformations of α5β1 (12). However, the fairest comparison is between αVβ8 and αVβ6 binding to the same ligand, pro-TGF-β1. This comparison suggests that we cannot consider the closed conformation of αVβ8 to be a high-affinity conformation because αVβ6, which can access an open conformation, binds to pro-TGF-β1 with 30-fold higher affinity in Mg2+ and 500-fold higher affinity in Mn2+.
Thus, although the 100-nM affinity of αVβ8 for pro-TGF-β1 appears high for a closed integrin conformation, it is far lower than the affinity for pro-TGF-β1 that αVβ6 achieves with headpiece opening. The affinity of αVβ8 for pro-TGF-β1 is also ∼100-fold lower than achieved with binding of the open headpiece conformation of α5β1 to fibronectin. Studies with α5β1 suggest that Mn2+ both shifts the conformational equilibrium toward the open headpiece conformation and raises its intrinsic affinity (12). An increase in intrinsic affinity of the closed conformation must account for the two- to threefold higher affinity in Mn2+ compared with Mg2+ of αVβ8 for pro-TGF-β1 found here because EM showed that αVβ8 had a closed conformation when bound to pro-TGF-β1 in both Mn2+ and Mg2+. On the other hand, the more profound increase in affinity of 55-fold in Mn2+ for αVβ6 suggests that Mn2+ additionally shifts the equilibrium so that the αVβ6 conformational ensemble contains a higher proportion of the open headpiece in Mn2+ than in Mg2+. Additionally, the presence of a small proportion of the open headpiece in the αVβ6 conformational ensemble in Mg2+ is likely to account for the 30-fold higher affinity for pro-TGF-β1 in Mg2+ of αVβ6 compared with αVβ8. Given the ∼1,000-fold higher affinity of integrin open than closed states, a small percentage of the open conformation can make a major contribution to ensemble affinity (12).
We found unusual effects of metal ions on αVβ8 affinity for ligand. In integrin βI domains, the MIDAS metal ion forms a direct coordination to an acidic residue in the ligand. The MIDAS is flanked on opposite sides by the synergistic metal ion-binding site (SyMBS) and the ADMIDAS. Physiologically, it appears that Mg2+ is bound to the MIDAS and that Ca2+ is bound to the SyMBS and ADMIDAS. Mn2+ can replace the metals at all three sites (22, 23). In conformational change from closed to open, the ADMIDAS metal ion moves ∼6 Å toward the MIDAS, and its coordination sphere alters from pentagonal bipyramidal that favors Ca2+ to octahedral that favors Mn2+ and Mg2+ (23, 24). Mutations of metal ion-coordinating residues in integrin α4β7 showed that the SyMBS was required for synergistic enhancement of ligand binding by Ca2+ and Mg2+ (16). In contrast, in integrins α4β7 and α5β1, the ADMIDAS was required for inhibition at higher concentrations by Ca2+ and appeared to be the site at which Mn2+ competed with Ca2+ to enhance ligand binding (16, 25). Integrin αVβ8 resembled other integrins in showing synergism between Ca2+ and Mg2+ (16, 26, 27). Thus, ligand binding by αVβ8 in 1 mM Ca2+ was dependent on Mg2+ with an EC50 of 75 μM, and binding in 1 mM Mg2+ was dependent on Ca2+ with an EC50 of 19 μM. Similarly, binding in 0.2 mM Ca2+ was dependent on Mn2+ with an EC50 of 1.6 μM. These results may reflect synergism between Ca2+ at the ADMIDAS and Mg2+ or Mn2+ at the MIDAS. Most unusual was inhibition of ligand binding to αVβ8 by Mn2+ at higher concentrations with an IC50 of 13 mM. Such inhibition was not seen with αvβ6 or reported to our knowledge for any other integrin. Inhibition by Mn2+ may reflect substitution for Ca2+ at the SyMBS or ADMIDAS metal ion site in the closed conformation. In other integrins, Mn2+ is thought to selectively replace the ADMIDAS metal ion in the open conformation, which is distinctive in both metal ion location and coordination geometry (23, 24).
Our characterization here of αVβ8 bound to its biological ligand pro-TGF-β1 has definitively shown that ligand binding does not stabilize an open conformation of αVβ8 (13). Our measurements here of the affinity of αVβ8 for ligand and comparative measurements of αVβ6 provide unique information. Although previous studies have attempted to address whether αVβ8 has high or low affinity for ligand, ligand-binding affinity has not been measured. Quantitation shows that the closed conformation of αVβ8 has higher affinity than the closed conformation of some integrins, but is certainly lower in affinity than the open conformation of integrin α5β1 for fibronectin (12) or the affinity of the basal ensemble of conformations of αVβ6 for pro-TGF-β1. Currently, we can only measure a population-weighted average affinity for pro-TGF-β1 of the αVβ6 conformational ensemble; its closed conformation might well have a similar affinity to that of αVβ8. The pro-TGF-β1–binding integrins αVβ6 and αVβ8 do appear under basal conditions to have unusually high affinity for ligand compared with other integrins, including three other αV integrins. High affinity for ligand may reflect the requirement that these integrins not only bind but also transmit force from the cytoskeleton to the prodomain to release TGF-β, as suggested in at least some cellular systems by the requirement for a covalent linkage between pro-TGF-β1 and a milieu molecule for activation by αVβ6 and αVβ8 (10; but see ref. 9). Evidence is mounting that activation of most integrins requires force exertion by the actin cytoskeleton and resistance by the ligand to stabilize the extended-open integrin conformation compared with the bent-closed and extended-closed conformations (11, 28, 29). As αVβ8 couples to a distinct type of cytoskeletal network through the DAL-1/Band 4.1B adaptor (6), it may either not be subjected to and regulated by cytoskeletal force or be subjected to force of a different magnitude than applied by the actin cytoskeleton. Our studies provide a quantitative framework for comparisons among integrins that are activated by distinctive cellular mechanisms.
Materials and Methods
The cDNA encoding the signal sequence and mature residues of the αv ectodomain (1–960) or headpiece (1–594) with the M400C mutation and Gly inserted prior to residue 400 (15) was cloned into pcDNA3.1-Hygromycin(-) vector. The cDNA encoding the mature residues of the β8 ectodomain (1–639) or headpiece (1–456) with the V259C mutation was cloned into the ET10 vector (14). The expression constructs were cotransfected in HEK293S Gnt1−/− cells. Clonal cell lines were selected and protein was purified as described (14). Human pro-TGF-β1 with a R249A cleavage site mutation was prepared as described (15).
Negative-stain EM was as described (19, 30, 31). Purified fresh protein from gel-filtration peaks was loaded on glow-discharged carbon grids and fixed with uranyl formate. Low-dose images were acquired with an FEI Tecnai-12 transmission electron microscope at 120 kV and a nominal magnification of 52,000×. Image processing was performed with SPIDER (32) and EMAN (33) as described (30). About 5,000 particles were picked manually and subjected to multireference alignment and K-means classification.
SPR studies were performed using a Biacore3000 instrument (GE Healthcare). The pro-TGF-β1 with a R249A cleavage site mutation was immobilized on a CM5 chip through amine coupling. Purified αvβ8 ectodomain or headpiece was injected at 20 μL/min in HBS buffer (20 mM HEPEs, pH 7.5, 150 mM NaCl) containing either 1 mM Mg2+ and 1 mM Ca2+ or 1 mM Mn2+ and 0.2 mM Ca2+. The surface was regenerated with a pulse (50 μL/min, 30 s) of 15 mM HCl at the end of each cycle. Kinetics were analyzed with Biacore evaluation software. A 1:1 Langmuir binding model was applied for experimental data fitting, and kinetic parameters were fit globally to sensorgrams at different analyte concentrations. Low χ2 values (0.6–1.7) indicated good fits.
Fluorescence polarization was in HBS buffer with FITC-labeled pro-TGF-β3 RGD peptide (FITC-Aminocaproic acid-GRGDLGRLKK). In saturation binding assay, αvβ8 and αvβ6 were serially diluted in 1.4-fold decrements and mixed with 10 nM of probe (for αvβ6 ectodomain and headpiece in the presence of Mn2+, 5, 10, and 20 nM of probe were used for global fitting to account for the ligand depletion effect for high-affinity binding) in the presence of 1 mM Mg2+ and 1 mM Ca2+ or 1 mM Mn2+ and 0.2 mM Ca2+ at 20 °C for 30 min. Fitting FP as a function of integrin concentration to [S1] (Fig. S9) at fixed probe concentrations yielded the Kd values for fluorescent pro-TGF-β3 peptide. When multiple probe concentrations were used, FP was globally fit to both probe and integrin concentrations using [S1] (Fig. S9) to yield Kd values. In competitive binding assays, unlabeled pro-TGF-β1 and pro-TGF-β3 peptides were serially diluted in 1.4-fold decrements and mixed with 10 nM of probe and 100 nM of αvβ8 headpiece in HBS buffer containing 1 mM Mg2+ and 1 mM Ca2+ and incubated at 20 °C for 30 min. FP and competitor concentrations at fixed integrin and probe concentrations were fit to [S2] (Fig. S9) using the known integrin affinity for the probe to yield the competitor peptide Kd values. To test the effects of cations, 100 mM of Mg2+, Ca2+, or Mn2+ were serially diluted in twofold decrements and mixed with 100 nM αvβ8 or 20 nM αvβ6 and 10 nM of probe in HBS buffer with other indicated cations. To avoid interference by background Ca2+ present in laboratory water, concentrations of free Ca2+ of 100 μM and below were achieved by including 1 mM of EGTA using a total Ca2+ concentration calculated as described (34). The mixture was equilibrated at 20 °C for 30 min, and FP data were recorded on a Synergy NEO HTS plate reader. FP and cation concentrations were fit to [S3] (Fig. S9) to yield the EC50 values for cations that only activated binding. For cations that both activated and inhibited, FP and cation concentrations were fit to [S4] (Fig. S9) to obtain EC50 and IC50 values.
Fig. S9.
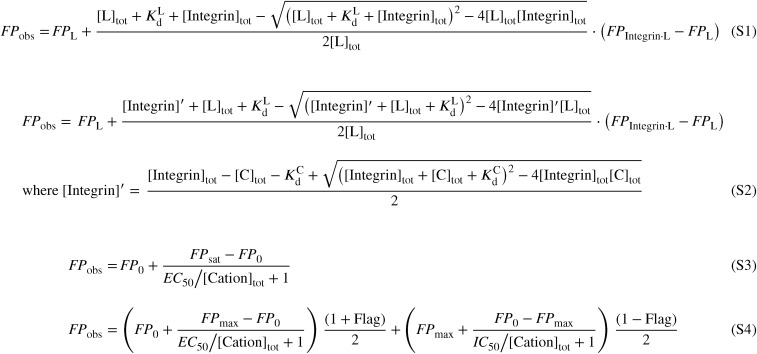
Fitting functions. [S1] Fitting function for FP saturation binding assay. [S2] Fitting function for FP competitive binding assay. [S3] Fitting function for cation activating effect. [S4] Ftting function for cation activating and inhibiting effect at different concentrations. FPobs is the measured FP. FPL and FPIntegrin·L are FP of free FITC-labeled pro-TGF-β3 peptide and integrin-bound complex, respectively. [Integrin]tot, [L]tot, and [C]tot are total concentrations of integrin, FITC-labeled pro-TGF-β3 peptide, and competitor, respectively. [Integrin]′ is the concentration of competitor-free integrin, either with or without bound FITC-labeled pro-TGF-β3 peptide. and are affinities of integrins for pro-TGF-β3 peptide and competitors, respectively. In [S3] and [S4], FP0 is the FP without added cation; FPmax is highest value of FP reached at any cation concentration; [Cation]tot is total concentration of cation or total concentration of Ca2+ not bound to EGTA; EC50 and IC50 are the cation concentrations at the inflection point where half-maximum change in FP was observed. Flags are assigned to data points depending on whether they are used to fit EC50 (Flag = 1) or IC50 (Flag = −1).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705129114/-/DCSupplemental.
References
- 1.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:a004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24:107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 4.Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 6.McCarty JH, Cook AA, Hynes RO. An interaction between αVβ8 integrin and Band 4.1B via a highly conserved region of the Band 4.1 C-terminal domain. Proc Natl Acad Sci USA. 2005;102:13479–13483. doi: 10.1073/pnas.0506068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinck AP, Mueller TD, Springer TA. Structural biology and evolution of the TGF-β family. Cold Spring Harb Perspect Biol. 2016;8:a022103. doi: 10.1101/cshperspect.a022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson IB, Rifkin DB. Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb Perspect Biol. 2016;8:a021907. doi: 10.1101/cshperspect.a021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu D, et al. The integrin αVβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R, et al. GARP regulates the bioavailability and activation of TGFβ. Mol Biol Cell. 2012;23:1129–1139. doi: 10.1091/mbc.E11-12-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schürpf T, Springer TA. Regulation of integrin affinity on cell surfaces. EMBO J. 2011;30:4712–4727. doi: 10.1038/emboj.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, et al. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 2017;36:629–645. doi: 10.15252/embj.201695803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minagawa S, et al. Selective targeting of TGF-β activation to treat fibroinflammatory airway disease. Sci Transl Med. 2014;6:241ra79. doi: 10.1126/scitranslmed.3008074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X, Hudson NE, Lu C, Springer TA. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat Struct Mol Biol. 2014;21:1091–1096. doi: 10.1038/nsmb.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X, et al. Force interacts with macromolecular structure in activation of TGF-β. Nature. 2017;542:55–59. doi: 10.1038/nature21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Salas A, Springer TA. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat Struct Biol. 2003;10:995–1001. doi: 10.1038/nsb1011. [DOI] [PubMed] [Google Scholar]
- 17.Xia W, Springer TA. Metal ion and ligand binding of integrin α5β1. Proc Natl Acad Sci USA. 2014;111:17863–17868. doi: 10.1073/pnas.1420645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, et al. Structural specializations of α4β7, an integrin that mediates rolling adhesion. J Cell Biol. 2012;196:131–146. doi: 10.1083/jcb.201110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu P, Luo BH. Integrin αVβ8 adopts a high affinity state for soluble ligands under physiological conditions. J Cell Biochem. 2016 doi: 10.1002/jcb.25780. [DOI] [PubMed] [Google Scholar]
- 22.Xiong JP, et al. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ. Role of ADMIDAS cation-binding site in ligand recognition by integrin α5β1. J Biol Chem. 2003;278:51622–51629. doi: 10.1074/jbc.M306655200. [DOI] [PubMed] [Google Scholar]
- 26.Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Chen J. The regulation of integrin function by divalent cations. Cell Adhes Migr. 2012;6:20–29. doi: 10.4161/cam.18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Z, Guo SS, Fässler R. Integrin-mediated mechanotransduction. J Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordenfelt P, Elliott HL, Springer TA. Coordinated integrin activation by actin-dependent force during T-cell migration. Nat Commun. 2016;7:13119. doi: 10.1038/ncomms13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Yu Y, Mi LZ, Walz T, Springer TA. Molecular basis for complement recognition by integrin αXβ2. Proc Natl Acad Sci USA. 2012;109:4586–4591. doi: 10.1073/pnas.1202051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi J, Springer TA. Integrin activation and structural rearrangement. Immunol Rev. 2002;186:141–163. doi: 10.1034/j.1600-065x.2002.18613.x. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 34.Schoenmakers TJ, Visser GJ, Flik G, Theuvenet AP. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques. 1992;12:870–874, 876–879. [PubMed] [Google Scholar]