Significance
Product template (PT) domains from fungal nonreducing polyketide synthases (NR-PKSs) are responsible for controlling the aldol cyclizations of poly-β-ketone intermediates during polyketide biosynthesis. Our ability to understand the high regioselective control that PT exerts is hindered by the inaccessibility of unstable poly-β-ketones for in vitro studies. We describe here the crystallographic application of “atom replacement” mimetics in which isoxazole rings linked by thioethers mimic the alternating sites of carbonyls in the poly-β-ketone intermediates. The probe contains a heptaketide mimetic tethered to a modified 4′-phosphopantetheine, which provides important empirical evidence for the PT-catalyzed cyclization mechanism. These findings afford a view of a polyketide “atom-replaced” mimetic in a NR-PKS active site that could prove general for other PKS domains.
Keywords: polyketide synthase, polyketide mimetics, product template, aflatoxin, atom replacement
Abstract
Product template (PT) domains from fungal nonreducing polyketide synthases (NR-PKSs) are responsible for controlling the aldol cyclizations of poly-β-ketone intermediates assembled during the catalytic cycle. Our ability to understand the high regioselective control that PT domains exert is hindered by the inaccessibility of intrinsically unstable poly-β-ketones for in vitro studies. We describe here the crystallographic application of “atom replacement” mimetics in which isoxazole rings linked by thioethers mimic the alternating sites of carbonyls in the poly-β-ketone intermediates. We report the 1.8-Å cocrystal structure of the PksA PT domain from aflatoxin biosynthesis with a heptaketide mimetic tethered to a stably modified 4′-phosphopantetheine, which provides important empirical evidence for a previously proposed mechanism of PT-catalyzed cyclization. Key observations support the proposed deprotonation at C4 of the nascent polyketide by the catalytic His1345 and the role of a protein-coordinated water network to selectively activate the C9 carbonyl for nucleophilic addition. The importance of the 4′-phosphate at the distal end of the pantetheine arm is demonstrated to both facilitate delivery of the heptaketide mimetic deep into the PT active site and anchor one end of this linear array to precisely meter C4 into close proximity to the catalytic His1345. Additional structural features, docking simulations, and mutational experiments characterize protein–substrate mimic interactions, which likely play roles in orienting and stabilizing interactions during the native multistep catalytic cycle. These findings afford a view of a polyketide “atom-replaced” mimetic in a NR-PKS active site that could prove general for other PKS domains.
Product template (PT) domains are a structural feature of the nonreducing class of iterative polyketide synthases (NR-PKSs). They both control the high reactivity of poly-β-ketone intermediates that are rapidly assembled upstream in the β-ketoacyl synthase (KS) domain (1) and catalyze their intramolecular closure to cyclic and fused cyclic products. How both of these tasks are carried out, and how competing self-condensations to more thermodynamically favored products are avoided, are central functional questions of NR-PKS catalysis. We address them here with the single PT domain for which an X-ray crystal structure exists and report the organized structure of a stable mimetic of the native but synthetically inaccessible poly-β-ketone substrate bound in the PksA PT domain.
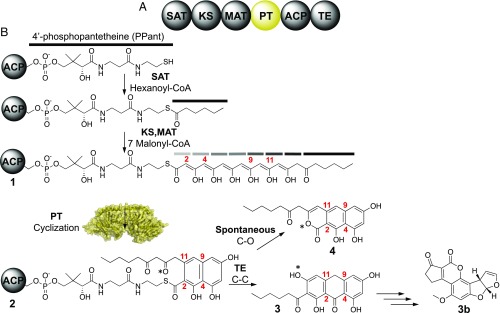
Biosynthesis of the environmental carcinogen aflatoxin B1 is initiated by a fungal, iterative NR-PKS known as PksA (Fig. 1B) (2, 3). This polypeptide consists of six covalently linked enzyme domains, where the function of each has been characterized by its specialized role in the controlled polymerization of ketide units and cyclization to a particular product (Fig. 1A) (2, 3). Polyketide elongation is initiated by the starter unit, ACP transacylase (SAT) domain, which selectively primes the acyl carrier protein (ACP) with a hexanoyl starter unit (4). The KS then catalyzes seven iterative rounds of decarboxylative Claisen condensations using malonyl building blocks supplied by the malonyl-CoA:ACP transacylase (MAT) domain to yield a mature 20-carbon poly-β-keto intermediate 1 for which direct evidence has been obtained by mass spectrometry (Fig. 1B) (2, 3). This reactive 20-carbon intermediate 1 is then cyclized via two regioselective intramolecular aldol reactions by the PT domain to yield a C4–C9/C2–C11 cyclized intermediate 2 (5). Next, the thioesterase domain (TE) conducts a Claisen/Dieckmann cyclization to release the product from the 4′-phosphopantetheine (PPant)-tethered ACP to afford noranthrone 3 (6). The growing polyketide intermediate remains attached to the ACP-tethered PPant group until the release of noranthrone 3 from PksA by a TE domain. In the absence of the TE domain a spontaneous cyclization gives rise to pyrone 4 (6, 7). Further postsynthase tailoring of 3 yields aflatoxin B1 (3b) (Fig. 1B) (2).
Fig. 1.
Aflatoxin biosynthesis. (A) Domain architecture of the aflatoxin NR-PKS. (B) Biosynthetic pathway of aflatoxin starts with a hexanoyl starter unit and undergoes seven rounds of elongation to yield a C20 linear intermediate 1. Gray and black bars represent diketone groups. The PT domain cyclizes a C4–C9 and C2–C11 reaction 2 that is subsequently aromatized and released by the TE domain 3. Further biosynthetic processing yields aflatoxin B1 3b. Absence of the TE yields pyrone product 4.
The first-ring cyclization (from 1 to 3 in Fig. 1B) is pivotal to the formation of stable polyketide intermediate 2, which can be further processed toward the final product 3b. To date, there are at least six classes of PT domains that differentially cyclize poly-β-ketones to unique ring patterns in a highly selective manner (SI Appendix, Figs. S1 and S2) (8, 9). To achieve this selectivity, a high degree of control must be maintained between the PT and the ACP-tethered poly-β-ketone intermediate. To date, only the PT domain of PksA has yielded a crystal structure (Protein Data Bank: 3HRQ and 3HRR) (5). Fortuitously, the structure was obtained with the methyl terminus of palmitate at its deepest recess and extending to the carboxylate in the PT reaction chamber. Using this 16-carbon chain as a template, superimposition of the 20-carbon polyketide intermediate 1 and energy minimization with limited constraints gave a revealing model of PPant–substrate binding and led to a proposal for selective activation for controlled cyclization (5). As unanticipated and appealing as this view of PT function was, it lacked experimental support.
Two problems exist that have blocked our interrogation of PT activity. First, the high intrinsic propensity to self-reaction of poly-β-ketones precludes them as practically useful substrates for structural studies on PKSs. Second, the high reactivity of the native 20-carbon poly-β-keto intermediate 1 prevents its de novo synthesis. Recently, a general strategy for the design and synthesis of stable mimics was developed using the concept of “atom replacement” (10). Conceptually, isoxazoles linked by thioethers are visualized to parallel the alternating pattern of methylene and carbonyl groups of NR-PKS intermediates. Furthermore, the native thioester within these substrates was replaced by an isosteric amide bond to further stabilize the pantetheine–substrate connection (10, 11). In this study, we prepared 4′-phosphorylated and unphosphorylated atom-replaced mimetics selected to represent substrate chain lengths of 8–16 carbon units of the corresponding linear and proposed monocyclic intermediates. Here, we report the 1.8-Å cocrystal structure of the PksA PT domain bound to a 4′-phosphopantetheinylated linear heptaketide mimetic in the crystallographic application of isoxazole–thioether mimetics in PKS structural investigations.
Results and Discussion
Design and Rationale of the Atom Replacement Mimetics.
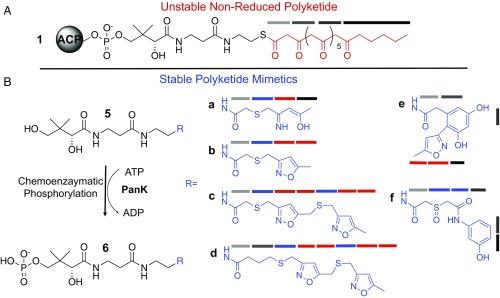
To understand the chain-length selectivity of the PksA PT, we designed stable atom-replaced mimics of different chain lengths and tethered them to a nonhydrolizable Pant group (Fig. 2). Our current set of mimetics includes: a linear ring-opened 4c-mimetic 5a, and the corresponding linear isoxazole 4c-mimetic 5b; the bis-isoxazole 7c and 8c-mimetics 5c and 5d (10). In addition, to probe cyclization specificity, we also prepared the cyclized and aromatized polyketide mimics 5e and 5f (10) (Fig. 2) to mirror hexaketide shunt products of PksA (Fig. 1). Our selection of these materials was based on the fact that the tetraketides (5a and 5b) with eight carbons are the shortest chain length that a fungal PT has been shown to accept (2). Each of these materials can be optionally phosphorylated at the 4′-hydroxyl group of the pantetheine using a promiscuous pantetheine kinase (PanK) (Fig. 2) as given by the respective conversions 5a–5f into 6a–6f (12–15).
Fig. 2.
Atom replacement mimetics. (A) Structure of the ACP-tethered natural substrate (1) containing an elongated polyketone tail (red) and PPant unit (black). (B) Structures of linear mimetics 5a–5d and cyclic mimetics 5e and 5f. The corresponding phosphates 6a–6f can be prepared chemoenzymatically from 5a–5d using the pantetheine kinase CoaA. Various shaded gray bars denote natural ketide units (with the exception of the hexanoyl-starter unit, shown in black). Ketide mimetics containing N and S atom replacement are shown in blue bars and isoxazole-derived ketides are shown in red bars.
Chemoenzymatic Phosphorylation and Isolation Of Pantetheine Mimetics.
To obtain the phosphorylated mimetics 6a–6f, we incubated 5a–5f with PanK from the pantothenate (vitamin B5) biosynthetic pathway in Escherichia coli (15). Previous work on one-pot chemoenzymatic preparation of CoA analogs provided a standard methodology for pantetheine phosphorylation (15). Because we only required the PanK for pantetheine phosphorylation and not the remaining CoA ligating and modifying enzymes, we first explored optimizing conditions to conduct pantetheine phosphorylation by evaluating varying concentrations of recombinant PanK and mimetics 5a–5f in diverse buffers, pHs, concentrations of ATP, salts, and temperatures. The phosphorylation of mimetics 5a–5f into 6a–6f was monitored by analytical HPLC analysis and electrospray ionization mass spectrometry (ESI-MS). Time points were taken at 1, 3, 6, 12, and 24 h. Overall, no significant improvements were found in PanK phosphorylation. We found that the condition giving the highest yield of phosphorylated mimetics 6a–6f was after incubating in 2 μM PanK, 25 mM potassium phosphate pH 7.5, 10 mM MgCl2, and 8 mM ATP at 37 °C for 3 h (SI Appendix, Figs. S5–S7). Once complete phosphorylation was observed, 6a–6f were purified by semipreparative HPLC. This approach offered an effective protocol to prepare PPant mimics in purity and amounts sufficient for X-ray crystallography and enzyme assays.
The Crystallization and Crystal Diffraction of PksA PT with Phosphorylated and Unphosphorylated Atom-Replaced Mimetics: Importance of Chain Length and PPant Phosphate.
To date, the crystallization of ligands with PksA PT had proven to be extremely difficult due to the presence of endogenous palmitate in the active site, which blocked substrate binding. We have found that the best way to perform this procedure involves first the incubation of PksA PT with XAD-2 or XAD-4 hydrophobic resins to remove the palmitate, followed by addition of the substrate or substrate mimetic at a concentration that does not cause immediate protein precipitation (5). To explore for the substrate binding and cyclization motifs of PT, we rigorously screened for conditions that allowed the cocrystallization of PksA PT and other PT construct mimetics 5a–5f and phosphorylated mimetics 6a–6f (Fig. 2). For PksA PT, we found that XAD-2 resins often resulted in protein precipitation, and hence, we screened 1,200 conditions for each mimetic at 4 °C and 23 °C. From this set, we selected the most promising conditions and explored them for crystal growth in refinement trays. In addition to PksA PT, we also applied the screen to three other PT domains from NR-PKSs (Pks1, Pks4, and ACAS) that represent different clades of PT (SI Appendix, Table S1) (8).
Heptaketide-PT Structure Reveals Extensive Substrate–Enzyme Interactions in PPant Binding Region and Cyclization Chamber.
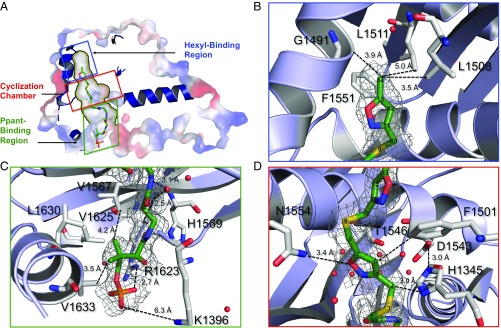
The 6c-bound PksA PT structure (6c • PksA PT) revealed a dimer with two active sites, with each monomer containing a double hotdog (DHD) fold (SI Appendix, Fig. S3A). Within each monomer the substrate-binding pocket contained a hexyl-binding region, a cyclization chamber, and a PPant-binding region (Fig. 3) (5). Mimetic 6c interacted with residues in only the cyclization chamber and the PPant-binding region (Fig. 3 C and D and SI Appendix, Fig. S3B). The PPant segment of 6c served to orient the heptaketide mimetic by electrostatic and hydrophobic interactions (Fig. 3C and SI Appendix, Fig. S3). Here, the terminal PPant phosphate of 6c is anchored by strong electrostatic interaction with Arg1623 of PT near the entrance of the substrate-binding pocket (Fig. 3C). Further, the gem-dimethyl group of PPant was oriented toward three Val and one Leu near the entrance of the catalytic pocket (Val1567, Val1625, Leu1630, and Val1633). These previously unidentified amino acids defined an important hydrophobic pocket within the PPant binding region (Fig. 3C). In addition, the backbones of Leu1398 and Cys1353, along with a crystalline water, formed hydrogen bonds with the hydroxyl and carbonyl groups of 6c. The water molecule was further coordinated to the backbone of Leu1398 and Cys1353, both of which are well conserved among most clades of PT domains (SI Appendix, Fig. S2) (8, 9). These observations suggest that the PPant binding region interacts extensively with the heptaketide substrate, and PPant binding is crucial for substrate orientation.
Fig. 3.
Structure of the 6c • PksA PT complex. (A) Surface rendition depicting the hexyl-binding chamber, crystallization chamber, and Ppant binding region within the 1.8 Å crystal structure of the 6c • PksA PT complex. The PPant-tethered heptaketide outlines various residues in the catalytic pocket. Schematic representation illustrates the close contacts in the (B) hexyl-binding region, (C) PPant binding region, and (D) cyclization chamber. Residue numbers and distances between the substrate and protein are provided.
In the cocrystal structure of 6c with PksA PT, the hexyl-binding region was largely unoccupied, which is consistent with its selectivity to bind the hexyl starter unit that is not present in 6c (Fig. 3B). A 7.0-Å gap between the back of the hexyl-binding region and the methyl terminus of the 6c only contained one detectable water molecule. This observation is in accord with the hydrophobic nature of the hexyl-binding region, such that in the absence of the hexyl group, 6c binds PT without contacting the hexyl-binding region (Fig. 3B). The fact that the structures of 6c • PksA PT and the previously reported palmitate–PT were highly similar (rmsd = 0.154 Å) also supports the view that the lack of hexyl group in 6c does not affect overall protein conformation of PksA PT.
The placement of the heptaketide mimetic 6c in the cyclization chamber confirmed the importance of the cyclization chamber for polyketide orientation and catalysis, as proposed earlier (Figs. 3D and 4) (5). From the 6c • PksA PT structure, we identified previously uncharacterized residues in the cyclization chamber that are important for polyketide recognition. The catalytic dyad of PksA PT is composed of His1345 and Asp1543 (5). Regiospecific C4–C9 first-ring formation depends on the ability of His1345 to deprotonate C4 of 1 and initiate first-ring cyclization. In the 6c • PksA PT structure, the catalytic His1345 is 3.0 Å from C4 of 6c, thus strongly supporting C4 deprotonation and subsequent enolate formation for proper ring closure (Fig. 3D). The oxyanion hole was located in the back of the cyclization chamber, constrained by Pro1355. It has been previously proposed that a network of organized water molecules bound to Ser1356, Asp1543, Asn1568, and Thr1546 stabilize the oxyanion and provide a highly structured, polar environment that favors aldol cyclization (Fig. 4) (5, 9). Now, the 6c • PksA PT structure provides concrete evidence for the maintenance of a polar environment through a water network that is, indeed, coordinated with Ser1356, Asp1543, Asn1568, and Thr1546 (Fig. 3D). In the bis-isoxazole moiety of 6c (Fig. 2), a bridging sulfur atom replaces the C9 carbonyl of the actual polyketide intermediate 1 (Fig. 1B). This weaker hydrogen bond acceptor shows no interaction with the water network, but is constrained intrinsically to a 90° bond angle and was oriented away from the active site by its placement between the two rigid flanking isoxazole rings. Nonetheless, it was remarkable that the location of C4 near His1345 (3.0 Å) and the C9 carbonyl mimetic, the sulfur atom in 6c, are in close proximity to the water network, which strongly supports the proposed mechanism that PT activates a particular set of nucleophile/electrophile partners to catalyze a single regiospecific aldol cyclization between C4 and C9.
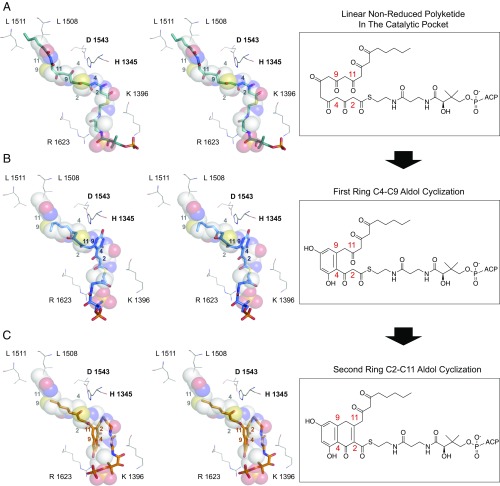
Fig. 4.
Docking studies. Stereoimages of (A) the docked linear polyketide intermediate (1), (B) the monocyclic C4–C9 intermediate, and (C) the bicyclic C4–C9/C2–C11 (3). The docking ligands were rendered in stick format and heptaketide ligand in space-filled representations. Colored numbers indicate the carbon registry of the docked linear polyketide intermediate, and the gray numbers indicate the carbon registry for the 6c ligand.
Catalytic Activity and Mutagenesis of the PksA PT.
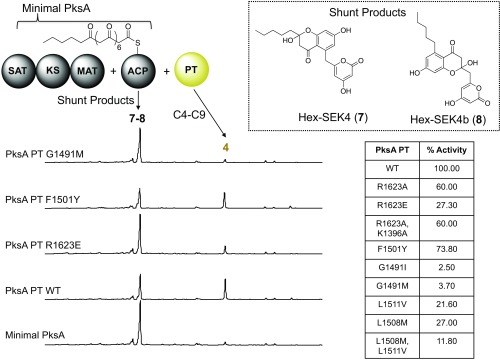
We then turned our attention to identify residues important for substrate binding. Using our 6c • PksA PT structure as a guide, we generated mutants in the PPant binding region, cyclization chamber, and hexyl-binding regions. The PT mutants were reconstituted with holo-PksA ACP, the tridomain PksA SAT–KS–MAT, hexanoyl-CoA, and malonyl-CoA. The PksA PT cyclizes the linear intermediate 1 to biosynthesize norpyrone 4 in vitro. Products of the enzymatic reactions were determined using time course HPLC analysis to monitor the reaction course (Fig. 5).
Fig. 5.
In vitro reconstitution analysis of PksA PT WT and mutants. The minimal PKSs consist of the basic enzymes required to generate linear polyketide products, which include the SAT–KS–MAT and the ACP. Minimal PKSs generate various shunt products 7 or 8. Addition of the PT generates a C4–C9/C2–C11 norpyrone 4. PksA PT mutants display a decrease in 4 formation compared with wild type. Mutants include: R1623A, R1623E, F1501Y, G1491I, G1491M, L1511V, and L1508M; double mutants include R1623A, K1396A, L1508M, and L1511V.
Effects on pocket entrance.
From these data, we were able to access the importance of Arg1623 and Lys1396, two positively charged surface residues located at the entrance of the PPant binding region that interact with the phosphate terminus of 6c. R1623A and K1396A mutants were 60% as efficient as wild-type PT in their production of norpyrone 4, the in vitro product of PksA. The K1396Q–R1623Q double mutant also displayed a similar efficacy (60% efficiency as compare with wild-type PT). The moderate decrease in norpyrone formation for R1623A, K1396A, and K1396Q–R1623Q indicates that the positively charged side chains of Arg1623 and Lys1396 were not essential for product formation. Substitution of the negatively charged residue, R1623E, did show further reduction of activity and generated 30% production of norpyrone. However, the mutational result showed that the positive charges of Arg1623 and Lys1396 are not the sole determinant for substrate binding. These observations were consistent with the 6c • PksA PT structure where the PPant binding region interacts with PPant with both charge–charge and hydrophobic interactions.
Interactions within the cyclization chamber.
The Ala mutants of the active site dyad His1345 and Asp1535 completely abolished catalyzed norpyrone 4 formation (5). The result confirmed the crucial catalytic role of both residues for cyclization. Previous Ala mutations of Thr1546 and Asn1548 also resulted in a diminished activity (5), which is consistent with the observed interaction between 6c and residues in the cyclization chamber (Fig. 3D).
Interactions with the hexyl-binding region.
Sequence alignment suggests that most PT domains, which typically incorporate shorter acetyl starter units, have a Met at the position Leu1508, located near the beginning of the hexyl-binding region. This observation suggested that the methionine residue may serve to block the hexyl-binding region and prevents the loading of a hexanoyl starter unit (SI Appendix, Fig. S2). Further, Gly1491 defines the bottom of the pocket, which is otherwise a bulky, hydrophobic residue in the nonhexyl loading PTs (SI Appendix, Fig. S2). To decrease the volume of the hexyl-binding region, we generated G1491I and G1491M. Both of these mutants returned less than 5% of the production of 4 compared with that of wild type (5). The fact that truncation of the hexyl-binding region can greatly reduce product formation for PksA PT is in accord with the proposal that the overall size and shape of the substrate binding pocket was important in chain-length selectivity.
PT-Polyketide Docking Simulations Are Consistent with the Cocrystal Structure.
Each isoxazole moiety (Fig. 2) is aimed to mimic a β-diketone/keto-β-enol in a linear polyketide (Fig. 2). Whereas the isoxazole ring within 5a–6f serves to provide stability, it also restricts the number of potential enolization isomers sampled by the natural polyketone in 1. We conducted in silico docking using GOLD (16) to further explore the potential issues arising from the selection of distinct enol-keto tautomers of 1. Hexanoyl-containing polyketide intermediates with 14 and 20 carbons were docked into the substrate binding pocket of PksA PT. We found that the binding motifs of the native mature 20-carbon or nascent 14-carbon intermediates are highly similar to the cocrystal structure of mimetic 6c (Fig. 4A), especially in the following features: First, the electrostatic interactions between the PPant phosphate and Arg1623/Lys1396 remained in close proximity, further supporting the importance of the PPant phosphate for substrate localization and binding. Second, the location and orientation of C4 and its proximity to the catalytic His1345 was highly similar in the docked and observed 6c • PksA PT structure, supporting the view that the key residue for deprotonation is correctly placed in the cocrystal structure of 6c • PksA PT (Fig. 4A). Finally, in the catalytic chamber and hexyl-binding region, the 20-carbon intermediate and 14-carbon mimetic had highly similar interactions (both polar and nonpolar) with the active site residues, especially surrounding the carbonyl moieties flanking C4 and C2 that will be deprotonated to initiate the first- and second-ring cyclizations. The overall orientation of the PPant linked to the endogenous 20-carbon unreduced polyketide kinks inside the 30-Å PksA PT pocket. The previously identified Val/Leu PPant channel residues (5) along with the Arg/Lys surface residues help orient C4 toward His1345 (Fig. 4A). Deprotonation and enolate formation at C4 by His1345, followed by aldol condensation at C9 generates the first C4–C9 ring intermediate (SI Appendix, Fig. S8). In this mechanism, the C4–C9 ring is directed toward the cyclization chamber, allowing for a second His1345 deprotonation at C2, which undergoes reaction at C11. The second aldol reaction of C2–C11 generates intermediate 2, which is displaced further into the cyclization chamber (Fig. 4 B and C). The proposed position of the bicyclic intermediate 2 in the cyclization chamber is further supported by the similar position observed for the HC8 cyclization analog used in the first PksA PT domain structure analysis (5). Overall, both the mature 20-carbon and nascent 14-carbon substrates align in orientations similar to the heptaketide mimetic 6c, thus confirming that the atom replacement mimics are indeed a valid mimetic for the catalysis, specificity, and mechanism of the PKS PTs.
Conclusions
We report the X-ray crystallographic application of atom-replacement mimetics to elucidate the cyclization specificity of fungal NR-PKS PT domains. Using a combination of structural biology, structure-based mutagenesis, and enzymatic assays, we have shown how these mimetics help identify key residues involved in substrate recognition that are not otherwise identifiable. The inherent instability and reactivity of linear, unreduced polyketide intermediates has hindered previous efforts to accurately visualize the interaction between a poly-β-ketone intermediate and a PKS. Whereas no structure can exactly mimic the chemical properties of natural substrate, the atom replacement mimetics have facilitated our ability to structurally visualize these linear intermediates in a PT domain by providing the polyketide mimicry while blocking their intrinsic reactivity. We demonstrate that attachment of the PPant moiety to atom-replaced mimetics can allow for further structural insight into the role of the PPant arm on the delivery and accurate placement of the reactive polyketide intermediate in the active site. The 6c • PksA PT cocrystal structure provides critical visual confirmation for the role of PT domains in regiospecific cyclization(s) during fungal polyketide biosynthesis. In these studies, the 14-carbon mimetic 6c delivered high-resolution structural data that further validated the proposed substrate-binding residues. The binding region proximal to the PPant arm in 6c contains several key residues previously unidentified that are responsible for coordinating the hydroxyl and stabilizing the gem-dimethyl moieties of the PPant arm. The nature of 6c’s binding to the catalytic chamber provided further evidence for residue flexibility to accommodate interactions with the rigid isoxazole rings. In an important observation, the heptaketide C4 carbon is positioned proximal to the catalytic His1345, providing further experimental evidence to the previously proposed mechanism of polyketide cyclization by PT. Such atom-replacement mimetics can serve as powerful tools to further investigate distinct type I, II, and III PKS enzymes that would otherwise be inaccessible by conventional polyketides, which would be subject to rapid aldol and Claisen condensations in media used for X-ray cystallography. Overall, this study demonstrates that medicinal chemical techniques, such as structure mimicry, can have an immediate impact in the study of natural product biosynthesis. We found that the atom replacement concept not only offers sufficiently stable materials, but also offers materials with sufficient mimicry to support high-resolution analyses of structural biology.
Materials and Methods
Procedures and methods for protein expression and purification, site-directed mutagenesis, protein X-ray crystallography, in silico docking, preparation and phosphorylation of the mimetics, in vitro reconstitution assay, and circular dichroism assay can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank J. R. Luo, T. Poulos, A. G. Newman, P. A. Storm, and S. Opella for helpful discussions. These studies were conducted through funding from the National Institutes of Health (NIH) Grants GM100305, GM100738, and GM076330 (to S.-C.T.); GM095970 and GM094924 (to M.D.B.); and ES001670 (to C.A.T.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences under Contract DE-AC02-76SF00515, and by the NIH, National Institute of General Medical Sciences Grant (P41GM103393). Portions of this research were carried out at the Advanced Light Source, which is supported by the Office of Basic Energy Sciences of the US DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KBZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609001114/-/DCSupplemental.
References
- 1.Vagstad AL, Bumpus SB, Belecki K, Kelleher NL, Townsend CA. Interrogation of global active site occupancy of a fungal iterative polyketide synthase reveals strategies for maintaining biosynthetic fidelity. J Am Chem Soc. 2012;134:6865–6877. doi: 10.1021/ja3016389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Townsend CA. Aflatoxin and deconstruction of type I, iterative polyketide synthase function. Nat Prod Rep. 2014;31:1260–1265. doi: 10.1039/c4np00092g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford JM, et al. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korman TP, et al. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Natl Acad Sci USA. 2010;107:6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman AG, Vagstad AL, Storm PA, Townsend CA. Systematic domain swaps of iterative, nonreducing polyketide synthases provide a mechanistic understanding and rationale for catalytic reprogramming. J Am Chem Soc. 2014;136:7348–7362. doi: 10.1021/ja5007299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem. 2010;285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, et al. Rational reprogramming of fungal polyketide first-ring cyclization. Proc Natl Acad Sci USA. 2013;110:5398–5403. doi: 10.1073/pnas.1301201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shakya G, et al. Modeling linear and cyclic PKS intermediates through atom replacement. J Am Chem Soc. 2014;136:16792–16799. doi: 10.1021/ja5064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring AM, Lambalot RH, Vogel KW, Drueckhammer DG, Walsh CT. Ability of Streptomyces spp. acyl carrier proteins and coenzyme A analogs to serve as substrates in vitro for E. coli holo-ACP synthase. Chem Biol. 1997;4:17–24. doi: 10.1016/s1074-5521(97)90233-7. [DOI] [PubMed] [Google Scholar]
- 12.Haushalter RW, Worthington AS, Hur GH, Burkart MD. An orthogonal purification strategy for isolating crosslinked domains of modular synthases. Bioorg Med Chem Lett. 2008;18:3039–3042. doi: 10.1016/j.bmcl.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa F, Haushalter RW, Lee DJ, Finzel K, Burkart MD. Sulfonyl 3-alkynyl pantetheinamides as mechanism-based cross-linkers of acyl carrier protein dehydratase. J Am Chem Soc. 2013;135:8846–8849. doi: 10.1021/ja4042059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worthington AS, Burkart MD. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org Biomol Chem. 2006;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- 15.Nazi I, Koteva KP, Wright GD. One-pot chemoenzymatic preparation of coenzyme A analogues. Anal Biochem. 2004;324:100–105. doi: 10.1016/j.ab.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.