Significance
Chromatin factors generally act within large, multisubunit complexes; thus, identifying both their normal and aberrant interactors in cancer should provide important information regarding potential targets for therapeutic intervention. Here, we apply this principle to analysis of BRD4–NUT, a fusion oncoprotein that drives an aggressive subtype of squamous cell cancer. We identify ZNF532 as a prominent BRD4–NUT–interacting protein in an established NUT midline carcinoma patient cell line, and independently discover ZNF532 fused directly to NUT in a newly analyzed patient. Like BRD4–NUT, ZNF532–NUT forms unusually large (100-kb to 1-Mb) domains of hyperactive chromatin, including at the MYC locus, and drives self-reinforcing regulatory loops that are likely to be a powerful strategy for the growth advantage of cancer cells.
Keywords: BioTAP-XL, hyperacetylation, ZNF532–NUT, topological domains, BRD4
Abstract
To investigate the mechanism that drives dramatic mistargeting of active chromatin in NUT midline carcinoma (NMC), we have identified protein interactions unique to the BRD4–NUT fusion oncoprotein compared with wild-type BRD4. Using cross-linking, affinity purification, and mass spectrometry, we identified the EP300 acetyltransferase as uniquely associated with BRD4 through the NUT fusion in both NMC and non-NMC cell types. We also discovered ZNF532 associated with BRD4–NUT in NMC patient cells but not detectable in 293T cells. EP300 and ZNF532 are both implicated in feed-forward regulatory loops leading to propagation of the oncogenic chromatin complex in BRD4–NUT patient cells. Adding key functional significance to our biochemical findings, we independently discovered a ZNF532–NUT translocation fusion in a newly diagnosed NMC patient. ChIP sequencing of the major players NUT, ZNF532, BRD4, EP300, and H3K27ac revealed the formation of ZNF532–NUT–associated hyperacetylated megadomains, distinctly localized but otherwise analogous to those found in BRD4–NUT patient cells. Our results support a model in which NMC is dependent on ectopic NUT-mediated interactions between EP300 and components of BRD4 regulatory complexes, leading to a cascade of misregulation.
Mutations in the subunits of chromatin regulatory complexes are found at high frequencies in cancer cells. Thus, the comprehensive identification of the components of chromatin complexes implicated in disease via their protein–protein interactions is an important avenue toward finding potential targets for therapeutic intervention. In NUT midline carcinoma (NMC), a subtype of squamous cell cancer, the transcriptional machinery is hijacked to drive expression of progrowth, antidifferentiation genes (1–3). NMC is defined by chromosomal rearrangement of the NUT (NUTM1) gene, which is most commonly fused to the BRD4 gene (4, 5). The resulting BRD4–NUT chimeric oncoprotein forms large nuclear foci (6), proposed to form through tethering of the BRD4 bromodomains to acetylated chromatin and the acetylation of neighboring histones by EP300 via its interaction with NUT (7). Our recent genomic analysis of NMC patient cell lines provides strong evidence that the dual properties of acetyl-histone binding and EP300 recruitment result in a feed-forward expansion of acetylated chromatin and BRD4–NUT over massive genomic domains, often filling entire topologically associating domains (TADs) (3). The number and magnitude of these “megadomains” correlate with the characteristic nuclear foci seen in diagnostic patient tumor samples or in cultured NMC cells stained with a NUT-specific antibody (3, 6, 7).
Megadomains encompassing the MYC and TP63 regulatory regions are common to all NMCs examined to date, and RNAi knockdown of either of these genes in patient cells blocks growth and, in the case of MYC, leads to differentiation in culture (2, 3). Similarly, small-molecule BET inhibitors such as JQ1, which disengage BRD4–NUT from chromatin, diminish megadomain-associated transcription, including at MYC enhancers and TP63, and also result in differentiation and growth arrest of NMC cells (3, 8). Thus, it appears that BRD4–NUT directly misregulates these two key genes, and potentially many others, to drive one of the most aggressive tumors known in cancer biology. BET inhibitors, specifically targeting the BRD4 bromodomains, exhibit on-target activity in NMC patients (9), albeit with significant dose-limiting toxicity. Multiple BET inhibitor clinical trials are currently enrolling NMC patients (NCT01587703, NCT01987362, NCT02431260, and NCT02259114).
Given the potency of aberrant chromatin regulation by BRD4–NUT, and reasoning that identifying additional therapeutic targets will be necessary to defeat this disease, we have undertaken a comprehensive analysis of BRD4–NUT protein interactions on chromatin in TC-797 NMC patient cells and when ectopically expressed in 293TRex cells. Rather than taking a candidate approach, we have used comprehensive proteomics to probe how fusion to NUT alters normal BRD4 function. Our cross-linking approach revealed that EP300 is indeed the acetyltransferase uniquely attracted by the NUT portion of the fusion oncoprotein. We further identified ZNF532 and a short list of additional factors as candidates associated with BRD4–NUT complexes specifically in patient cells. Remarkably, our identification of ZNF532 through BRD4–NUT proteomics coincided with our independent discovery of a translocation of ZNF532 to NUT in a newly identified NMC patient. We found that ZNF532–NUT fusion protein forms megadomains of hyperacetylated chromatin, similar to those formed by BRD4–NUT, suggesting a common feed-forward mechanism for megadomain formation.
Results
EP300 Acetyltransferase Is Specific to BRD4–NUT Affinity Purification.
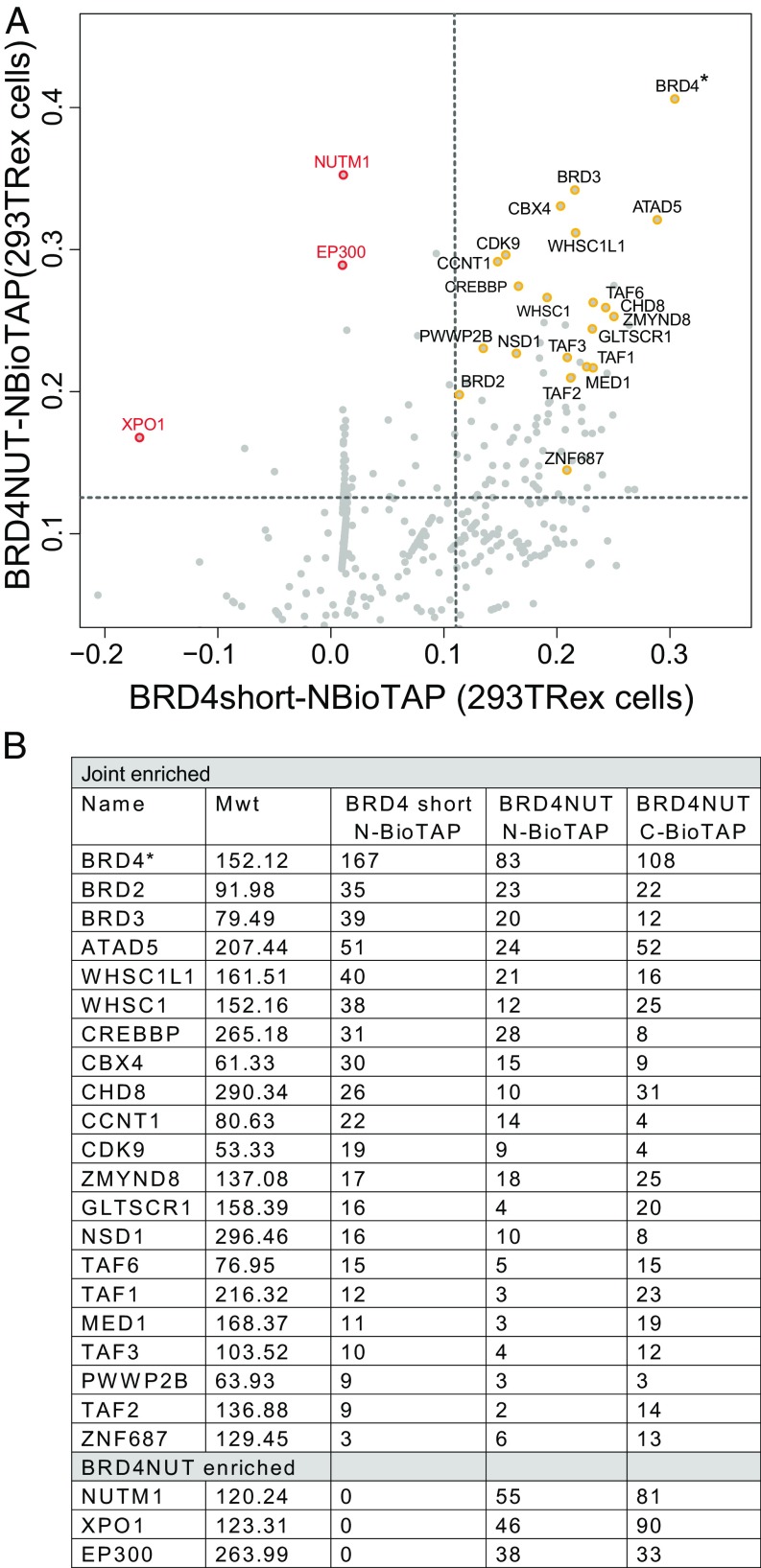
To identify factors that may contribute to BRD4–NUT–driven oncogenesis, we sought to affinity purify the fusion oncoprotein and compare a comprehensive list of its interacting proteins with factors copurified with BRD4 lacking NUT. To this end, we induced expression of BioTAP-tagged BRD4–NUT (BRD4–NUT–BioTAP) or the short isoform of BRD4 (BRD4short–BioTAP), encoding only the portion of BRD4 included in the BRD4–NUT fusion oncoprotein (10). We expressed the epitope-tagged proteins from single-copy transgenes integrated in a non-NMC cell line, 293T, the derivative of which we term 293TRex (3, 11). 293TRex cells serve as a useful model, as they do not normally harbor the oncogenic fusion but, when induced to express BRD4–NUT, form de novo nuclear foci and hyperacetylated megadomains (3). Chromatin cross-linking, affinity purification, and mass spectrometry (BioTAP-XL) (3, 12) allowed stringent purification of N- and C-terminally BioTAP-tagged BRD4–NUT and N-BioTAP–BRD4–associated proteins from 293TRex cells. Enrichment over input chromatin was calculated for each identified interaction (Materials and Methods). To illustrate the extent of shared and unique protein interactions across our experiments, we plotted each protein identified in our dataset as an individual point whose coordinates along a specific axis correspond to its relative enrichment (or depletion) from the specified pulldown (for the full list of proteins recovered, please see Dataset S1). Fig. 1A shows a pairwise comparison of N-terminally tagged BRD4 and BRD4–NUT pulldowns from 293TRex cells. Proteins jointly enriched by both baits are found in the plot (Fig. S1, Upper Right Quadrant). In addition, the top interactors, consistently recovered in both N- and C-tagged BioTAP-XL pulldowns, are listed in Fig. 1B. Such shared interactors include other double-bromodomain–containing proteins (BRD3, BRD2) and numerous transcriptional and chromatin regulators, including ATAD5, NSD3 (WHSC1L1), NSD2 (WHSC1), CBX4, CHD8, and pTEFb (CDK9 + CCNT1) (13, 14). Interestingly, we identified the acetyltransferase CBP (CREBBP) as enriched in both pulldowns, consistent with its previous strong association with BRD4–NUT nuclear foci (7). Thus, the findings of our unbiased mass spectrometry strongly validate the known role for BRD4 as a transcriptional coactivator, and provide an expanded list of potential interactors to help understand its critical function in cell-type specificity and oncogenesis.
Fig. 1.
BRD4 vs. BRD4–NUT protein interactions. (A) Scatterplot comparing BioTAP–BRD4short and BioTAP–BRD4–NUT pull-down enrichments in 293TRex cells. Each point represents an individual protein with coordinates (x, y) corresponding to its enrichment in each respective pulldown relative to input. Dashed lines represent the 85th percentile of BioTAP–BRD4short enrichment (vertical line) and 95th percentile of BioTAP–BRD4–NUT enrichment (horizontal line). See Dataset S1 for full results. (B, Top) Joint enriched: The top proteins and the number of peptides recovered in both the BRD4 and BRD4–NUT pulldowns, based on reproducibility and the total peptide enrichment over input. Note that both N- and C-terminally tagged BRD4–NUT were analyzed. (Bottom) BRD4–NUT enriched: The top three proteins and the number of peptides recovered uniquely in BRD4–NUT vs. BRD4 pulldowns. Asterisks denote bait used for pulldown. Mwt, molecular weight of protein in kilodaltons.
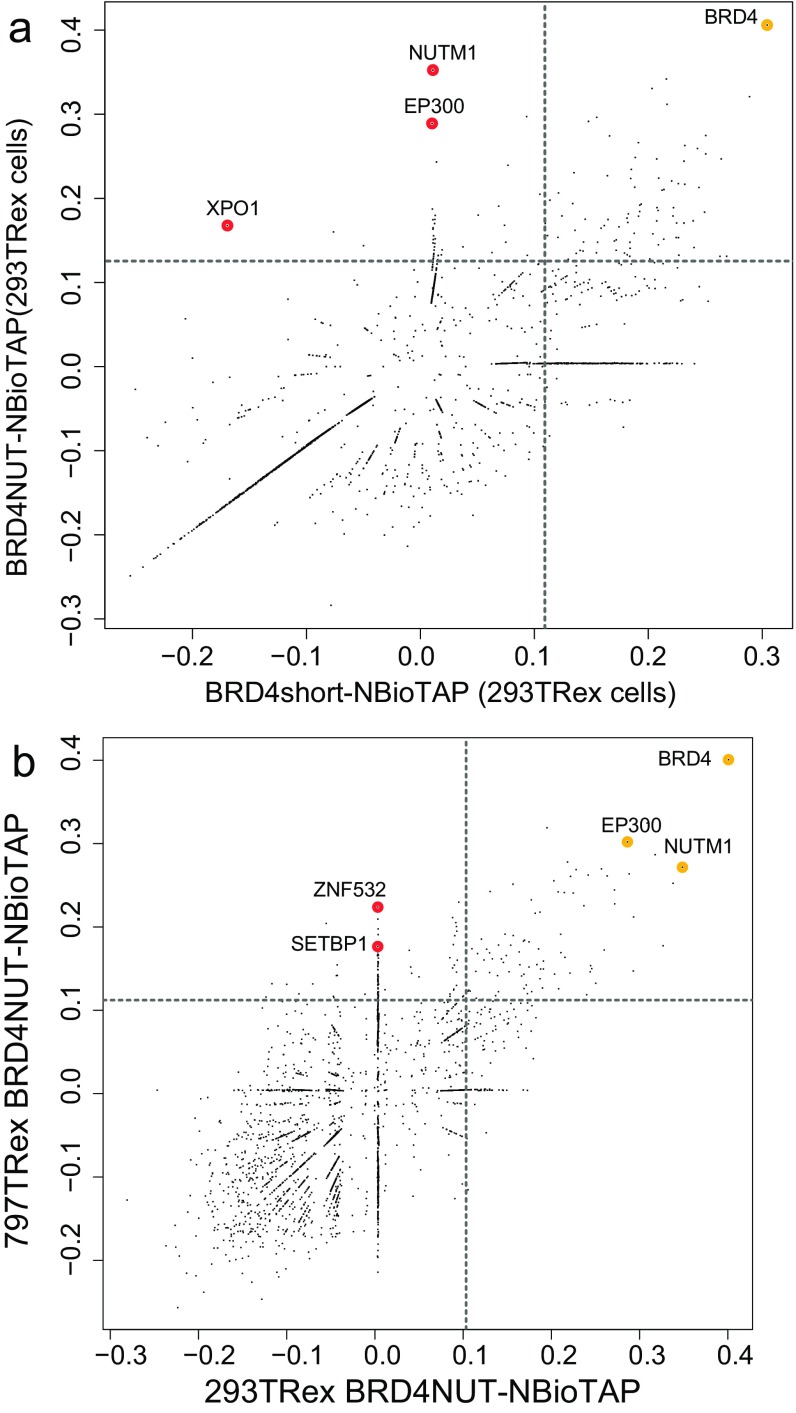
Fig. S1.
Full view of BRD4–NUT protein interactions. (A) Scatterplot comparing log enrichment of proteins identified in the NBioTAP–BRD4 pulldown and in the NBioTAP–BRD4–NUT pulldown from 293TRex cells. The same data as shown in Fig. 1A, but with an expanded view of the entire protein set. (B) Scatterplot comparing log enrichment of proteins identified in the NBioTAP–BRD4–NUT pulldown from 293TRex cells and in the NBioTAP–BRD4–NUT pulldown from 797TRex cells. The same data as shown in Fig. 2A, but with an expanded view of the entire protein set.
In contrast, proteins enriched in the BRD4–NUT pulldown and not in the BRD4 pulldown are found in the upper left corner of Fig. 1A and listed in Fig. 1B, Bottom. Besides NUT and its putative transport factor XPO1 (6), the most highly enriched interacting protein is the EP300 acetyltransferase, affirming its previous identification through a candidate approach (7) and highlighting its unique status when BRD4–NUT interactors are analyzed comprehensively. This discovery lends strong support to a model in which BRD4–NUT formation of hyperacetylated megadomains occurs through sequential acetylation by EP300 and interaction with acetylated residues via the bromodomains of BRD4.
Positive Autoregulation of BRD4–NUT Complexes in Patient Cells.
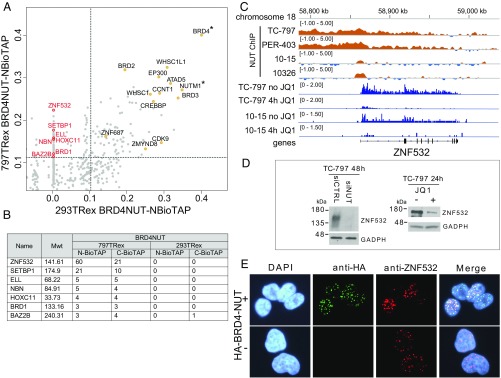
We next used cross-linking, affinity pulldown, and mass spectrometry to identify BRD4–NUT interactions specifically enriched in NMC patient cells, reasoning that these might help explain the disease state. We compared the proteins associated with BRD4–NUT–BioTAP in non-NMC 293TRex cells with those identified in 797TRex NMC patient cells expressing BRD4–NUT–BioTAP from an inducible, single-copy transgene in the presence of endogenous BRD4–NUT. Robust interactions common to both cell types (Fig. 2A and Fig. S1B) include BRD2, BRD3, WHSC1L1, WHSC1, ATAD5, CCNT1, CREBBP, and EP300. In addition, we found several proteins that were enriched in 797TRex pulldowns and absent from 293TRex affinity purifications (Fig. 2B). The two proteins consistently enriched to the highest levels only in 797TRex cells were SETBP1 and ZNF532. Mutations that stabilize SETBP1 protein are implicated in proliferation in leukemia cells (15). ZNF532 is a putative DNA-binding protein containing 12 C2H2 zinc-finger domains (www.uniprot.org/uniprot/Q9HCE3). The absence of SETBP1 and ZNF532 in 293TRex pulldowns may be due to their low expression levels (Dataset S1).
Fig. 2.
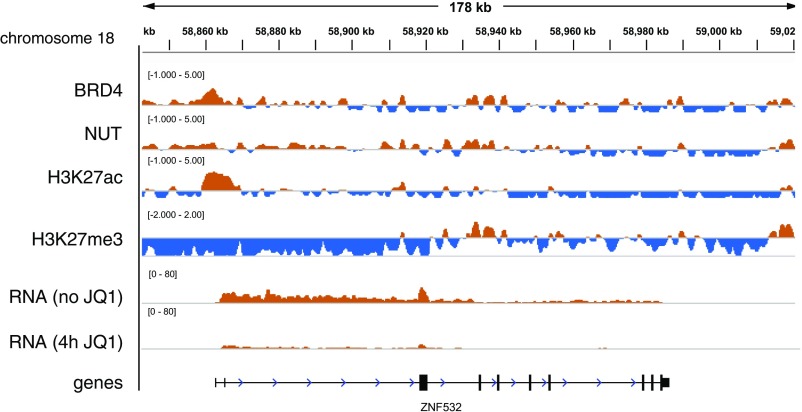
BRD4–NUT interactions in 293TRex cells vs. 797TRex NMC patient cells. (A) Scatterplot of affinity pull-down enrichments comparing BioTAP–BRD4–NUT from 293TRex and 797TRex cells. Thresholds for enrichment in both pulldowns are at the 95th percentile. Commonly enriched hits are depicted as yellow dots in the upper right quadrant, whereas hits enriched in 797TRex patient cells are depicted as red dots in the upper left quadrant. Asterisks denote bait used for pulldown. (B) Top hits unique to 797TRex patient cells that were enriched in both N- and C-terminally tagged BRD4–NUT pulldowns. Mwt, molecular weight of protein in kilodaltons. (C) BRD4–NUT (NUT ChIP) binds the 5′ promoter region of the ZNF532 gene in different NMC patient-derived cells. ZNF532 transcription is strongly reduced after 4 h of 0.5 μM JQ1 treatment as measured by nascent RNA-seq (3). (D) Immunoblot of siRNA (48 h) (Left) or JQ1-treated (24 h) (Right) TC-797 lysates with ZNF532 antibody. GAPDH was used as a loading control for protein normalization. (E) Immunofluorescence of 797TRex NMC cells induced to express HA-tagged BRD4–NUT. Antibodies used were anti-HA and anti-ZNF532. (Magnification: 400×.)
Interestingly, in addition to the association of BRD4–NUT and ZNF532 proteins in 797TRex patient cells, we found that the ZNF532 locus is a binding target for BRD4–NUT. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis using anti-NUT antibodies revealed strong enrichment over or adjacent to the ZNF532 locus in a variety of NMC cells, including TC-797, 797TRex, 10-15 (16), and PER-403 (17), all of which harbor a BRD4–NUT fusion, and also 10326 cells, which carry a BRD3–NUT translocation (7) (Fig. 2C). Moreover, treatment of NMC cells with the bromodomain inhibitor JQ1 rapidly reduced nascent ZNF532 transcription in both cell lines tested (Fig. 2C). Consistent with the decrease in mRNA production, ZNF532 protein levels were also reduced by JQ1 treatment (Fig. 2D). JQ1 sensitivity of ZNF532 expression might be due to inhibition of endogenous BRD4, rather than of the BRD4–NUT fusion protein. Therefore, we tested for a specific dependency of ZNF532 expression on the BRD4–NUT fusion and found a strong reduction of ZNF532 protein following siRNA knockdown directed against NUT in TC-797 cells (Fig. 2D). BET inhibition also leads to a reduction in ZNF532 expression in various non-NMC tissues (18–22). These results demonstrate that ZNF532 expression is strongly dependent on BET bromodomain proteins such as BRD4, and especially on BRD4–NUT association with the locus in NMC patient cells.
To test whether ZNF532 indeed colocalizes with BRD4–NUT in patient cells, either broadly or within specific megadomains, we performed immunofluorescent staining of TC-797 patient cells with anti-ZNF532 antibodies. We found broad colocalization of ZNF532 with BRD4–NUT in the hyperacetylated nuclear foci corresponding to megadomains and characteristic of NMC (Fig. 2E), consistent with our proteomic analyses. Collectively, these findings provide strong evidence that ZNF532 in complex with BRD4–NUT positively regulates its own expression in a self-reinforcing regulatory loop in NMC. This feed-forward effect on expression, along with positive regulation of the Mediator subunit MED23 (3), is likely to boost the amounts of oncogenic NUT complexes available to help perpetuate the iterative cycle of acetylation and binding that leads to megadomain formation.
ZNF532–NUT Is a Novel Fusion Oncogene in NUT Midline Carcinoma.
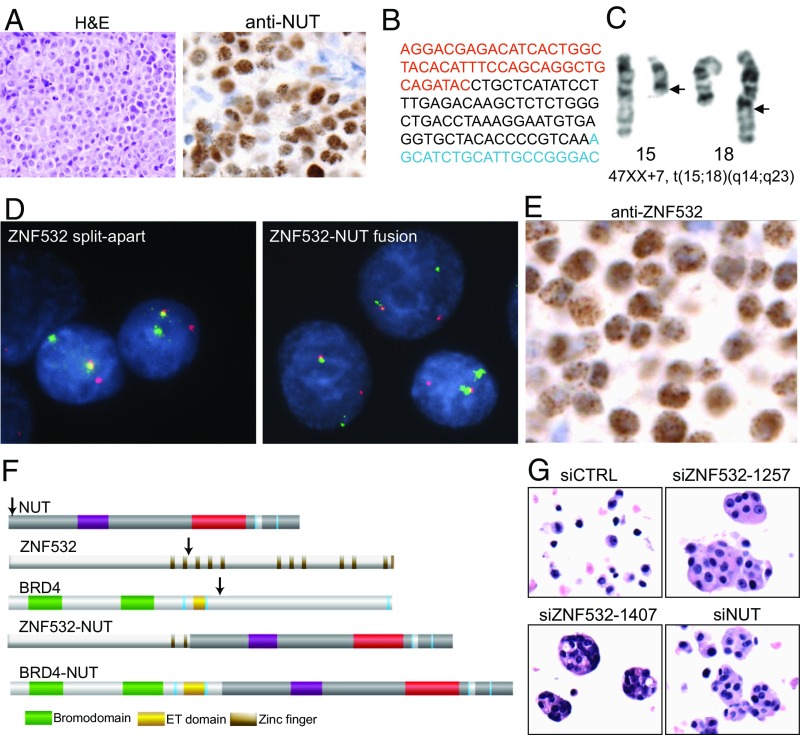
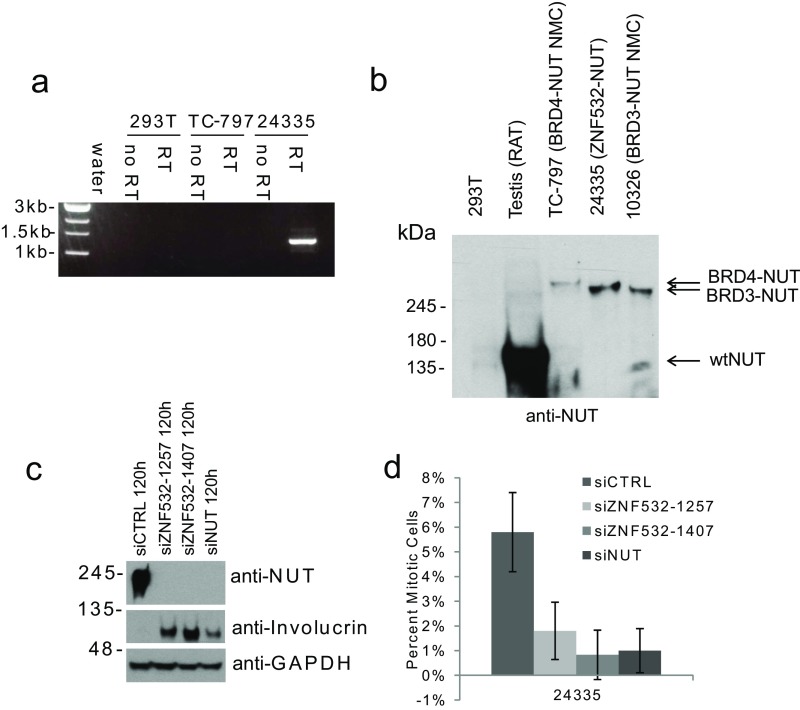
During the course of these studies, we encountered a 62-y-old female patient with NMC involving the right upper lobe of her lung and pleura diagnosed based on biopsy and positive nuclear immunohistochemical staining with the NUT antibody (Fig. 3A). The patient was enrolled in the NUT Midline Carcinoma Registry (www.NMCRegistry.org) and we established a cell line, 24335, from her malignant pleural fluid posthumously through internal review board approval. Whole-transcriptome sequencing (RNA-seq) on this cell line, 24335, identified a ZNF532–NUT fusion gene (Fig. 3B) that was confirmed by cytogenetics (Fig. 3C), RT-PCR (Fig. S2A), fluorescence in situ hybridization (Fig. 3D), Western blotting (Fig. S2B), and immunohistochemistry (Fig. 3E), demonstrating nuclear foci similar to those of BRD4–NUT. The resulting predicted 1,500-amino acid protein includes the N-terminal 778 amino acids of ZNF532, encoding only the first 2 of 12 zinc fingers and a large unstructured domain, part of intron 1 of NUT, and its remaining exons 2 to 7 (Fig. 3F). The first zinc finger included in the ZNF532–NUT fusion encodes a putative zinc-ribbon domain that is predicted to bind nucleic acids directly. Exons 2 to 7 of NUT are recurrently included in previously documented NUT fusions (10, 23–25), strongly suggesting that the ZNF532–NUT fusion protein is likely to be a primary driver of this malignancy.
Fig. 3.
Discovery and characterization of a human NMC harboring a ZNF532–NUT fusion oncogene. (A) H&E stain (Left) and diagnostic anti-NUT immunohistochemical stain (Right) of a resected NMC of the lung of a 64-y-old woman. (Magnification: 340×, Left; 700× Right.) (B) cDNA sequence of the patient’s tumor cell line, 24335, reveals the fusion of ZNF532 (red) to NUT intron 1 (black) and NUT exon 2 (blue). (C) Partial karyotype taken from 24335 was 47, XX, +7 t(15;18)(q14;q23). Arrows denote breakpoints. (Magnification: 4,700×.) (D) Dual-color FISH split-apart assay of ZNF532 (red, centromeric 5′; green, telomeric 3′) and fusion assay of ZNF532 (red) to NUT (green). (Magnification: 3,230×.) (E) Immunohistochemical stain of the patient’s primary tumor using anti-ZNF532 antibodies. (Magnification: 1,380×.) (F) Schematic of NUT fusions with ZNF532 and BRD4 (arrows denote fusion breakpoints). (G) H&E staining of 24335 cells 120 h following duplicate sequential siRNA transfection at 0 and 48 h. (Magnification: 400×.)
Fig. S2.
Characterization of an NMC patient cell line, 24335, harboring a ZNF532–NUT fusion oncogene. (A) RT-PCR using ZNF532 forward and NUT reverse primers. 293T and TC-797 serve as negative control cell lines. RT, reverse transcription. (B) Immunoblot of wild-type NUT (rat testis) and NMCs with different fusions. (C) Immunoblot of 24335 lysates harvested after 120 h from the start of sequential siRNA transfections at 0 and 48 h. Involucrin is a marker of squamous differentiation. GAPDH is the loading control. (D) Quantification of mitotic cells, treated as in C, by anti–phospho-histone H3 (Ser10) immunohistochemistry (rabbit polyclonal; Cell Signaling Technology). Three hundred cells were counted per transfection in technical triplicates. Error bars represent the standard deviation of each technical triplicate.
To test the requirement for ZNF532–NUT in proliferation and in the blockade of differentiation in 24335 patient cells, we treated the cells with siRNAs against either ZNF532 or NUT. We found that in each case, ZNF532–NUT knockdown resulted in morphological changes indicative of differentiation (Fig. 3G) as well as induction of a terminal squamous differentiation marker, involucrin (Fig. S2C). Furthermore, proliferation of 24335 patient cells was attenuated after ZNF532–NUT knockdown (Fig. S2D). Together, these results confirm that the novel ZNF532–NUT translocation causes a variant form of NMC.
ZNF532–NUT Forms Megadomains of Hyperacetylated Chromatin, Including at the MYC Locus.
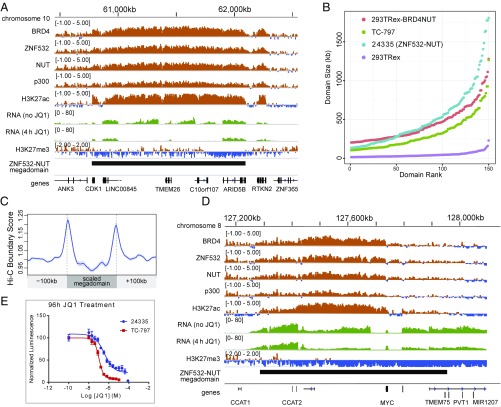
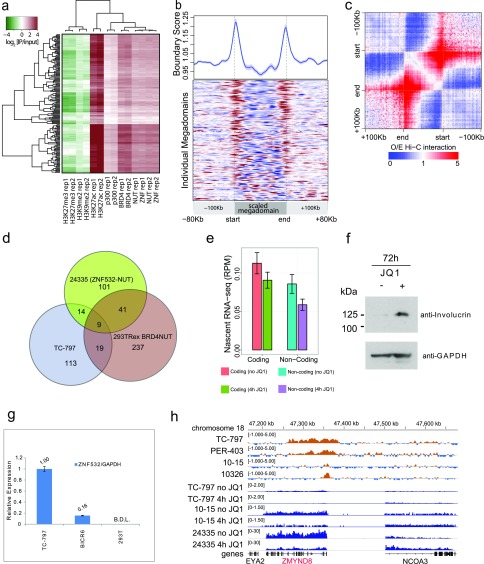
Our finding that ZNF532–NUT forms nuclear foci (Fig. 3A) led us to test whether ZNF532–NUT forms transcriptionally active megadomains similar to those of BRD4–NUT. Indeed, ChIP-seq analyses of 24335 cells, expressing endogenous ZNF532–NUT, revealed 100-kb- to 2-Mb-sized contiguous regions highly enriched with NUT, ZNF532, BRD4, EP300, and H3K27Ac (Fig. 4A). Consistent with a transcriptionally active state, the silencing mark H3K27me3 was excluded from ZNF532 megadomains (Fig. S3A). Thus, the ZNF532–NUT megadomains closely resemble BRD4–NUT megadomains, with an even larger size distribution (Fig. 4B). ZNF532 megadomains, like those of BRD4–NUT (3), typically are delimited by topologically associating domain boundaries, often filling entire TADs (Fig. 4C and Fig. S3 B and C). Interestingly, despite these similarities, there was little overlap in the genomic location of ZNF532–NUT megadomains with those of BRD4–NUT in TC-797 and 293TRex cells (Fig. S3D), consistent with the idea that NUT-associated megadomains are cell type-specific (3).
Fig. 4.
ZNF532–NUT forms megadomains and is JQ1-sensitive. (A) An example of a ZNF532–NUT domain in proximity to the CDK1 locus in the 24335 cell line. The domain shows strong enrichment for ZNF532, BRD4, NUT, EP300, and H3K27ac. H3K27me3 depletion is shown for contrast. Nascent RNA read density with and without the JQ1 treatment is in green. (B) ZNF532–NUT domain size in 24335 cells, compared with BRD4–NUT megadomains in TC-797 and 293TRex cells. The dot plots show the top 150 enhancer-state domains (as defined by continuous regions of H3K27ac enrichment; Materials and Methods) in the different cell lines. 293TRex is representative of a normal cell state, lacking a NUT-fusion protein. 293TRex BRD4–NUT shows the extent of megadomains formed after 7 h of BRD4–NUT induction. (C) Hi-C boundary score (Materials and Methods) shows pronounced peaks at the megadomain boundaries, indicating that they tend to coincide with topologically associating domain boundaries (see also Fig. S3 B and C). Hi-C data from the GM12878 cell line were used. (D) MYC locus megadomain in 24335 ZNF532–NUT cells. The RNA tracks illustrate the difference in nascent transcription following a 4-h JQ1 treatment. (E) Dose–response curve of 24335 and TC-797 cells to JQ1 scored by CellTiter-Glo (Promega Biosciences) 96 h following treatment with JQ1 was performed in biologic triplicates. The results are representative of one experiment; error bars are the standard deviation of quadruplet technical replicates for each concentration of JQ1. IC50, half-maximal inhibitory concentration, was 357 nM for 24335 cells and 104 nM for TC-797 cells.
Fig. S3.
ZNF532–NUT megadomains are depleted for H3K27me3 and delimited by TADs. (A) Average ChIP enrichment within ZNF532–NUT megadomains in 24335 cells. In addition to prominent enrichment for BRD4, NUT, H3K27ac, and EP300, the ZNF532–NUT megadomains show marked depletion of histone marks H3K27me3 and H3K9me2 that are associated with epigenetic silencing. (B) An extended version of Fig. 4C; the Hi-C boundary score profiles for the individual domains are shown as a heatmap under the average plot, with red colors marking high boundary scores and blue colors marking low scores. Hi-C data from GM12878 were used. (C) A combined observed/expected (O/E) Hi-C interaction frequency plot for the ZNF532–NUT megadomains. Rescaled megadomains and 100-kb flanking regions are shown. (D) The number of overlapping megadomains is shown for the ZNF532–NUT domains in 24335 cells and BRD4–NUT domains in TC-797 cells and 293TRex cells after 7 h of BRD4–NUT induction. (E) The average levels of nascent transcription (y axis, RPM) drop in 24335 cells following JQ1 treatment. Different bars show genic and intergenic nascent transcription rates before and after a 4-h JQ1 treatment. The error bars show the 95% error of the mean. RPM, reads per million. (F) Immunoblot of 24335 cells treated with 500 nM JQ1 BET inhibitor. (G) Quantitative RT-PCR of ZNF532 RNA in TC-797 NMC cells and BICR6, a non-NMC head and neck squamous cancer cell line. ZNF532 RNA was below detection limits (B.D.L.) in 293T cells. Error bars represent standard deviation. (H) BRD4–NUT (NUT ChIP) binds the 5′ promoter region of the ZMYND8 gene in different NMC patient-derived cells, along with levels of nascent transcription (blue tracks) with and without JQ1 treatment. In all tested cell lines, the rate of ZMYND8 transcription was decreased following JQ1 treatment.
Despite the very limited overlap of BRD4–NUT versus ZNF532–NUT megadomains, the MYC locus, including adjacent noncoding regions, is highly enriched with ZNF532–NUT (Fig. 4D). Moreover, most of the large regulatory region is transcriptionally active, similar to, and even more extensive than, all other NMC cells analyzed (3). These findings further support that MYC dysregulation is critical to the oncogenic function of all NUT-fusion proteins in NMC, including ZNF532–NUT.
Evidence That ZNF532–NUT Interacts with BET Proteins to Drive Proliferation.
One model to explain how megadomains form in patient cells harboring a ZNF532–NUT fusion is that linkage to ZNF532 allows NUT to ectopically interact with endogenous BRD4 complexes, forming complexes analogous to those found in BRD4–NUT patient cells. To test whether ZNF532–NUT is dependent on BET proteins for its oncogenic role, we asked whether 24335 patient cells are sensitive to JQ1, the acetyl-histone mimetic that specifically inhibits binding of BET-family bromodomains. We found that ZNF532–NUT megadomains are associated with transcription genome-wide, which decreases rapidly upon BET inhibition (Fig. 4 A and D and Fig. S3E). Most importantly, 24335 cells differentiate and arrest growth in the presence of JQ1, with IC50 values in the same range (357 nM) as that of TC-797 cells (104 nM) (Fig. 4E and Fig. S3F). These results support a model in which the ZNF532–NUT fusion protein is dependent on BET bromodomain proteins for its oncogenic properties.
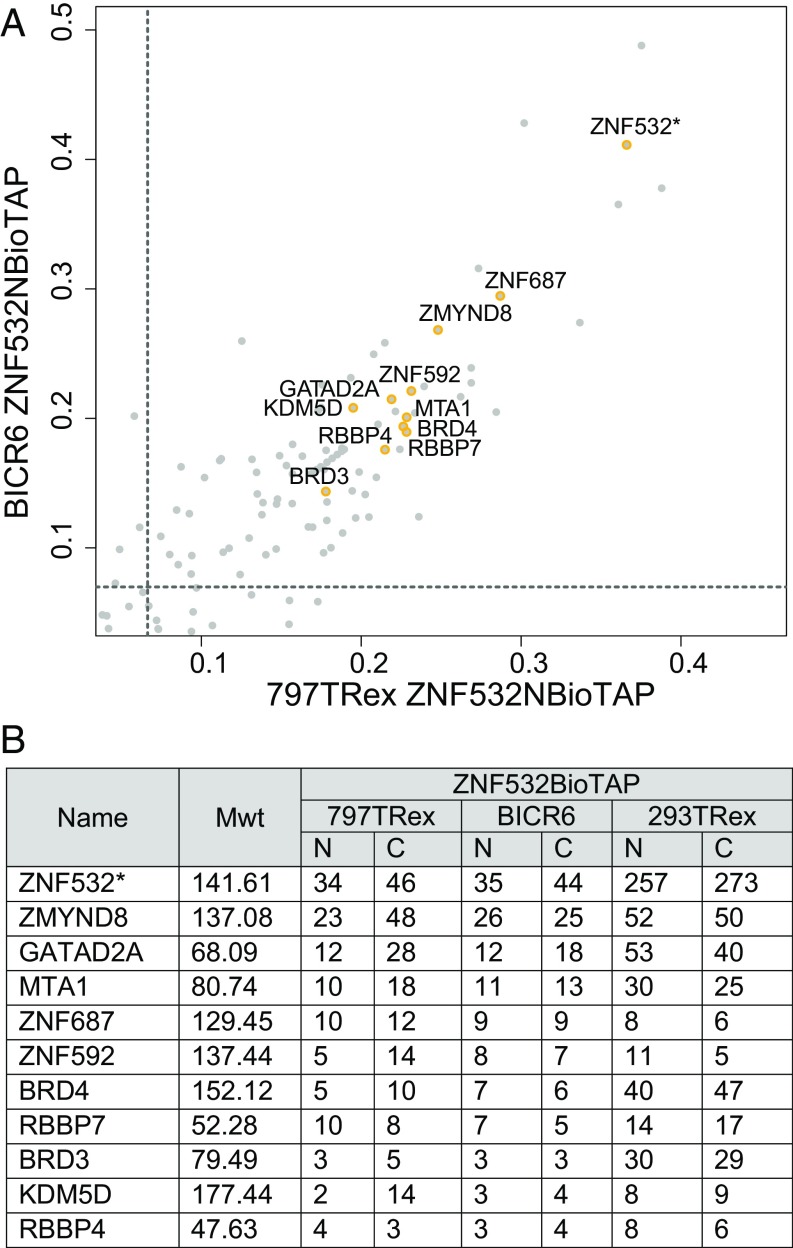
Based on our initial identification of ZNF532 in BRD4–NUT pulldowns from NMC patient cells, we tested whether ZNF532 might provide a physical bridge to BRD4 complexes. To accomplish this, we tagged ZNF532 and assessed its protein interactions in 797TRex NMC patient cells, BICR6 non-NMC cells, and 293TRex cells. BICR6 is a non-NMC head and neck squamous cell patient cell line that expresses endogenous ZNF532, whereas expression is not detectable in 293TRex cells (Fig. S3G). We found that in all three cell lines, BioTAP-tagged ZNF532 displays similar interactions (Fig. 5), including with ZMYND8, ZNF687, ZNF592, and BRD4. These proteins were also linked to ZNF532 in data from previous mass spectrometric analyses (26–28). Interestingly, the ZMYND8 locus, encoding a top interactor of ZNF532, is bound by NUT-fusion proteins in all NMC lines tested (selected examples are shown in Fig. S3H). Furthermore, ZMYND8 transcription is sensitive to BET protein inhibition in both BRD4–NUT and ZNF532–NUT patient cell lines (Fig. S3H), and in several additional non-NMC cell lines studied by others (19). The ZNF532 locus is also JQ1-sensitive in the 24335 patient cell line (Fig. S4). Taken together, our findings strongly support a model in which BET proteins, coupled directly or indirectly to NUT, form an oncogenic complex that drives transcription in multiple self-sustaining regulatory loops (Fig. 6).
Fig. 5.
BioTAP–ZNF532 pulldowns enrich for BET bromodomain proteins and additional conserved interactions. (A) Scatterplot of affinity pull-down enrichments, comparing BioTAP–ZNF532 in 797TRex NMC patient cells and in BICR6 non-NMC cells. Thresholds for enrichment from both pulldowns were set at the 95th percentile. (B) A list of the common hits, showing results from both N- and C-terminally tagged ZNF532 pulldowns in 797TRex, BICR6, and 293TRex cells. Asterisks denote bait used for pulldown. Mwt, molecular weight of protein in kilodaltons.
Fig. S4.
Transcription of the ZNF532 locus is JQ1-sensitive in 24335 cells. Moderate enrichment for BRD4, NUT, and H3K27ac together with exclusion of the silencing mark H3K27me3 at the 5′ promoter region correlate with a transcriptionally active state of the ZNF532 gene in 24335 cells. Transcription is strongly reduced after 4 h of 10 μM JQ1 treatment as measured by nascent RNA-seq.
Fig. 6.
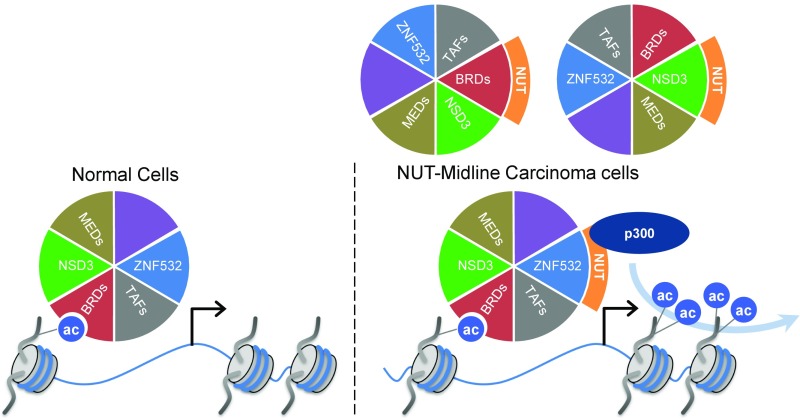
Model for how NUT-fusion proteins drive oncogenic megadomain formation. BRD complexes function at gene-specific locations to facilitate transcription in normal cells. In contrast, NMC patients harbor NUT translocations to BRD4, BRD3, NSD3, or ZNF532 that form large megadomains, often filling whole TADs. This is postulated to occur through NUT recruitment of EP300 to the aberrant BRD regulatory complexes, leading to iterative histone acetylation and NUT-fusion protein recruitment to newly acetylated chromatin. NUT fusions to a variety of BRD-complex members result in an analogous mechanism for megadomain formation. MEDs, mediator complex subunits; TAFs, TATA box-binding protein-associated factors.
Discussion
Mutations in chromatin regulatory proteins are prevalent in cancer. Chromatin factors generally act within large, multisubunit complexes; thus, identifying both their normal and aberrant interactors in cancers should provide important information regarding potential targets for therapeutic intervention. Here we apply this principle to analysis of BRD4–NUT, a fusion oncoprotein that drives an aggressive subtype of squamous cell cancer. We use chromatin cross-linking combined with affinity purification and mass spectrometry (BioTAP-XL) to affinity purify BRD4 vs. BRD4–NUT complexes and compare their protein–protein interactions. We present three principal findings from this approach. First, our proteomic analyses reveal that the NUT–EP300 interaction distinguishes BRD4–NUT from wild-type BRD4 complexes, providing strong support for a model in which a feed-forward acetylation and acetyl-binding cycle drives the formation of hyperacetylated megadomains. Second, we discover ZNF532 to be a BRD4–NUT interactor in 797TRex patient cells that is fused directly to NUT in 24335 patient cells to create another oncogenic fusion protein, ZNF532–NUT. Our results provide evidence that linkage of NUT to BRD4 complexes directly, via BRD4–NUT translocations, or indirectly, via fusion to other subunits (ZNF532, NSD3, BRD3) (6, 16), drives acetylated megadomains and oncogenesis in NMC patients (Fig. 6). Conceptually similar to the discovery that various oncogenic MLL fusions commonly interact in a “superelongation” complex in leukemia (29), our work supports a model in which distinct oncogenic translocations act interchangeably because they are able to force a common set of factors to interact aberrantly. Third, beyond the EP300/BRD4 acetylation and binding cycle posited to form megadomains, we have evidence for self-reinforcement of the oncogenic complex via several feed-forward regulatory loops. We find that multiple genetic targets of BRD4–NUT or ZNF532–NUT, including ZNF532 and ZMYND8, encode components of BRD4 regulatory complexes. Interestingly, it was previously reported that ZMYND8/ZNF532/ZNF592/ZNF687 interact with each other as a central hub in a large transcriptional network (26, 30). ZMYND8 has several important chromatin-binding domains (bromodomain, PWWP, PHD finger, MYND), and can function as a combinatorial reader of histone modifications such as H3K4me1 and H3K14ac (31). Whether ZNF532 and ZMYND8 perform essential functions in NMC is still under investigation. However, another transcriptional coregulator complex, Mediator, is clearly implicated in BRD4 function in both normal and NMC cells (3). Selection for these types of self-reinforcing regulatory loops may be a powerful strategy for driving the growth advantage of cancer cells.
The discovery of misregulation via protein–protein interactions will require a more comprehensive proteomic approach to cancer biology, but current studies have not kept pace with genomic analyses. This is likely in part due to the technical challenges inherent to biochemical purifications, especially of chromatin- and DNA-binding protein complexes. We believe that our cross-linking approach largely circumvents many of the technical issues and represents a powerful complementary approach to existing analyses. Beyond the specific relevance to NMC, our interaction lists obtained by cross-linking combined with affinity purification and mass spectrometry provide new associations with potentially broad relevance to BRD4-dependent cancers. BET inhibitor studies have revealed a growing spectrum of cancers dependent on nonmutated BRD4; thus, identifying the breadth of normal and aberrant BRD4 interactions should provide useful candidates for combined therapeutic approaches. Additional candidates with known inhibitory compounds include XPO1 (6, 32), EP300 (33), and the transcriptional elongation complex pTEFb, composed of CDK9 and CCNT1 (34). Interestingly, our study confirms that pTEFb is associated with BRD4–NUT as well as BRD4 (Fig. 1A), and flavopiridol, a known CDK9 inhibitor, was one of the most efficacious compounds among a large panel of drugs tested on NMC cells and xenografts (35). Furthermore, our results suggest that the putative coregulatory module consisting of ZNF532 and ZMNYD8 should be considered for small-molecule targeting, as ZMNYD8 carries multiple chromatin modification binding domains that could be subject to inhibition (31).
Materials and Methods
Cell Culture.
293TRex, TC-797 (36), and BICR6 (hypopharyngeal squamous cell carcinoma) (37) were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (S10350; Atlanta Biologicals), 1% penicillin/streptomycin, and 1× GlutaMAX (Invitrogen). NMC 24335 cells (ZNF532–NUT) were obtained under institutional IRB approval (Dana–Farber Cancer Institute protocol number 10-228) and were initially established in OCMI-E media (38) (purchased from the Live Tumor Culture Core, University of Miami; sylvester.org/shared-resources/Live-Tumor-Culture-Core). The patient was deceased at the time of fluid collection and thus informed consent was waived per this IRB protocol as defined by 45 CFR 46.116(d).
797TRex and 293TRex derivatives, which contain a single genomic FRT recombination site and constitutively express the tetracycline repressor (TR), were created as described (2) and maintained as above with blasticidin (Invitrogen) and zeocin (Invitrogen). 797TRex, 293TRex, and N- and C-BioTAP–BRD4–NUT lines were generated as described previously (2, 3) and maintained in blasticidin and hygromycin (Sigma).
Stable 797TRex and BICR6 ZNF532–BioTAP lines were generated by lentiviral transduction using the vectors N-BioTAP and C-BioTAP pHAGE-CMV-TET-ZNF532 (details on vectors and constructs will be provided upon request) followed by selection with puromycin.
Purification of RNA and Identification of ZNF532–NUT Fusion by RNA Sequencing.
RNA was extracted from live cultured 24335 cells using the RNeasy Plus Kit (Qiagen) followed by the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs). rRNA removal was performed using the Ribo-Zero Kit (Epicentre) following the manufacturer’s instructions. The library was prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs) following the manufacturer’s instructions and sequenced at the Partners HealthCare Personalized Medicine/Genetics and Genomics core facility (personalizedmedicine.partners.org) using the HiSeq 2500 (Illumina).
Samples were evaluated for quality and trimmed of adapters and low-quality bases using Trim Galore, version 0.3.3 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default settings to produce FastQC plots and trimmed read files. Alignment to the human genome (GRCh37/hg19) and identification of putative fusions was done with TopHat2 (39) and TopHat-Fusion (40) (TopHat, version 2.0.8) using UCSC Genome Browser annotations downloaded from the Illumina iGenome server as the known transcript reference (ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Homo_sapiens/UCSC/hg19/Homo_sapiens_UCSC_hg19.tar.gz; May 14, 2014). Default parameters were used except for the following: Bowtie (version 1.0.0, –bowtie1) was used as the aligner (bowtie-bio.sourceforge.net/index.shtml); inner mate–pair distance and SD were determined from a test alignment (–mate-inner-dist 116 –mate-std-dev 150); –max-intron-length was set to 1000000; and alignments to chrM were excluded (–fusion-ignore-chromosomes chrM).
BioTAP-XL Procedure and Mass Spectrometric Data Analysis.
Cells (7.5 × 108) grown as monolayer cultures in 150-mm dishes (50 plates total) were used for BioTAP-XL purification. Tetracycline was added (1 μg/mL) to 60 to 70% confluent culture to induce transcription of the N- or C-BioTAP–tagged BRD4–NUT cDNA for 24 h or for 4 d in the case of ZNF532 cDNA. The main steps of the BioTAP-XL procedure were as follows: cell harvesting, formaldehyde cross-linking, chromatin preparation, affinity purification, input, and IP protein recovery were performed as described (12, 41). Mass spectrometric files were searched with the SEQUEST algorithm against the human proteome (42). The mass spectrometric proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005786. We determined the normalized spectral abundance factor for each protein identified per LC-MS experiment essentially as described (43). The main difference in this report is that we proportionately scaled the pseudocount given for zero-spectral count proteins based on the sum of spectra sequenced in the input and pulldown. To illustrate using a designated base value of 0.5, if the IP had one-fourth the number of spectra as the input in one scenario and if the IP had an equal number of spectra as the input in another scenario, we would use a pseudocount of 0.1 for the IP and 0.4 for the input with the former and a pseudocount of 0.25 for both IP and input with the latter.
ChIP-Seq and Data Analysis.
Cross-linked chromatin for ChIP-seq experiments using antibodies against proteins and histone modifications was prepared from 5 × 108 24335 (ZNF532–NUT) NMC cells as described (41). Affinity purification (44) and input and IP DNA recovery and library preparation (41) were performed as described. The following antibodies against proteins and histone modifications were used for ChIP-seq experiments: anti-H3K27ac (10 μL per IP; Active Motif; 39133); anti-P300 (10 μL; Bethyl; A300-358A); anti-H3K27me3 (10 μL per IP; Cell Signaling Technology; 9733); anti-BRD4 (5 μL per IP; Bethyl; A301-985A); anti-ZNF532 (10 μL per IP; Bethyl; A303-329A); and anti-NUT (15 μL per IP; clone C52B1; Cell Signaling Technology; 3625). ChIP-seq reads were aligned to the human reference genome (GRCh38/hg38 assembly) using Bowtie (version 0.12.5), retaining only uniquely mapped reads. Smoothed enrichment profiles were generated using the SPP package (45), using a smoothing bandwidth of 500 bp.
Recovery and Analysis of Nascent RNA.
ZNF532–NUT 24335 cells (4 × 106) were grown in 100-mm dishes. Cells were split into two T-25 flasks, and JQ1 (10 μM final concentration) or DMSO was added to the media. After 4 h, cells were harvested and nascent RNA was purified as described (3). Each RNA library was constructed from 50 ng of nascent RNA following the manufacturer’s instructions (NEBNext Ultra Directional RNA Library Prep Kit; New England BioLabs).
Nascent RNA-seq reads were first aligned to the human genome reference using the procedure identical to that used for ChIP-seq processing. To compare nascent transcription rates between different samples, the smoothed read density profiles (500-bp bandwidth) were normalized to the total read density using the SPP package (45).
RNA Purification and Quantification by Quantitative Real-Time PCR.
Total RNA was extracted from 1 × 106 TC-797, BICR6, and 293TRex cells by a standard TRIzol extraction (Life Technologies), followed by RNA purification and removal of genomic DNA with the RNeasy Plus Kit (Qiagen). The VILO cDNA Synthesis Kit (Invitrogen) was used to reverse-transcribe RNA. To quantify transcript levels, real-time PCR was performed on an ABI PRISM 7000 machine as described (46). Quantitative (q)PCR primer sequences are provided in Table S1.
Table S1.
Oligonucleotides used for qRT-PCR and siRNAs
| Oligonucleotide | Sequence, 5′-3′ |
| qRT-PCR | |
| ZNF532-195-F | TTTGTCCGAAGATAGCGTGC |
| ZNF532-325-R | AACATACCTCGAGTGGAAGGAA |
| qGAPDH(1S) | CAATGACCCCTTCATTGACC |
| qGAPDH(1A) | TTGATTTTGGAGGGATCTCG |
| siRNA | |
| ZNF532-1407 | |
| Sense | GCAAAGAACCAGUGGCCAAUU |
| Antisense | UUGGCCACUGGUUCUUUGCUU |
| ZNF532-1257 | |
| Sense | ACAGAGUCCUAGAUGGGAAUU |
| Antisense | UUCCCAUCUAGGACUCUGUUU |
| ZNF532–NUT breakpoint oligos for RT-PCR | |
| bpZNF532-1951-Fa | GAAGCTCGCTAACAACACCAC |
| bpNUT-814-Ra | CGAATTGAGGGCAGTCTGAGT |
F, forward; R, reverse.
siRNA Transfections.
For ZNF532 and NUT knockdown in 24335 cells, 1.5 × 106 cells were transfected with 50 nM siRNA using the RNAiMAX (Invitrogen; 13778) reverse-transfection protocol, sequentially at times 0 and 48 h, and plated in suspension in a 100-mm cell-culture dish. siGENOME Non-Targeting siRNA no. 4 (Dharmacon; D-001210-04) was used as a negative control. All siRNAs are included in Table S1.
Immunofluorescence.
Immunofluorescence of TC-797 cells was performed as described previously (7), and nuclei were counterstained with ProLong Gold Antifade Reagent with DAPI (Life Technologies; P36935). Primary antibodies used were rabbit polyclonal anti-ZNF532 (1:1,000; Bethyl Laboratories; A305-442A) and mouse monoclonal anti-HA (1:1,000; clone 16B12; Covance Research Products; MMS-101R). Secondary antibodies included goat anti-rabbit Alexa Fluor 594 and goat anti-mouse Alexa Fluor 488 (1:1,000; Life Technologies; A-11012 and A-11001). Photographs were taken with a Nikon Eclipse E600 fluorescence microscope using a Spot RT Slider camera (Diagnostic Instruments) and Spot Advanced software (Diagnostic Instruments).
Histology and Immunohistochemistry.
Formalin-fixed, paraffin-embedded (FFPE) cell blocks of cultured cells were prepared as described (6, 47) using HistoGel (Richard-Allan Scientific/Thermo Fisher Scientific). Immunohistochemistry of the tumor from which the ZNF532–NUT cell line, 24335, was derived was performed as described (47) using anti-NUT (clone C52B1; 1:50) and anti-ZNF532 (1:4,000; Bethyl; A305-442A).
Quantification of Cell Viability.
TC-797 and 24335 cells were plated at a density of 3,000 per well in a 96-well plate, and CellTiter-Glo (Promega Biosciences) was used to determine cell viability as a measure of ATP content according to the manufacturer’s instructions.
Fluorescence in Situ Hybridization.
Dual-color FISH for ZNF532, BRD4, and NUT was performed on 5 µM FFPE sections of surgically removed tumor from which the 24335 cell line was derived as described (48). Probes used for NUT included the 3′ flanking telomeric BAC clones RP11-1H8 and -64O3 and the 5′ flanking centromeric probes RP11-145E17 and -92B21. Probes used for BRD4 included the 5′ centromeric probes RP11-207i16 and -3055m5 and the 3′ telomeric probes RP11-319010 and -681D10. Probes used for ZNF532 included the 3′ telomeric BAC probes RP11-350K6 and -1061A13 and the 5′ centromeric probes RP11-351N16 and -722P5. Two hundred nuclei were counted in four different areas of the tumor. Eighty percent positivity in interpretable nuclei was defined as positive for rearrangement.
Immunoblotting.
Western blots were prepared as described (6). Primary antibodies used were rabbit anti-ZNF532 (1:2,000; Bethyl; A305-442A), mouse anti-GAPDH (1:5,000; clone 6C5; Life Technologies), rabbit anti-NUT (1:1,000; clone C52B1; Cell Signaling Technology), and mouse anti-involucrin (1:5,000; clone GAPDH-71.1; Sigma-Aldrich).
Quantification of Mitoses.
Immunohistochemistry was performed on cells cultured on glass coverslips as follows. Following washing in PBS, coverslips were fixed in 10% phosphate-buffered formaldehyde and then washed and blocked in PBS, Triton X-100 (3%; PBT; Sigma) for 1 h. Coverslips were then incubated in rabbit phospho-histone H3 (Ser10) antibody (1:500; Cell Signaling Technology; 9701) with PBT containing 2% nonfat milk for 1 h. After washing in PBS, coverslips were incubated in secondary antibody (HRP-conjugated swine anti-rabbit; 1:250; Agilent/Dako; P0217) in PBT, 2% milk for 1 h. After washing in PBS, coverslips were incubated in 200 µL (one drop) of DAB peroxidase substrate per the manufacturer’s instructions (Dako). After washing in PBS, coverslips were counterstained in hematoxylin and mounted onto glass slides for manual quantification using an Olympus BX40 microscope. Three hundred cells were counted per technical triplicate.
Megadomain Identification and TAD Boundary Detection.
As in ref. 3, the megadomains were detected as regions of contiguous H3K27ac enrichment on the chromosome. The regions were detected using a three-state hidden Markov model (HMM) with Gaussian emissions, with the emission mean set to μ, μ + 2σ, and μ − 2σ for the neutral, enriched, and depleted states, respectively (μ, genome-wide enrichment mean; σ, genome-wide enrichment variance). The HMM was run on an enrichment (log2 ChIP/input ratio) profile estimated in 500-bp windows tiled along the chromosome. The state-transition probability was set to 10 to 50. For the purposes of comparison (e.g., Fig. S3D), BRD4–NUT domains were identified using the same method. Analysis of Hi-C data, including Hi-C boundary scores and combined observed/expected (O/E) estimates, were performed as in ref. 3.
Supplementary Material
Acknowledgments
We are grateful to Dr. Peter Hammerman for providing the BICR6 cell line and Ross Tomaino for help with protein samples (Taplin Mass Spectrometry Facility, Harvard Medical School). This work was supported by a St. Baldrick’s Foundation research grant (to C.A.F.), National Institutes of Health Grants NCI R01CA124633 (to C.A.F.) and R01GM101958 (to M.I.K.), Ellison Medical Foundation Grant AG-NS-0965-12 (to T.P. and P.V.K.), and The Jane Coffin Childs Memorial Fund fellowship (to B.M.Z.).
Footnotes
Conflict of interest statement: T.A.I. has a pending patent application relating to the OCMI-E medium.
Data deposition: The ChIP-seq and nascent RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE96775). The RNA-seq data detecting the ZNF532–NUT fusion transcript reported in this paper have been deposited in the Sequence Read Archive (accession no. PRJNA361284). The mass spectrometric proteomics data reported in this paper have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (accession no. PXD005786).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702086114/-/DCSupplemental.
References
- 1.Wang R, et al. Activation of SOX2 expression by BRD4-NUT oncogenic fusion drives neoplastic transformation in NUT midline carcinoma. Cancer Res. 2014;74:3332–3343. doi: 10.1158/0008-5472.CAN-13-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson AR, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014;33:1736–1742. doi: 10.1038/onc.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekseyenko AA, et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015;29:1507–1523. doi: 10.1101/gad.267583.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 5.French C. NUT midline carcinoma. Nat Rev Cancer. 2014;14:149–150. doi: 10.1038/nrc3659. [DOI] [PubMed] [Google Scholar]
- 6.French CA, et al. BRD-NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 7.Reynoird N, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 2010;29:2943–2952. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stathis A, et al. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French CA, et al. BRD4-NUT fusion oncogene: A novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 11.Schwartz BE, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011;71:2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci USA. 2014;111:2488–2493. doi: 10.1073/pnas.1400648111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue D, et al. SETBP1 mutations drive leukemic transformation in ASXL1-mutated MDS. Leukemia. 2015;29:847–857. doi: 10.1038/leu.2014.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French CA, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov. 2014;4:928–941. doi: 10.1158/2159-8290.CD-14-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kees UR, Mulcahy MT, Willoughby ML. Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19) Am J Pediatr Hematol Oncol. 1991;13:459–464. doi: 10.1097/00043426-199124000-00011. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, et al. HEXIM1 as a robust pharmacodynamic marker for monitoring target engagement of BET family bromodomain inhibitors in tumors and surrogate tissues. Mol Cancer Ther. 2017;16:388–396. doi: 10.1158/1535-7163.MCT-16-0475. [DOI] [PubMed] [Google Scholar]
- 19.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henssen A, et al. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin Cancer Res. 2016;22:2470–2481. doi: 10.1158/1078-0432.CCR-15-1449. [DOI] [PubMed] [Google Scholar]
- 21.Fiskus W, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014;13:1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- 22.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haruki N, et al. Cloned fusion product from a rare t(15;19)(q13.2;p13.1) inhibit S phase in vitro. J Med Genet. 2005;42:558–564. doi: 10.1136/jmg.2004.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson-Wicking K, et al. Novel BRD4-NUT fusion isoforms increase the pathogenic complexity in NUT midline carcinoma. Oncogene. 2013;32:4664–4674. doi: 10.1038/onc.2012.487. [DOI] [PubMed] [Google Scholar]
- 25.Stirnweiss A, et al. A novel BRD4-NUT fusion in an undifferentiated sinonasal tumor highlights alternative splicing as a contributing oncogenic factor in NUT midline carcinoma. Oncogenesis. 2015;4:e174. doi: 10.1038/oncsis.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloet SL, et al. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 2015;282:1774–1785. doi: 10.1111/febs.12972. [DOI] [PubMed] [Google Scholar]
- 27.Hein MY, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 28.Gong F, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 30.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li N, et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol Cell. 2016;63:470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etchin J, et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia. 2017;31:143–150. doi: 10.1038/leu.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammitzsch A, et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci USA. 2015;112:10768–10773. doi: 10.1073/pnas.1501956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newcomb EW. Flavopiridol: Pleiotropic biological effects enhance its anti-cancer activity. Anticancer Drugs. 2004;15:411–419. doi: 10.1097/01.cad.0000127332.06439.47. [DOI] [PubMed] [Google Scholar]
- 35.Beesley AH, et al. Comparative drug screening in NUT midline carcinoma. Br J Cancer. 2014;110:1189–1198. doi: 10.1038/bjc.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toretsky JA, et al. Translocation (11;15;19): A highly specific chromosome rearrangement associated with poorly differentiated thymic carcinoma in young patients. Am J Clin Oncol. 2003;26:300–306. doi: 10.1097/01.COC.0000020960.98562.84. [DOI] [PubMed] [Google Scholar]
- 37.Edington KG, Loughran OP, Berry IJ, Parkinson EK. Cellular immortality: A late event in the progression of human squamous cell carcinoma of the head and neck associated with p53 alteration and a high frequency of allele loss. Mol Carcinog. 1995;13:254–265. doi: 10.1002/mc.2940130408. [DOI] [PubMed] [Google Scholar]
- 38.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, Salzberg SL. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alekseyenko AA, et al. BioTAP-XL: Cross-linking/tandem affinity purification to study DNA targets, RNA, and protein components of chromatin-associated complexes. Curr Protoc Mol Biol. 2015;109:21.30.1–21.30.32. doi: 10.1002/0471142727.mb2130s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 43.Zee BM, Dibona AB, Alekseyenko AA, French CA, Kuroda MI. The oncoprotein BRD4-NUT generates aberrant histone modification patterns. PLoS One. 2016;11:e0163820. doi: 10.1371/journal.pone.0163820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Haack H, et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol. 2009;33:984–991. doi: 10.1097/PAS.0b013e318198d666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.French CA, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19) Am J Pathol. 2001;159:1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.