Abstract
HIV-1 envelope spike (Env) is a heavily glycosylated, type I membrane protein that mediates fusion of viral and cell membranes to initiate infection. It is also a primary target of neutralizing antibodies and thus an important candidate for vaccine development. We have recently reported an NMR structure of the transmembrane (TM) domain of HIV-1 Env reconstituted in a membrane-like environment. Taking HIV-1 as an example, we discuss here how a TM domain can anchor, stabilize and modulate a viral envelope spike and how its high-resolution structure can contribute to understanding viral membrane fusion and to immunogen design.
HIV-1 (human immunodeficiency virus type 1) envelope spike [Env; trimeric (gp160)3, cleaved to (gp120/gp41)3] is a type I membrane protein that fuses viral and host cell membranes to initiate viral infection [1]. Sequential binding of gp120 to receptor (CD4) and coreceptor (e.g., CCR5 or CXCR4) triggers large conformational changes in both gp120 and gp41, leading to fusion and viral entry [2-4]. The mature and functional Env spikes, (gp120/gp41)3, are the sole antigens on the virion surface and thus the sole candidates for B-cell based vaccine development [5, 6]. The native prefusion conformation of HIV-1 Env is recognized by most broadly neutralizing antibodies (bnAbs) [7-9], and it is generally believed to have the potential for inducing such antibody responses. Thus, production of a recombinant form that closely resembles the conformation of Env spikes on the surface of virions is an important objective in immunogen design.
Gp140, the uncleaved ectodomain of (gp160)3 with both the transmembrane segment (TM) and the cytoplasmic tail (CT) truncated, has often been considered as a likely surrogate for the native Env spike. Most HIV-1 gp140 preparations, however, are unstable and conformationally heterogeneous [10]. We have previously screened many HIV-1 primary isolates and identified two (clade A 92UG037.8 and clade C C97ZA012) that yield stable and homogeneous gp140 trimers when produced recombinantly in human cells [11, 12]. Other strategies to produce stable soluble Env trimers include the “SOSIP” modifications, which require an added disulfide bond between gp120 and gp41, an Ile-to-Pro mutation in gp41, and deletion of the membrane proximal external region (MPER) [9, 13]. While these approaches have yielded some useful Env trimer immunogens [8, 14, 15], they cannot explain why most soluble HIV-1 Env trimers are unstable. Moreover, we have shown recently that truncation of the CT in the context of the full-length Envs has an unexpectedly large impact on the antigenic structure of the ectodomain [7]. These data suggest that the transmembrane domain (TMD), the only direct link between the CT and the ectodomain, may have a greater role in stabilizing the entire Env and in maintaining its native antigenic structure than previously recognized.
As a type I membrane protein, HIV-1 Env has a single TM segment with a GxxxG motif, implicated in oligomeric assembly of TM helices [16], and a conserved, positively charged residue (usually arginine). The TM is also more conserved than a typical transmembrane anchor, suggesting it may have functions other than just spanning a lipid bilayer. Indeed, TM helices of many cell surface receptors are not merely inert anchors but play essential roles in receptor assembly and signal transmission. Examples include EGFR, integrins and Fas receptor [17-20], for which association of TM helices correlates the conformation of the extracellular domain with the conformations of the membrane proximal regions and the cytoplasmic signaling motifs. A similar mechanism could also underlie our observation that CT truncation affects the antigenic surface of the ectodomain of HIV-1 Env on the opposite side of the membrane [7].
Structure of the transmembrane domain of HIV-1 Env
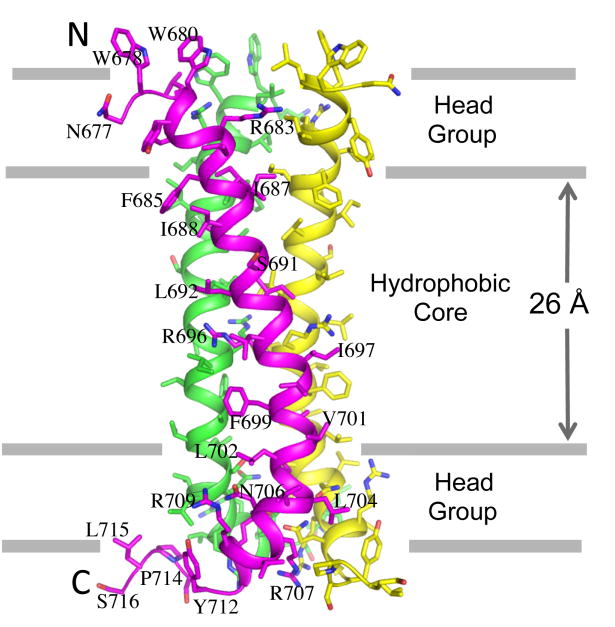
To understand the physical coupling (conformation and/or dynamics) between the CT and the ectodomain, we have determined a structure of the TMD of HIV-1 Env (residues 677-716), reconstituted in bicelles, by NMR ([21]; Fig. 1). The TMD is a well-structured trimer. It shows two unusual features not seen with other known oligomeric transmembrane helices (TMHs). The first is that there is an arginine (R696) near the middle of each of the TMHs, suggesting three unbalanced charges in the hydrophobic core of the membrane if the Arg remains protonated. In our NMR structure, the tips of the long sidechains of these arginines are facing lipids, and each of them is surrounded by three hydrophobic residues (L692 and L695 from the same TMH and I697 from the neighboring TMH). A precise physical basis for the stability of R696 in the highly hydrophobic environment remains unclear, but the underlying mechanism must tolerate a Lys residue, which is present in some viral isolates at this position. In our nuclear Overhauser enhancement (NOE) experiment for arginines, the R696 epsilon protons showed clear water NOE even with a short NOE mixing time of 60 ms, indicating bound water molecule(s) at this Arg position. While a high-resolution crystal structure would be needed to reveal atomic details of the structured water molecule(s) in the surrounding region, we speculate that the charge of R696 can be partially stabilized by the water dipole moments.
Figure 1. Structure of the HIV-1 Env TMD trimer in bicelles.
The protein construct used for structure determination is a fragment of gp41 (residues 677-716), derived from a clade D HIV-1 isolate 92UG024.2. The bicelles are formed with 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine (DMPC) and 1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine (DHPC) at a molar ratio of 0.5. The approximate placement of the trimer structure in the presumed DMPC bilayer, which is slightly thinner than the characterized thickness of DOPC bilayer (35), was based on the solvent paramagnetic relaxation enhancement of the four arginines (R683, R696, R707, and R709) [21].
The TMD trimer structure is obviously necessary for protecting R696 in the middle of the hydrocarbon core of lipid bilayer. A positively-charged Arg or Lys in the TM segment of viral fusion protein is present in some related enveloped viruses, including simian immunodeficiency virus, caprine arthritis and encephalitis virus, equine infectious anemia virus, visna virus, and foamy virus, as well as in hepatitis C virus [22-27], but absent in many others. It is therefore not a prerequisite for viral membrane fusion in general [28]. Our functional studies indicate that the R696A mutant of HIV-1 Env showed some defect in cell-cell fusion, but it could be fully compensated by high Env expression [21]. Moreover, this mutant has wildtype viral infectivity. In vitro infectivity and cell-cell fusion do not, however, mimic all the conditions under which the virus moves from one cell to another in an infected individual, nor are those assays particularly sensitive to physiologically relevant kinetic parameters. We note that R696 occupies a “d” position in the coiled-coil, with its Cβ facing inwards toward the trimer interface, unlike its configuration in previously proposed models [29, 30]. Its interactions in the trimeric coiled-coil are probably those of optimal stability, once having paid the energetic cost of burying its charge. Thus, R696 would contribute to the kinetic barrier for any rearrangement and to restoration of the trimeric structure after any perturbation. One such rearrangement indeed occurs during membrane fusion, when the TMD trimer must break and reform [3, 31], but a more sensitive fusion assay would be needed to test whether R696 contributes to fusion kinetics.
The second unusual feature is that the TMD trimer appears to be stabilized by two separate packing modes. The N-terminal half encompassing the GxxxG motif forms a coiled coil trimer, whereas the C-terminal half is held together by a network of polar contacts, which we named the hydrophilic core. For the N-terminal coiled coil, a helical wheel representation of the trimer clearly indicates packing of hydrophobic residues such as Ile and Val at the “a” and “d” positions of the heptad motif, forming a hydrophobic core. The GxxxG is a well-known motif that drives TMH dimerization [32-34]. In the classic example of the glycophorin A TMD dimer structure, the two glycines of one TMH allow close packing with the GxxxG face of another TMH, resulting in a very strong TMH dimeric complex [32, 34]. There has been no previous report, however, of GxxxG involvement in TMH trimerization. In the coiled coil region of the HIV-1 Env TMD, only G690 is involved in the trimer assembly, i.e., its small sidechain allows a close VDW contact with V689 of the adjacent TMH. The other glycine, G694, is on the periphery of the trimer facing outwards and its mutation to alanine or valine has essentially no effect on TMD trimerization or Env functions [21]. Therefore, the key difference in the structural role of the GxxxG motif between the glycophorin A TMD dimer and HIV-1 Env TMD trimer is that both the glycines are required for forming inter-monomer contacts in the dimer, while only one glycine is important for the trimeric assembly.
TM segments of many viral fusion proteins contain a GxxxG motif or “SmallxxxSmall” motifs (“Small” refers to residues with a small side chain, such as, glycine, alanine, serine or cysteine) [35-37], suggesting that oligomerization of their TMDs may be a common property. For example, recent biochemical evidence has shown that the TMDs of hepatitis C virus envelope glycoproteins E1 and E2 form stable dimers or trimers that are also resistant to SDS [36]. No high-resolution structure of any TMD oligomer from other viral fusion proteins has been reported, due to technical challenges for structural studies of such constructs in the context of lipid bilayer. Our NMR structure of the HIV-1 Env TMD may provide some clues for how other viral fusion proteins oligomerize.
Less obvious is the physical basis of the hydrophilic core formation. Our current hypothesis is that inter-monomer polar contacts among S703, N706, R707, and Q710 in the otherwise lipophilic environment are energetically favorable, which can enforce trimerization. Nevertheless, our mutational studies show that the TMD trimer cannot be disrupted by simple mutations in the trimer interface. Both the coiled-coil and the hydrophilic core are the structural determinants critical for stabilizing the TMD trimer. Major changes in both of them together are necessary to disrupt the trimer structure.
Conformational dynamics of the HIV-1 Env TMD
Although the HIV-1 Env TMD is a strongly associated trimer in bicelles, the NMR data suggest internal dynamics as indicated by the variation in the amide proton resonance linewidth across the TMD structure. In particular, the N-terminal half preceding the intramembrane arginine (R696) showed significantly more line broadening due to chemical exchange than the C-terminal half, suggesting that the N-terminal half is structurally less stable. One plausible cause is that the truncated MPER on the N-terminal side might contribute to stabilizing the TM coiled coil region. The more homogeneous linewidth of the C-terminal hydrophilic core suggests a more stable assembly, which is consistent with the results of mutational studies, showing that the hydrophilic core plays a more important role in TMD trimerization than does the coiled coil region [21]. Furthermore, this result also agrees with our previous observation that deletions in the Env CT, which is directly linked to the hydrophilic core of the TMD, can alter the antigenic structure of the ectodomain, probably by weakening the TMD trimerization.
Homology models of the Env TMDs of HIV-2 and SIV
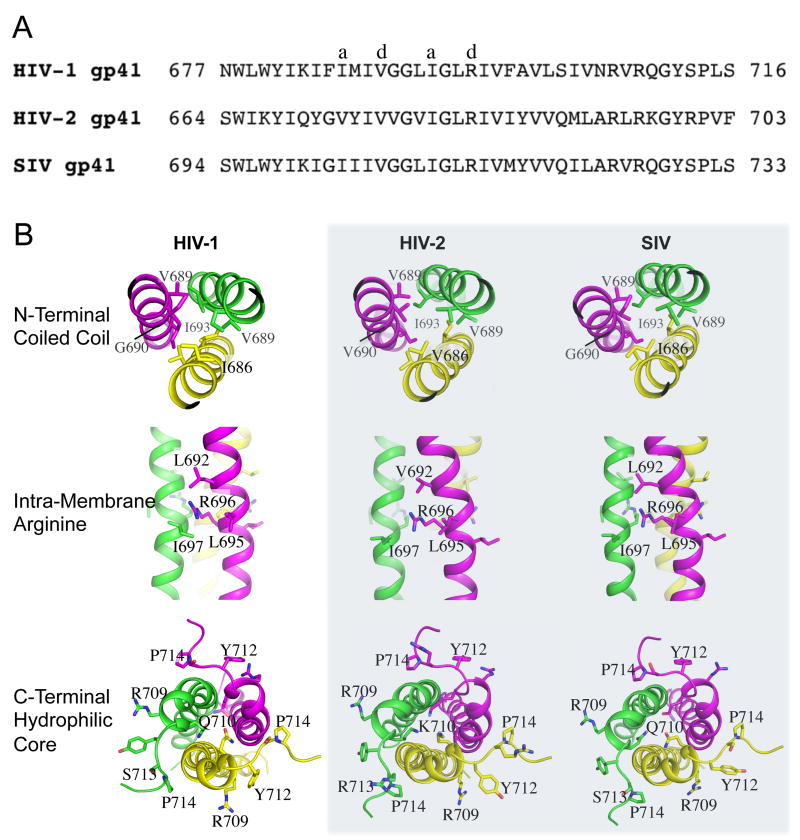
The sequences of Env TMDs of closely related human immunodeficiency virus type 2 (HIV-2) and simian immunodeficiency virus (SIV) are quite different from that of HIV-1 (Fig. 2A). The sequence identity is ∼45% between HIV-1 and HIV-2, and ∼77% between HIV-1 and SIV (calculated with the consensus sequences of HIV-1, HIV-2, and SIV Env TMDs using Clustal 2.1 [38]). To determine whether our trimer structure of HIV-1 Env TMD is compatible with HIV-2 and SIV sequences, we have generated homology models of HIV-2 and SIV TMDs, based on the sequence alignment using the SWISS-MODEL server [39]. Shown in Fig. 2B, the models are compared in the context of the three defining features of the TMD structure: the N-terminal coiled coil, the intramembrane arginine, and the C-terminal hydrophilic core. The key residues in and around these three structural regions are very similar among the three types, with SIV more similar to HIV-1 than to HIV-2. For the SIV TMD, the relevant residues (labeled) in the three regions are identical to those of HIV-1. In the case of the HIV-2 TMD, G690 in the coiled coil is replaced with other residues, such as Val or Leu, which may reduce the trimer stability in this region assuming that the trimer assembly remains the same as that of the HIV-1 TMD. Additionally, in the C-terminal hydrophilic core, Q710 is replaced by Lys, which maintains the hydrophilicity but might introduce a destabilizing factor due to charge repulsion. Overall, these homology models suggest that the SIV and HIV-2 Env TMDs can indeed adopt the observed trimer structure of the HIV-1 Env TMD.
Figure 2. TMD homology between HIV-1, HIV-2 and SIV.
(A) Consensus amino acid sequences of the gp41 TMDs of HIV-1, HIV-2 and SIV. The number of sequences used to generate the consensus sequence is ∼5000, 113, and 172 for HIV-1, HIV-2, and SIV, respectively. (B) Comparison of the gp41 TMD of HIV-1 with the homology models (shaded) of the TMDs from HIV-2 and SIV generated using the SWISS-MODEL server. The models are compared for the three important structural regions: the N-terminal coiled coil, the intramembrane arginine, and the C-terminal hydrophilic core.
Implications for immunogen design
To test whether the Env TMD can modulate the antigenic structure of the ectodomain and whether mutations in the TMD can affect antibody sensitivity of the functional Env spike, we used a cell-cell fusion assay and a pseudovirus-based neutralization assay to measure inhibition, by monoclonal antibodies, of full-length Env variants containing mutations in the TMD [21]. We analyzed one such mutant, named 704-713, in which we had mutated an entire stretch of residues in the hydrophilic core to either serine or alanine (except for a glycine), against a panel of both neutralizing and nonneutralizing antibodies. For cell-cell fusion, the wildtype Env is sensitive to trimer-specific bnAbs and resistant to non-neutralizing antibodies, as expected. The antibody inhibition pattern is reversed, however, for the fully functional mutant 704-713, which becomes sensitive to the non-neutralizing antibodies, but resistant to the potent bnAbs. Similar results were obtained with the pseudovirus neutralization assay, indicating that the hydrophilic core of the TMD plays an important role in stabilizing and modulating the antigenic structure of Env.
These findings are highly relevant to vaccine development as they can guide Env-based immunogen design. Our data show that mutations that destabilize the hydrophilic core of the TMD trimer resemble the CT deletion in altering the sensitivity of the functional Env to both non-neutralizing and trimer-specific neutralizing antibodies. The trimer-specific bnAbs, which recognize the native conformation of Env [9, 40], fail to neutralize the virus when the TMD of the Env spike is destabilized. Thus, the TMD trimer structure seen by NMR probably represents the conformation of the TMD present in a native Env spike in a membrane. The observation that the TMD is a structural component critical for the stability of the functional Env spike provides a reasonable explanation for why most recombinant soluble Env preparations with the TMD deleted are unstable and conformationally heterogeneous. To design immunogens to better mimic the native and functional viral spikes on the surface of virion, we need to consider structural constraints imposed by the TMD on the ectodomain. To produce soluble Env immunogens in quantities suitable for clinical studies, future work will require clever protein engineering to solubilize the TMD by mutating surface-exposed, lipid-interacting hydrophobic residues while maintaining its trimeric structure. The high-resolution structure of the HIV-1 Env TMD trimer can be a valuable guide toward next-generation Env immunogens for inducing effective antibody responses.
Acknowledgments
We thank S. Harrison for critical reading of the manuscript. This work was supported by NIH grants AI127193 (to B.C. and J.J.C.), AI106488 (to B.C.), HL103526 (to J.J.C.), and AI112489 (to B.C.).
References
- 1.Harrison SC. Viral membrane fusion. Nature structural & molecular biology. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 3.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 4.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–61. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 6.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–5. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacs JM, Nkolola JP, Peng H, Cheung A, Perry J, Miller CA, Seaman MS, Barouch DH, Chen B. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12111–6. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffs SA, Goriup S, Kebble B, Crane D, Bolgiano B, Sattentau Q, Jones S, Holmes H. Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine. 2004;22:1032–46. doi: 10.1016/j.vaccine.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 11.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105:3739–44. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs JM, Noeldeke E, Ha HJ, Peng H, Rits-Volloch S, Harrison SC, Chen B. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18542–7. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89:3380–95. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep. 2016;16:2327–38. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teese MG, Langosch D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry. 2015;54:5125–35. doi: 10.1021/acs.biochem.5b00495. [DOI] [PubMed] [Google Scholar]
- 17.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–56. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18:984–9. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q, Fu TM, Cruz AC, Sengupta P, Thomas SK, Wang S, Siegel RM, Wu H, Chou JJ. Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor. Mol Cell. 2016;61:602–13. doi: 10.1016/j.molcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–61. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dev J, Park D, Fu Q, Chen J, Ha HJ, Ghantous F, Herrmann T, Chang W, Liu Z, Frey G, Seaman MS, Chen B, Chou JJ. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–5. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West JT, Johnston PB, Dubay SR, Hunter E. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity. J Virol. 2001;75:9601–12. doi: 10.1128/JVI.75.20.9601-9612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowles DP, Jr, Cheevers WP, McGuire TC, Brassfield AL, Harwood WG, Stem TA. Structure and genetic variability of envelope glycoproteins of two antigenic variants of caprine arthritis-encephalitis lentivirus. J Virol. 1991;65:5744–50. doi: 10.1128/jvi.65.11.5744-5750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice NR, Henderson LE, Sowder RC, Copeland TD, Oroszlan S, Edwards JF. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–8. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonigo P, Alizon M, Staskus K, Klatzmann D, Cole S, Danos O, Retzel E, Tiollais P, Haase A, Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–82. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 26.Pietschmann T, Zentgraf H, Rethwilm A, Lindemann D. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J Virol. 2000;74:4474–82. doi: 10.1128/jvi.74.10.4474-4482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciczora Y, Callens N, Montpellier C, Bartosch B, Cosset FL, Op de Beeck A, Dubuisson J. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J Gen Virol. 2005;86:2793–8. doi: 10.1099/vir.0.81140-0. [DOI] [PubMed] [Google Scholar]
- 28.Long Y, Meng F, Kondo N, Iwamoto A, Matsuda Z. Conserved arginine residue in the membrane-spanning domain of HIV-1 gp41 is required for efficient membrane fusion. Protein & cell. 2011;2:369–76. doi: 10.1007/s13238-011-1051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotem E, Reuven EM, Klug YA, Shai Y. The Transmembrane Domain of HIV-1 gp41 Inhibits T-Cell Activation by Targeting Multiple T-Cell Receptor Complex Components through Its GxxxG Motif. Biochemistry. 2016;55:1049–57. doi: 10.1021/acs.biochem.5b01307. [DOI] [PubMed] [Google Scholar]
- 30.Miyauchi K, Curran R, Matthews E, Komano J, Hoshino T, Engelman DM, Matsuda Z. Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Japanese journal of infectious diseases. 2006;59:77–84. [PubMed] [Google Scholar]
- 31.Harrison SC. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–3. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 33.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 34.Trenker R, Call ME, Call MJ. Crystal Structure of the Glycophorin A Transmembrane Dimer in Lipidic Cubic Phase. Journal of the American Chemical Society. 2015;137:15676–9. doi: 10.1021/jacs.5b11354. [DOI] [PubMed] [Google Scholar]
- 35.Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009;18:1343–58. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falson P, Bartosch B, Alsaleh K, Tews BA, Loquet A, Ciczora Y, Riva L, Montigny C, Montpellier C, Duverlie G, Pecheur EI, le Maire M, Cosset FL, Dubuisson J, Penin F. Hepatitis C Virus Envelope Glycoprotein E1 Forms Trimers at the Surface of the Virion. J Virol. 2015;89:10333–46. doi: 10.1128/JVI.00991-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleverley DZ, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci U S A. 1998;95:3425–30. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4351–6. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]