Indoleamine 2,3-dioxygenase (IDO) is a nonhepatic intracellular heme-containing enzyme.1 It catalyzes the conversion of L-tryptophan (L-Trp) to N-formylkynurenine (NFK), the initial and rate-determining step of the kynurenine pathway, by inserting both atoms of dioxygen across the C2=C3 bond of the indole moiety of L-Trp.2 IDO was first discovered by Hayaishi et al. more than four decades ago.3 Since then, its structural and functional properties were extensively studied until the early 1990s. This field of research was invigorated recently due to the discoveries that IDO is linked to a variety of immune-related pathophysiological conditions and that IDO is a potential target for pharmacological intervention against cancer.4 Here we report evidence supporting the presence of an inhibitory substrate binding site (Si site) in human IDO (hIDO), which is capable of binding substrates (L-Trp and 1-methyL-tryptophan), an effector (3-indole ethanol), and an uncompetitive inhibitor (Mitomycin C). The structures of these molecules are illustrated in Scheme 1.
Scheme 1.
Molecular Structures of 3-Indole Ethanol (IDE), Mitomycin C (MtoC), and 1-Methyl-tryptophan (1MTrp)
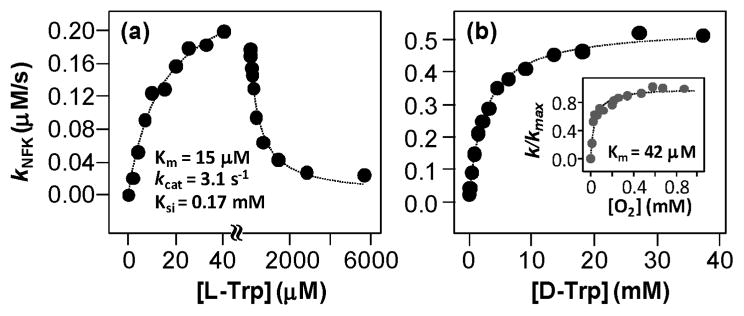
The activity of hIDO was first examined as a function of L-Trp concentration, following the mixing of the ferric enzyme with a methylene blue-ascorbate reducing system.3 As shown in Figure 1a, the activity of hIDO follows typical Michaelis–Menten behavior at [L-Trp] < 50 μM; further increase in [L-Trp] caused a decrease in the activity, signifying substrate-inhibition behavior, well-known for rabbit IDO (rIDO).3,5 It was generally believed that, at high [L-Trp], the substrate can bind to the ferric enzyme, thereby retarding the turnover of the enzyme by inhibiting its reduction to the active ferrous state.5 This scenario, however, can be excluded on the basis of two new observations: (1) the dissociation constant of L-Trp for the ferric IDO (Kd = 0.9 mM; see Figure S1) is significantly higher than the self-inhibition constant, Ksi (0.17 mM, vide infra), and (2) the redox potential of the L-Trp-bound ferric hIDO is ~46 mV higher than that of the substrate-free enzyme,6 indicating L-Trp binding to the ferric enzyme does not prevent its reduction.
Figure 1.
Michaelis–Menten plots of the dioxygenase reaction of hIDO (91nM) at pH 7.4 with respect to [L-Trp] (a) and [D-Trp] (b) under air-saturated conditions. The inset in (b) shows the corresponding plot for the reaction of hIDO with 36 μM L-Trp with respect to [O2] at pH 7.4.
We propose that the substrate-inhibition behavior of hIDO is a result of the binding of a second L-Trp in an inhibitory Si site of the enzyme. This hypothesis is consistent with recent computational data showing that the distal pocket of hIDO is flexible enough to accommodate both L-Trp and an indole derivative.7 On the basis of this scenario, the data shown in Figure 1a were fitted with the following equation8 by assuming L-Trp can bind to the active site, as well as the Si site.
| (1) |
Here, [S] is the substrate concentration; Vmax and Km are Michaelis–Menten constants. The best-fitted parameters are listed in Table 1. The data indicate that, under steady-state conditions, the probability of L-Trp binding to the ferric enzyme, followed by its reduction to the active ferrous state, is negligible (similar to that concluded by Sono et al. for rIDO5), as the L-Trp dissociation constant of the ferric enzyme (Kd = 0.9 mM) is much higher than the Km (15 μM) and Ksi (0.17 mM). It is noteworthy that the physiological concentration of L-Trp, ~50–100 μM,9 lies right between the observed Km and Ksi values.
Table 1.
Michaelis–Menten and Inhibition Constants of hIDO Associated with Its Reaction with L-Trp in the Presence and Absence of IDE and Those Associated with D-Trp and L-1MTrp (kcat Values Calculated from Vmax/[hIDO])
| L-Trp | +IDE2.5mM | +IDE5.1mM | D-Trp | L-1MTrp | |
|---|---|---|---|---|---|
| kcat (s−1) | 3.1 ± 0.2 | 3.2 ± 0.4 | 3.5 ± 0.6 | 5.9 ± 0.3 | 0.064 ± 0.003 |
| Km (μM) | 15 ± 2 | 16 ± 3 | 15 ± 3 | (2.6 ± 0.2) × 103 | 62 ± 9 |
| Ksi (mM) | 0.17 ± 0.2 | 0.48 ± 0.05 | 1.0 ± 0.1 | – | 5.0 ± 0.8 |
Similar activity studies were carried out with D-Trp. No substrate inhibition was observed (Figure 1b), indicating substrate inhibition is stereospecific to the L isomer, as reported for rIDO.3 The Michaelis–Menten fit of the data shows, as compared to the L-Trp reaction, the kcat is 2-fold faster (5.9 s−1), while the Km value is 170-fold larger (2.6 mM). The data demonstrate that the substrate stereoselectivity of hIDO is a result of its preferential binding of the L-isomer. Additional activity studies were conducted as a function of [O2] in the presence of 36 μM L-Trp (see the insert in Figure 1b). The kcat value was determined to be 2.9 s−1, similar to that determined by the data shown in Figure 1a. Km for O2 was determined to be 42 μM, which is near the physiological level of oxygen, ~50–76 μM,10 and is comparable to that of an analogous enzyme, tryptophan 2,3-dioxygenase, (40 μM)11 and those of monooxygenases, such as NOSs (6–24 μM)12 or P450s (1–15 μM).13
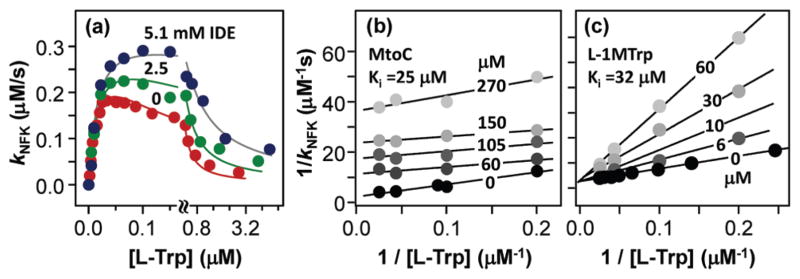
IDE has been long recognized as an effector for rIDO.14 It was believed IDE enhances the activity of rIDO by improving its kcat via binding to an accessory binding site located near the catalytic site of IDO.14 To investigate if the IDE binding site coincides with the aforementioned Si site, we examined the hIDO activity in the presence of 2.5 and 5.1 mM IDE. As shown in Figure 2a, the presence of IDE led to higher activity and less pronounced substrate inhibition. The best fit of data with eq 1 indicates IDE does not affect kcat and Km but causes the elevation of Ksi from 0.17 to 0.48/1.0 mM (for 2.5/5.1 mM IDE, see Table 1), indicating IDE competes with L-Trp for the Si site. As IDE bound to the Si site, unlike L-Trp, does not retard the active site catalysis, the apparent activity is higher than that in its absence. The data clearly demonstrate that IDE acts as an effector by binding to the Si site, thereby blocking it from L-Trp binding, instead of by facilitating kcat, as generally believed.
Figure 2.
Michaelis–Menten plots of the hIDO reaction at pH 7.4, in the absence or presence 2.5 or 5.1 mM IDE (a) and the Lineweaver–Burk plots of the steady-state activities of hIDO in the presence of various concentrations of MtoC (b) and L-1MTrp (c).
MtoC, a quinone-containing natural product from Streptomyces, has been widely used as a chemotherapy drug for several types of cancer.15 It is believed that the antitumor activity of MtoC is a result of its ability to generate oxygen radicals upon reduction, thereby inhibiting DNA synthesis. The recent findings that hIDO is a potential therapeutic target for cancer treatment and that quinone is a potent pharmacophore for hIDO inhibition16–18 prompted us to investigate if hIDO could function as a pharmacological target of MtoC. We examined the hIDO activity as a function of MtoC concentration. The Lineweaver–Burk plot of the data (Figure 2b) shows MtoC indeed inhibits hIDO in an uncompetitive fashion with an inhibition constant (Ki) of ~25 μM. The data indicate that, as an uncompetitive inhibitor, MtoC binds only to the L-Trp-bound enzyme, not the substrate-free enzyme. We propose that L-Trp binding in the active site induces structural changes to the Si site to accommodate the MtoC molecule and that the occupancy of the Si site by MtoC inhibits the active site catalysis.
As a control experiment, the inhibitory effect of 1MTrp was examined. 1MTrp has been widely used as an hIDO inhibitor.19 As shown in Figure 2c, the L-isomer acts as a competitive inhibitor, with a Ki of 32 μM, while the D-isomer exhibits no inhibitory effect (data not shown), consistent with that reported by Hou et al.20 Additional studies show, in the absence of L-Trp, L-1MTrp itself can act as a substrate, although kcat is 50-fold lower than that of L-Trp (Figure S2), similar to that reported by Chauhan et al.21 In addition, our data show that L-1MTrp exhibits substrate-inhibition behavior, indicating, like L-Trp, L-1MTrp binds to the Si site at elevated concentrations, thereby inhibiting hIDO activity. The 29-fold higher Ksi value, as compared to that of L-Trp (Table 1), indicates the bulky methyl group on the indole nitrogen significantly lowers its affinity toward the Si site.
In summary, our data demonstrate that hIDO possesses two substrate binding sites, an active binding site and a Si site (as illustrated in a cartoon shown in Figure S3). The binding of L-Trp in the substrate binding site introduces a conformational change to the Si site to allow it to accommodate molecules with a wide variety of structures, including L-Trp, 1MTrp, IDE, and MtoC. We demonstrated that, by binding to the Si site, these molecules can act as either an effector or an inhibitor. It is important to note that L-Trp-binding induced conformational changes to hIDO have been implicated in the crystallographic data,22 as well as molecular dynamics simulations,7 while the allosteric structural transition induced by the binding of L-Trp, L-1MTrp, or MtoC in the Si site, which gives rise to the inhibition of the active catalysis, remains to be further investigated. Nonetheless, the data presented in this work offer new mechanistic insights into the substrate-inhibition behavior of hIDO, as well as the functional mechanisms of its effector, IDE, and inhibitor, MtoC.
Recently, hIDO has emerged as a therapeutic target for cancer,4 leading to an active search for potent inhibitors. Our data show MtoC effectively inhibits hIDO, calling for re-evaluation of the action mechanism of this commonly used antitumor chemotherapeutic agent. They also suggest the newly discovered quinone-containing hIDO inhibitors,16–18 like MtoC, may selectively bind to the Si site. Taken together the data presented in this work provide the first glimpse of the Si site that offers potential guidelines for future development of more efficient hIDO inhibitors.
Supplementary Material
Acknowledgments
We would like to thank Dr. Denis L. Rousseau for valuable discussions.
Footnotes
Supporting Information Available: The Materials and Methods, the absorption spectra of the substrate-free and L-Trp-bound ferric hIDO and the associated L-Trp titration data, as well as the Michaelis–Menten plot with respect to L-1MTrp and a cartoon illustrating the Si site of hIDO. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Takikawa O. Biochem Biophys Res Commun. 2005;338:12–19. doi: 10.1016/j.bbrc.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 2.Sono M, Roach MP, Coulter ED, Dawson JH. Chem Rev. 1996;96(7):2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto S, Hayaishi O. J Biol Chem. 1967;242(22):5260–6. [PubMed] [Google Scholar]
- 4.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Nat Med. 2005;11(3):312–9. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 5.Sono M, Taniguchi T, Watanabe Y, Hayaishi O. J Biol Chem. 1980;255(4):1339–45. [PubMed] [Google Scholar]
- 6.Papadopoulou ND, Mewies M, McLean KJ, Seward HE, Svistunenko DA, Munro AW, Raven EL. Biochemistry. 2005;44(43):14318–28. doi: 10.1021/bi0513958. [DOI] [PubMed] [Google Scholar]
- 7.Macchiarulo A, Nuti R, Bellocchi D, Camaioni E, Pellicciari R. Biochim Biophys Acta. 2007;1774(8):1058–68. doi: 10.1016/j.bbapap.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Copeland RA. Enzymes. 2. Wiley-VCH; New York: 2000. [Google Scholar]
- 9.Torres MI, Lopez-Casado MA, Lorite P, Rios A. Clin Exp Immunol. 2007;148(3):419–24. doi: 10.1111/j.1365-2249.2007.03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunn HF, Poyton RO. Physiol Rev. 1996;76(3):839–85. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 11.Ishimura Y, Nozaki M, Hayaishi O. J Biol Chem. 1970;245(14):3593–602. [PubMed] [Google Scholar]
- 12.Rengasamy A, Johns RA. J Pharmacol Exp Ther. 1996;276(1):30–3. [PubMed] [Google Scholar]
- 13.Jones DP, Mason HS. J Biol Chem. 1978;253(14):4874–80. [PubMed] [Google Scholar]
- 14.Sono M. Biochemistry. 1989;28(13):5400–7. doi: 10.1021/bi00439a013. [DOI] [PubMed] [Google Scholar]
- 15.Carter SK, Crooke ST. Mitomycin C: current status and new developments. Academic Press; New York: 1979. [Google Scholar]
- 16.Kumar S, Malachowski WP, DuHadaway JB, LaLonde JM, Carroll PJ, Jaller D, Metz R, Prendergast GC, Muller AJ. J Med Chem. 2008;51(6):1706–18. doi: 10.1021/jm7014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr G, Chung MK, Mauk AG, Andersen RJ. J Med Chem. 2008;51(9):2634–7. doi: 10.1021/jm800143h. [DOI] [PubMed] [Google Scholar]
- 18.Volgraf M, Lumb JP, Brastianos HC, Carr G, Chung MK, Munzel M, Mauk AG, Andersen RJ, Trauner D. Nat Chem Biol. 2008;4(9):535–7. doi: 10.1038/nchembio.107. [DOI] [PubMed] [Google Scholar]
- 19.Cady SG, Sono M. Arch Biochem Biophys. 1991;291(2):326–33. doi: 10.1016/0003-9861(91)90142-6. [DOI] [PubMed] [Google Scholar]
- 20.Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, Mellor AL, Prendergast GC, Munn DH. Cancer Res. 2007;67(2):792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 21.Chauhan N, Thackray SJ, Rafice SA, Eaton G, Lee M, Efimov I, Basran J, Jenkins PR, Mowat CG, Chapman SK, Raven EL. J Am Chem Soc. 2009;131(12):4186–7. doi: 10.1021/ja808326g. [DOI] [PubMed] [Google Scholar]
- 22.Sugimoto H, Oda S, Otsuki T, Hino T, Yoshida T, Shiro Y. Proc Natl Acad Sci USA. 2006;103(8):2611–6. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.