Abstract
Objective
Forkhead box (FOX) proteins are important regulators of the epithelial-to-mesenchymal transition (EMT), which is the main mechanism of cancer metastasis. Different studies have shown their potential involvement in progression of cancer in different tissues such as breast, ovary and colorectum. In this study, we aimed to analyze the expression of genes encoding two FOX proteins in gastric adenocarcinoma.
Materials and Methods
In this experimental case-control study, the expression of FOXC2 and FOXQ1 was examined in 31 gastric adenocarcinoma tumors and 31 normal adjacent gastric tissues by reverse transcription polymerase chain reaction (PCR).
Results
The expression of both genes was significantly up-regulated in gastric adenocarcinoma tumors compared with the normal tissues (P<0.05). The differential expression of these two genes was also correlated with the grade of tumors (P<0.01).
Conclusion
We show that up-regulation of FOXC2 and FOXQ1 are likely to be involved in the progression of gastric adenocarcinoma.
Keywords: Gastric Cancer, Gene Expression, FOXC2, FOXQ1, Quantitative Polymerase Chain Reaction
Introduction
Gastric cancer is one of the most common malignancies worldwide with a high mortality rate. Interestingly, the vast majority of gastric cancers are adenocarcinomas. Therefore, an improved etiologic understanding of this cancer type seems to be essential. Although some classic and novel biomarkers such as carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA19- 9) (1) are known, finding effective biomarkers for early detection is yet to be identified. Also, developing new therapeutic options for this type of cancer is a necessity (2). The forkhead box (FOX) gene family encodes a large and diverse group of transcription factors. These factors share specific characteristics such as a conserved 100 amino acid long DNA-binding domain known as the forkhead or winged helix domain (3). There are 17 FOX subfamilies (FOXA-R) with a total of 41 genes currently identified in humans (4). Despite the high sequence conservation in the core forkhead motifs, FOX proteins control different cell fate decisions by regulating different gene networks involved in cell cycle progression, proliferation and differentiation. They contribute to different processes such as metabolism, senescence, survival and apoptosis (5). Multiple studies have therefore reported these genes and have shown their role in diseases such as cancer (6).
FOXC2, a member of the FOX family, is shown to be an essential regulator of vascular/ lymphatic vessel formation in cardiovascular development and disease (7). Furthermore, FOXC2 expression is directly associated with the epithelial mesenchymal transition (EMT) and cancer stem cell properties in breast cancer (1, 8). This gene has been also shown to be up-regulated in other cancers including those of the colorectum, glial cells and cervix (9-11). Independent studies have proposed that the encoded protein is involved in EMT of cancer cells, a critical process in tumor genesis and metastasis (10, 11). The studies have also described a key role for another member of the FOX family, namely forkhead box Q1 (FOXQ1), in regulating EMT and aggressiveness in some human cancers (12, 13). It has also been shown that this gene is aberrantly expressed in colorectal, breast, lung, glial cell and gastric cancers (3, 12, 14, 15). However, the clinical significance of FOXQ1 expression level in gastric adenocarcinoma has not been confirmed by an independent study. Due to the important role of EMT in the progression of gastric cancer (16, 17), we aimed to analyze the expression of these two genes in gastric adenocarcinoma. We suggest that protein encoded by these two genes are potentially involved in the clinicopathology of gastric adenocarcinoma tumors.
Materials and Methods
A total of 62 samples comprising 31 gastric adenocarcinoma tumors and their normal adjacent gastric tissues were obtained from Iran National Tumor Bank (Iran). Clinical features of the donor patients are given in Table 1. All patients had not received any medication prior. Also, all had signed a written informed consent prior to surgery and all specimens were evaluated by two pathologists. The tumor samples were grouped under two category of grade [low grade (n=17) and high grade (n=14)] and stage [T1-T2 (n=10) and T3-T4 (n=21)]. The design of the experiment was approved by the Medical Ethics Committee of Tarbiat Modares University. All tissue specimens were frozen in liquid nitrogen after collection and then stored in -80˚C.
Total RNA extraction and cDNA synthesis
Total RNA was extracted by acid guanidinium-phenol-chloroform procedure using the RNX™-plus solution (CinnaGen, Iran) according to the manufacturer’s protocol. The quality and integrity of extracted RNA were assessed by gel electrophoresis (1% agarose) and spectrophotometry respectively. To remove genomic DNA contamination, total RNA was treated with DNase I (Sigma, USA) at 37˚C for 30 minutes. Three microgram of total RNA was reverse transcribed with oligo dT and random hexamers (MWG, Germany) by RevertAidTM Reverse Transcriptase (Fermentas, Canada) in a total volume of 20 μl according to the manufacturer’s protocols.
Table 1.
Clinical features of donor patients with gastric adenocarcinoma
| Clinical parameter | Number of individuals | |
|---|---|---|
| Samples | 31 | |
| Gender | ||
| Male | 18 | |
| Female | 13 | |
| Age | ||
| <64 | 15 | |
| ≤64 | 16 | |
| Tumor site | ||
| Cardia | 10 | |
| Non cardia | 21 | |
| Grade of tumors | ||
| Low grade | 17 | |
| High grade | 14 | |
| Staging | ||
| T1-T2 | 10 | |
| T3-T4 | 21 | |
| Lymphatic invasion | ||
| Yes | 18 | |
| No | 13 | |
| Preineural invasion | ||
| Yes | 9 | |
| No | 22 | |
Real-time quantitative polymerase chain reaction
Expression of FOXQ1 and FOXC2 transcripts was quantified by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) technique by a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR primers were designed using the Allele ID software and are given in Table 2. The expression of each transcript was normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcripts as an internal control. The reaction mixture consisted of 10 ng cDNA, 10 μl of 2X SYBR Green I master mix (Takara, Japan) and 200 nM of forward and reverse primers in a total volume of 20 μl according to the manufacturer’s instructions. The cycling conditions were an initial denaturation for 30 seconds at 94˚C, followed by 40 cycles of denaturation at 95˚C for 10 seconds and annealing/ extension at 60˚C for 30 seconds. All reactions of q-PCR were run in triplicate. The specificity of the PCR products was examined by melting curve analysis, followed by electrophoresis on a 12% polyacrylamide gel and digestion pattern. In order to obtain the standard curve for each primer set, a serially diluted cDNA sample was used. The Livak method (ΔΔCT) was used to analyze differential gene expression (18).
Table 2.
Sequence of primers used for real time polymerase chain reaction (PCR)
| Gene | Primer sequence (5ˊ-3ˊ) | PCR product length |
|---|---|---|
| FOXQ1 | F: TGCTATTGACCGATGCTTCAC | 152 |
| R:CCAAGGAGACCACAGTTAGAG | ||
| FOXC2 | F: CGGCCCAGCAGCAAACTTTCC | 139 |
| R:AGAGGCGGCGTGGATCTGTAG | ||
| GAPDH | F: CCATGAGAAGTATGACAAC | 115 |
| R: GAGTCCTTCCACGATACC | ||
Statistical analysis
Statistical analysis was based on paired Student’s t test. Categorical data including grade and stage of tumors were analyzed by t test through GraphPad Prism software. Pearson correlation coefficient was also calculated between normalized expressions of both genes. The P<0.05 were considered as significant. Multiple testing corrections were done by Bonferroni adjustment procedure.
Results
FOXC2 and FOXQ1 are up-regulated in gastric adenocarcinoma
Gene expression analysis showed that FOXC2 and FOXQ1 transcript levels were significantly higher in gastric adenocarcinoma samples than in normal pair tissues. The average fold-change for FOXC2 and FOXQ1 expression levels in gastric adenocarcinoma samples was 3.076 (P=0.045) and 2.622 (P=0.03) respectively (Fig.1). However, Bonferroni correction did not find them as significant difference. Furthermore, correlation analysis showed that the normalized expression of FOXC2 and FOXQ1 are correlated in tumors compared to normal tissues (r=0.436, P<0.05).
Fig.1.
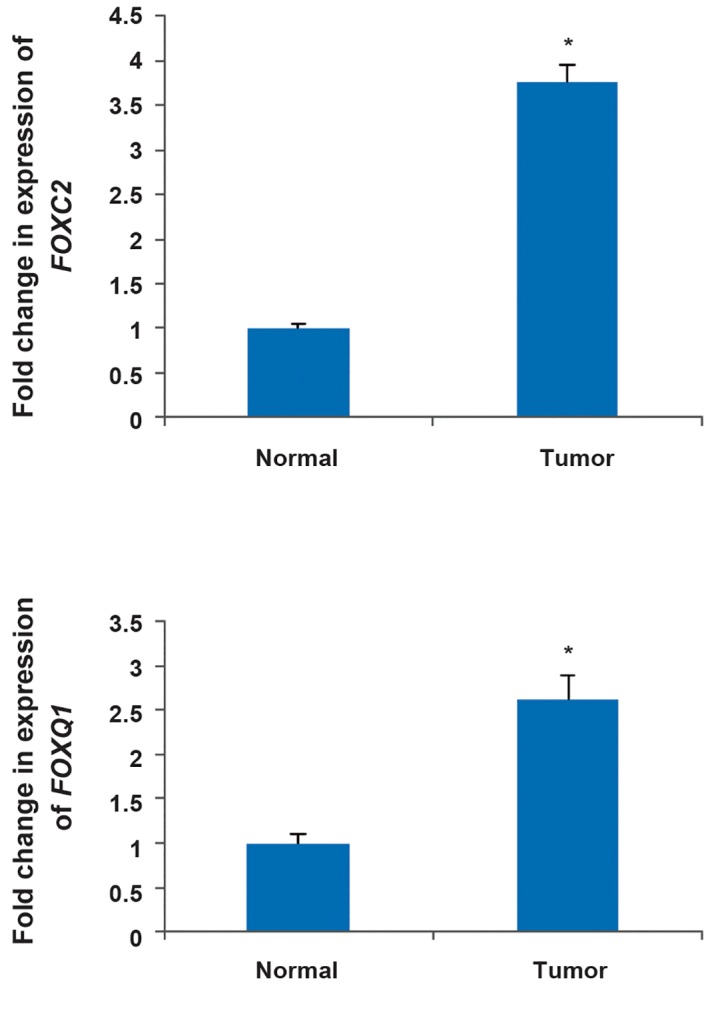
The up-regulation of FOXC2 and FOXQ1 in gastric adenocarcinoma tumors compared with normal tissues. The results are achieved by real time polymerase chain reaction (PCR). *; P<0.05.
FOXC2 and FOXQ1 have higher expression in high-grade tumors compared with low-grade tissues
In order to determine any potential association between the expression levels of either gene and progression of gastric tumor, we compared their expression levels between tumors of low and high grades. We found that FOXC2 and FOXQ1 are significantly over-expressed in high-grade tumors with fold changes of 5.44 and 3.953 respectively (Fig.2, P<0.01). Multiple testing also showed them as significant.
Fig.2.
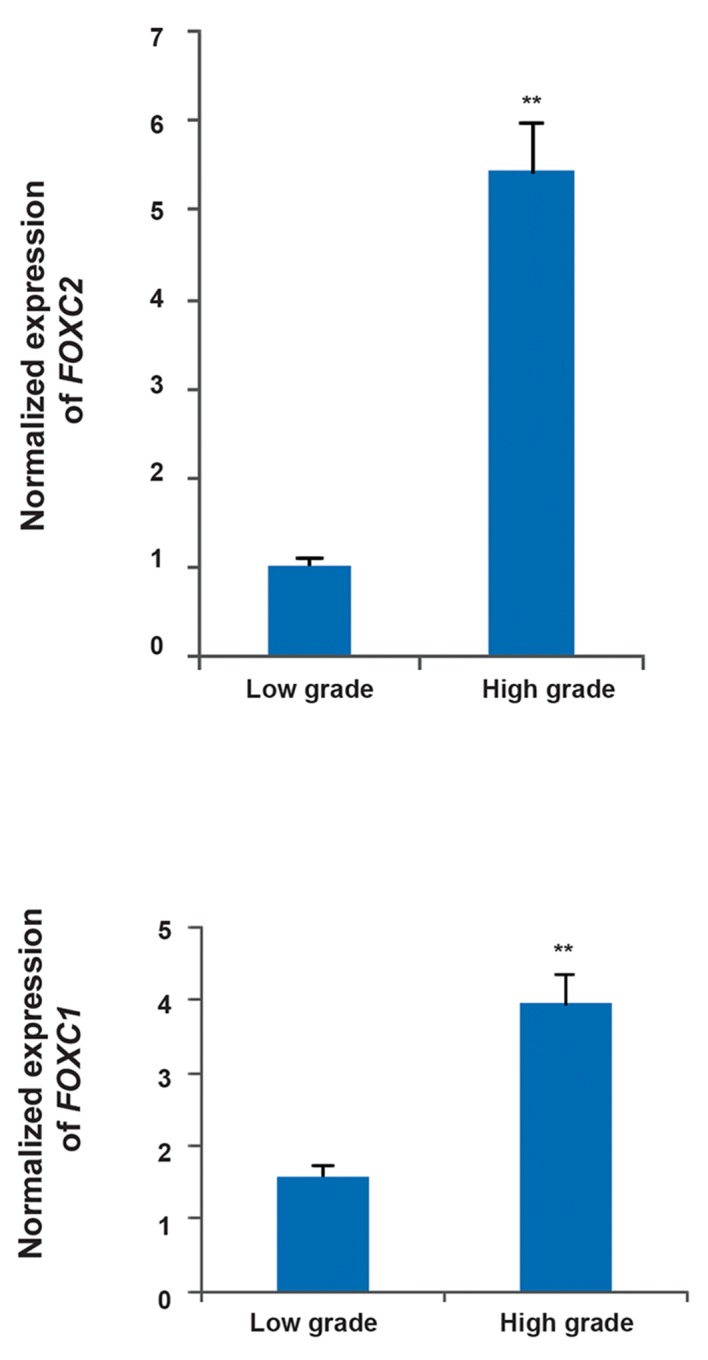
The significant difference of expression level of FOXC2 and FOXQ1 between tumors with different grades. The results show that the genes are up-regulated in high-grade tumors. The results are achieved by real time polymerase chain reaction (PCR). **; P<0.01.
FOXC2 and FOXQ1 expression analysis in high and low stage tumors
FOXC2 and FOXQ1 were not significantly differentially expressed between high (T3-T4)- and low (T1-T2)-stage tumors (Fig.3).
Fig.3.
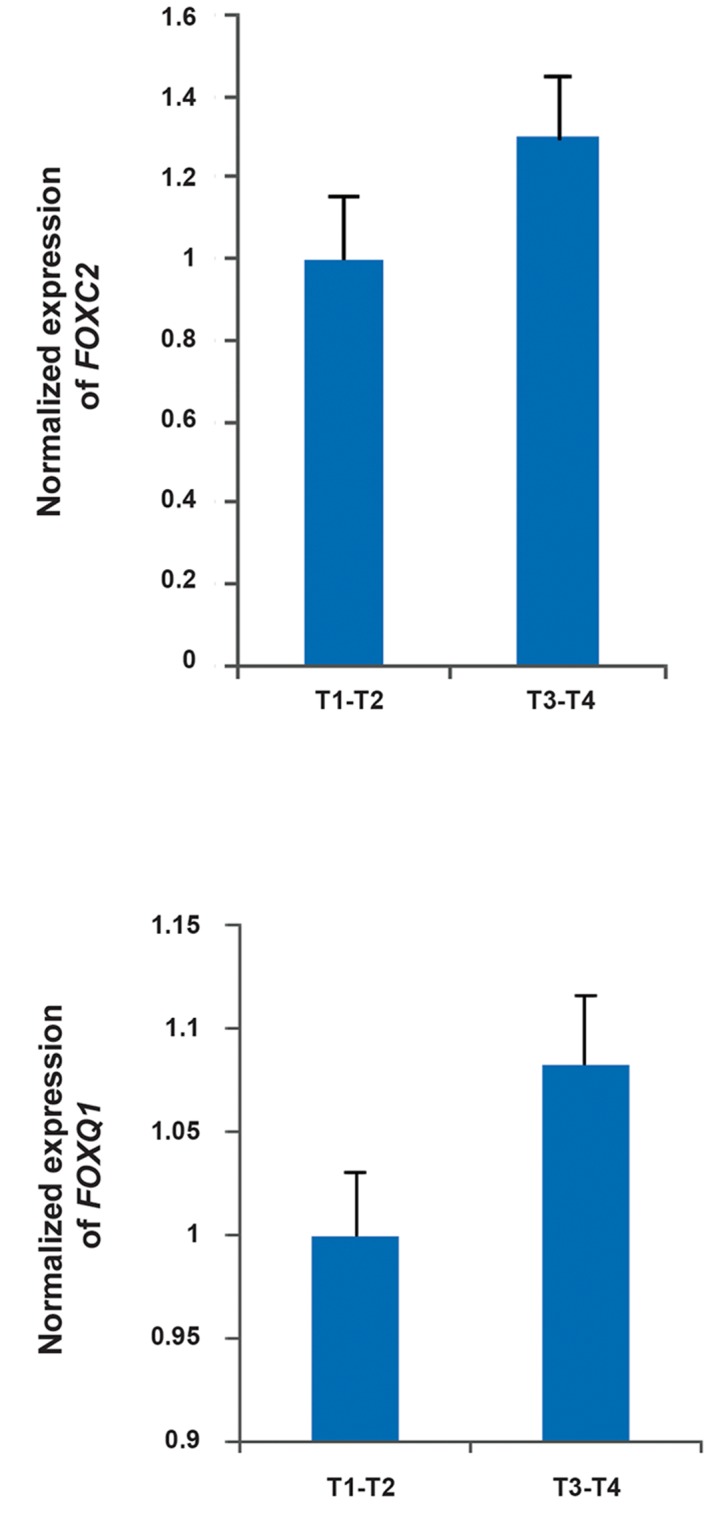
The expression of FOXC2 and FOXQ1 is not significantly different between different stages in gastric adenocarcinoma tissues. T1, 2, 3, 4; Different stage of tumor.
Discussion
Different members of the forkhead box (FOX) family of transcription factors regulate various biological processes including cellular development and differentiation (19). Based on this, multiple studies have identified their crucial role in the progression of different cancers. In this study, we have shown the potential role of FOXC2 and FOXQ1 in the progression of gastric adenocarcinoma. The results indicate that upregulation of these genes is associated with higher grades gastric tumors.
A number of studies have shown that FOXC2 expression is directly associated with EMT and cancer stem cell properties including disease recurrence, drug resistance, cell invasion, metastasis and poor prognosis (7, 8, 10). FOXC2 specifically promotes mesenchymal differentiation during the EMT and is associated with cancer metastasis in aggressive basal-like breast cancers (3, 6). Interestingly, FOXC2 expression is induced by a large number of known regulators of EMT, notably the Twist, Snail, and Goosecoid transcription factors as well as transforming growth factor-beta1 (TGF-β1). This suggests that FOXC2 is involved in a diverse array of EMT programs (20). Zhu et al. (13), in a study on gastric tumors, found that FOXC2 has higher expression at the protein level. The immunohistochemistry results showed that this protein can be a potential biomarker for gastric cancer. This is consistent with results herein showing that FOXC2 has also significantly higher transcript levels in high grade gastric tumors. Previous studies have also described a key role for FOXQ1 in regulating EMT and aggressiveness in human cancer (10, 12, 14). Unlike FOXC2, which does not seem to affect E-cadherin transcription, FOXQ1-induced EMT is accompanied by transcriptional repression of E-cadherin (21).
FOXQ1 transcript is highly expressed in murine tissues, particularly in stomach and bladder (22). Recently, it has been reported that it plays a role in stomach surface cells. This is because FOXQ1- deficient mice exhibit a lack of gastric acid secretion in response to various stimuli (3, 22). Furthermore, mucin expression and granule content in mucous cells of mouse stomach surface is also shown to be under the control of FOXQ1 (22). With respect to the important role of FOXQ1 in gastric development, we investigated its role in gastric tumorigenesis. We show that its up-regulation is associated with high-grade gastric tumors, thus indicating that abundance of its encoded protein may aid gastric cancer progression. Liang et al. (15), in a similar study, also found the up-regulation of FOXQ1 in gastric tumors at both transcript and protein levels. The current study was done on more samples and supports the potential role of FOXQ1 in gastric carcinogenesis.
The origin of gastric adenocarcinoma, which is a malignant epithelial tumor, is from the granular epithelium of the gastric mucosa. Studies have shown that the aberrant activation of EMT has an impact on adult epithelial cancer development (23). Since FOXC2 and FOXQ1 seem to be involved in EMT progression, we hypothesize that overexpression of these genes may be attributed to their role in regulation of EMT (24).
Conclusion
We demonstrate that FOXC2 and FOXQ1 are both associated with gastric cancer progression. Therefore, both may potentially be used as targets for prognosis of patients. Nevertheless, further investigation should be done for it to reach clinical trials.
Acknowledgments
The authors gratefully acknowledge the contribution of the patients and institutions in this study. The Iran National Science Foundation and the Department of Research Affairs of Tarbiat Modares University funded this work. The authors declare that they are no conflict of interest.
References
- 1.Jin Z, Jiang W, Wang L. Biomarkers for gastric cancer: Progression in early diagnosis and prognosis (review) Oncol Lett. 2015;9(4):1502–1508. doi: 10.3892/ol.2015.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70(5):2053–2063. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 4.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 5.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 6.Hajjari M, Behmanesh M, Zeinoddini M. The significant aberrant expression of FOXC1 as a high specific and sensitive potential biomarker in gastric adenocarcinoma tumor tissues. Gene Ther Mol Biol. 2013;15:1–7. [Google Scholar]
- 7.Hayashi H, Sano H, Seo S, Kume T. The Foxc2 transcription factor regulates angiogenesis via induction of integrin beta3 expression. J Biol Chem. 2008;283(35):23791–23800. doi: 10.1074/jbc.M800190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida N, Mimori K, Yokobori T, Sudo T, Tanaka F, Shibata K, et al. FOXC2 is a novel prognostic factor in human esophageal squamous cell carcinoma. Ann Surg Oncol. 2011;18(2):535–542. doi: 10.1245/s10434-010-1274-y. [DOI] [PubMed] [Google Scholar]
- 9.Cai L, Liu D, Lu S, Xu Y, Wang H. The relationship between gene expression of Forkhead box C2 and tumor progression in cervical carcinoma. Eur J Gynaecol Oncol. 2014;35(6):625–630. [PubMed] [Google Scholar]
- 10.Cui YM, Jiao HL, Ye YP, Chen CM, Wang JX, Tang N, et al. FOXC2 promotes colorectal cancer metastasis by directly targeting MET. Oncogene. 2015;34(33):4379–4390. doi: 10.1038/onc.2014.368. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Han SM, Tang XY, Han L, Li CZ. Overexpressed FOXC2 in ovarian cancer enhances the epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Oncol Rep. 2014;31(6):2545–2554. doi: 10.3892/or.2014.3119. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One. 2012;7(6):e39937–e39937. doi: 10.1371/journal.pone.0039937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu JL, Song YX, Wang ZN, Gao P, Wang MX, Dong YL, et al. The clinical significance of mesenchyme forkhead 1 (FoxC2) in gastric carcinoma. Histopathology. 2013;62(7):1038–1048. doi: 10.1111/his.12132. [DOI] [PubMed] [Google Scholar]
- 14.Sun HT, Cheng SX, Tu Y, Li XH, Zhang S. FoxQ1 promotes glioma cells proliferation and migration by regulating NRXN3 expression. PLoS One. 2013;8(1):e55693–e55693. doi: 10.1371/journal.pone.0055693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang SH, Yan XZ, Wang BL, Jin HF, Yao LP, Li YN, et al. Increased expression of FOXQ1 is a prognostic marker for patients with gastric cancer. Tumour Biol. 2013;34(5):2605–2609. doi: 10.1007/s13277-013-0808-x. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Wu RL, Xu AM. Epithelial-mesenchymal transition in gastric cancer. Am J Transl Res. 2015;7(11):2141–2158. [PMC free article] [PubMed] [Google Scholar]
- 17.Ghalandary M, Behmanesh M, Sadeghizadeh M. Evaluating of suppressor of zeste 12 and chromobox homolog 8 genes expression showed two possible origins of gastric cancer development. Indian J Cancer. 2015;52(1):27–31. doi: 10.4103/0019-509X.175566. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 20.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71(8):3076–3086. doi: 10.1158/0008-5472.CAN-10-2787. [DOI] [PubMed] [Google Scholar]
- 22.Verzi MP, Khan AH, Ito S, Shivdasani RA. Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology. 2008;135(2):591–600. doi: 10.1053/j.gastro.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natalwala A, Spychal R, Tselepis Ch. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J Gastroenterol. 2008;14(24):3792–3797. doi: 10.3748/wjg.14.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57(2):610–624. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]


