Abstract
Koolen de Vries syndrome (KDVS; MIM 610443) is a genomic disorder caused by a recurrent microdeletion derived from nonallelic homologous recombination mediated by flanking segmental duplications. Clinical manifestations of this syndrome are characterized by intellectual disability, hypotonia, a friendly behavior, distinctive facial features, and epilepsy. Herein, we report a case of 2 girls who revealed global developmental delay, mild facial dysmorphisms, friendly behavior, and epileptic seizure with a de novo 17q21.31 microdeletion detected by chromosomal microarray analysis (CMA). Conventional cytogenetics analysis by GTG-banding showed a female karyotype 46,XX for both girls. CMA revealed a microdeletion spanning approximately 500 kb in 17q21.31 in both girls, encompassing the following genes: CRHR1, MGC57346, CRHR1-IT1, MAPT-AS1, SPPL2C, MAPT, MAPT-IT1, STH, and KANSL1. Haploinsufficiency of one or more of these genes within the deleted region is the most probable cause of the probands' phenotype and is responsible for the phenotype seen in KDVS. CMA is a powerful diagnostic tool and an effective method to identify the de novo 17q21.31 microdeletion associated with KDVS in our probands.
Keywords: Chromosomal microarray analysis, Copy number variation, Microdeletion 17q21.31
Established Facts
Koolen-De Vries syndrome is a rare genetic condition caused by 17q21.31 microdeletion including the KANSL1 and MAPT genes.
The main clinical features are developmental delay, facial dysmorphism, and friendly behavior. The phenotype may vary comprising cryptorchidism, scoliosis, kidney and urologic anomalies as well as epilepsy.
Novel Insights
Our patients are 2 of 3 cases identified with a 17q21.31 microdeletion in Brazil and present with major features observed in Koolen De Vries syndrome. We believe that it is important to investigate rare syndromic disorders and try to identify the genetic causes of intellectual disability and that rare diseases demand to be published to make cases available for future studies.
Chromosomal microarray analysis is an effective method to establish adequate genomic diagnosis of idiopathic intellectual disability providing appropriate biological information which could be beneficial for patients and medical management.
Koolen de Vries syndrome (KDVS; MIM610443), also known as 17q21.31 microdeletion syndrome, is a genomic disorder defined by peculiar facial features, including a bulbous nasal tip, global developmental delay, moderate to severe intellectual disability, hypotonia, and friendly behavior [Koolen et al., 2006, 2008]. In addition, other phenotypic features were observed including scoliosis/kyphosis in 36% of the patients with this microdeletion, cryptorchidism (78%), kidney and urological anomalies (32%), as well as epilepsy described in more than 55% of the individuals [Bernardo et al., 2016]. The prevalence of KDVS varies from 1 in 13,000 and 1 in 20,000, identified as a de novo event [Egger et al., 2013]. This syndrome is caused by a recurrent microdeletion with sizes ranging from 500 to 650 kb at 17q21.31, derived from nonallelic homologous recombination (NAHR) mediated by flanking segmental duplications [Koolen et al., 2006, 2012]. The recurrent 17q21.31 microdeletion encompasses 5 genes CRHR1, KANSL1, MAPT, SPPL2C, and STH [Koolen et al., 2012; Egloff et al., 2014; Barone et al., 2015]. Additionally, point mutations in KANSL1 and MAPT cause the haploinsufficiency in these genes and were associated with the KDVS phenotype [Koolen et al., 2008, 2012; Zollino et al., 2012].
Herein, we report a case of 2 girls who presented with global developmental delay, mild facial dysmorphisms, friendly behavior, and epileptic seizure with a de novo 17q21.31 microdeletion detected by chromosomal microarray analysis (CMA).
Case Reports
Patient 1
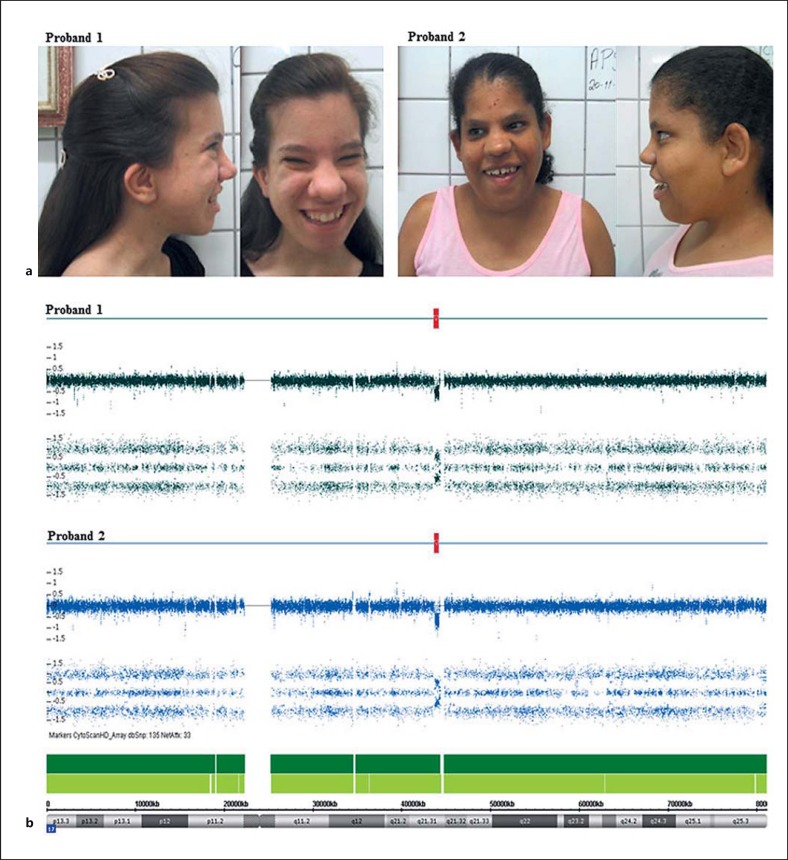
The proband, an 18-year-old female and the second child of nonconsanguineous parents, was born at 38 weeks' gestation to a 22-year-old mother and 26-year-old father by cesarean section. Her birth weight and size were 2,550 g and 45 cm, respectively. Her head circumference was 33 cm. She presented with epilepsy. The seizure episodes began in early infancy. Talking started at 3 years of age. Physical examination of the proband revealed global developmental delay, mild intellectual disability, and friendly behavior. She also had mild facial dysmorphisms, such as low-set ears, lop/cupped ears, a long face, and a tubular or ‘pear-shaped’ nose with a bulbous nasal tip. Her family history was unremarkable (Fig. 1a).
Fig. 1.
Phenotype and molecular features of the 2 probands with KDVS. a Proband 1: facial dysmorphisms such as low-set ears, long face, a bulbous nasal tip are shown. Proband 2: dysmorphic features including a long face, lop/cupped ears, and a bulbous nose are shown. b CMA of the 17q21.31 microdeletion. CMA data from proband 1 and proband 2 showing a de novo 0.56-Mb and 0.51-Mb microdeletion, respectively, indicated by red blocks.
Patient 2
The proband, a 22-year-old girl, was born to nonconsanguineous parents at 42 weeks' gestation to a 32-year-old mother and 48-year-old father. At birth, her weight was 2,770 g and her crown-heel length was 47 cm. Her head circumference was 60 cm. The child was delivered by cesarean section. She was born with a clubfoot, muscle weakness, and hypotonia. At 7 months, she manifested epilepsy. At physical examination, the patient revealed global developmental delay with cognitive and speech delay. Her facial dysmorphisms included a long face, lop/cupped ears, and a bulbous nose. The family history had no remarkable information (Fig. 1a).
Material and Methods
Karyotyping at >550-band resolution was carried out with peripheral blood samples from the patients using conventional cell culture, harvesting, and GTG-banding following standard procedures. Chromosomal analyses were done using the IKAROS® software (Metasystems Corporation, Germany).
Genomic DNA was isolated from whole blood using QIAamp® DNA Mini kit (Qiagen, Germany). CMAs were performed for both patients and their parents using GeneChip® CytoScanHD™ (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. The array was designed specifically for cytogenetic research, including approximately 2,696,550 CNV markers, 743,304 SNP markers, and >1,953,246 non-polymorphic markers. CEL files obtained by scanning the arrays were analyzed using the Chromosome Analysis Suite software (Affymetrix). The CNVs found in the 2 patients and their parents were compared with genomic variants in public databases, including DGV, DECIPHER, OMIM, and CytoScanHD™ Array Database.
17q21.31 Low-Copy Repeat Structure Analysis
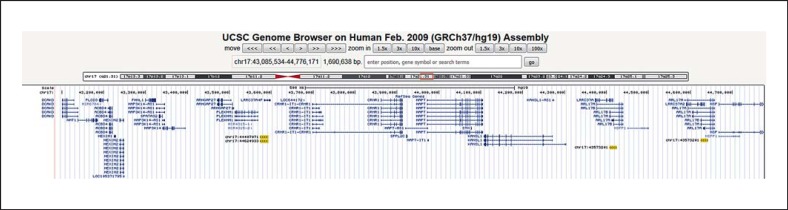
Using the Segmental Duplication track (http://genome.ucsc.edu/) from the genome browser (Human Genome Build 37.1), we performed an analysis of duplicated genomic sequences including known low-copy repeats (LCRs) (segmental duplication >1 kb of non-RepeatMasked sequence with over 98% similarity), comparing an approximately 1.69-Mb region surrounding the proximal 17q21.31 locus (chr17:43,085,534-44,776,171). This region represented 3 times of the CNV size.
Results
Karyotyping showed no visible numerical or structural alterations in the chromosomes. Both girls showed female karyotypes (46,XX). CMA revealed a de novo microdeletion in both girls spanning approximately 500 kb in 17q21.31, encompassing the following genes: CRHR1, MGC57346, CRHR1-IT1, MAPT-AS1, SPPL2C, MAPT, MAPT-IT1, STH, and KANSL1 (Fig. 1b; Table 1). Parental CMA analysis confirmed the deletions to be de novo events.
Table 1.
Clinical and molecular features of the 2 probands
| Case | Clinical features | Age, Years | Karyotype | CNV | Band | Size | Microarray nomenclature | Genes | Origin | LCR |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| classification | start/end | identity | position | size | ||||||||||
| 1 | GDD, FD, FB | 18 | 46,XX | loss | 17q21.31 | 0.56 Mb | 17q21.31 (43648662_44212416)×1 | LOC644172, CRHR1, MGC57346, C17orf69, MAPT-AS1, SPPL2C, MAPT, MAPT-IT1, STH, KANSL1 | de novo | 19A (chr17:44407071) 19B (chr17: 43573201) |
44,407,071–44,424,507 43,573,201–43,593,494 |
98.17% | direct | ~19,000 bp |
| 2 | GDD, FD, FB | 22 | 46,XX | loss | 17q21.31 | 0.51 Mb | 17q21.31 (43703801_44212416)x1 | CRHR1, MGC57346, CRHR1-IT1, MAPT-AS1, SPPL2C, MAPT, MAPTIT1, STH, KANSL1 | de novo | 19C (chr17:44624933) 19D (chr17:43573201) |
44,624,933–44,642,442 43,573,201–43,593,494 |
98.23% | direct | ~19,000 bp |
FB, friendly behavior; FD, facial dysmorphisms; GDD, global developmental delay; LCR, low-copy repeat.
Analyses of the rearranged 17q21.31 chromosomal region showed 2 major LCR families within the studied region. Each LCR family was named as 19A-19B and 19C-19D with approximately 19 kb flanking the disease locus. The LCRs were directly repeated and shared 98.2% identity (Fig. 2; Table 1).
Fig. 2.
Schematic representation of the 17q21.31 chromosomal region and summary of segmental duplication analyses. Two major LCR families [19A (chr17:44407071), 19B (chr17:43573201), 19C (chr17:44624933), and 19D (chr17:43573201)] in this region are represented by yellow blocks. Genes within this region are represented by blue lines.
Discussion
Intellectual disability is the most common developmental disorder, affecting 2–3% of the general population [Bernardini et al., 2010]. The frequency would probably be higher, if appropriate genetic testing and diagnostic procedures would be employed. According to Qiao et al. [2014], submicroscopic CNVs occur in 5–15% of the cases with intellectual disability. Thus, screening for submicroscopic chromosomal rearrangements using genomic microarrays in clinical practice have improved the diagnostic yield up to 10–25% in patients with intellectual disability [Battaglia et al., 2013].
Here, we report the cases of 2 girls presenting with several phenotypic traits commonly seen in KDVS, such as intellectual disability, dysmorphic facial features, friendly behavior, epilepsy, and a de novo 17q21.31 microdeletion harboring the genes KANSL1, CRHR1, and MAPT – characterized as the main genes associated with the syndrome.
Molecular mechanisms such as NAHR flanked by segmental duplications may collaborate to the formation of chromosomal rearrangements, leading to deleted or duplicated genomic segments. This finding demonstrates the importance of genomic instability in the recurrence of genomic disorders [Vlchova et al., 2014]. NAHR-mediated deletion has been observed as a potential mechanism in a considerable number of microdeletion syndromes, including the 17q21.31 microdeletion [Koolen et al., 2006]. We identified 2 major LCR families flanking the genomic rearrangement of 17q21.31 microdeletions. These findings show that LCRs may contribute to the increase in the susceptibility of CNV formation by NAHR, generating recurrent deletions in the 17q21.31 region.
The KANSL1 gene, a member of a histone acetyltransferase complex, encodes a nuclear protein which acts as a subunit of 2 protein complexes involved in histone acetylation, especially the H4 subunit. This gene is expressed in human tissues, including the central nervous system [Moreno-Igoa et al., 2015]. The haploinsufficiency of the KANSL1 gene is sufficient to cause a KDVS phenotype. Koolen et al. [2012] and Zollino et al. [2012] detected a de novo point mutation in KANSL1 in patients with failure to thrive in infancy, hypotonia, developmental delay with mild to moderate intellectual disability, and dysmorphic facial features.
The encoded microtubule-associated protein Tau (MAPT) is abundant in the axons of neurons and is highly expressed in the brain [Ballatore et al., 2007]. The Tau protein is a regulator of microtubule dynamics and plays a role in microtubule assembly and stabilization. This protein functions as an essential nuclear player in the protection of neuronal genomic DNA integrity under reactive oxygen species producing heat stress in primary neuronal cultures [Violet et al., 2014]. Besides, hyperphosphorylation and aggregation of the Tau protein leads to the formation of neurofibrillary tangles observed in neurodegenerative disorders (tauopathies) [Violet et al., 2014]. Mutations in the MAPT gene are involved in behavioral phenotypes and in certain neurodegenerative diseases [Koolen et al., 2008]. In addition, haploinsufficiency of the MAPT gene has been implicated in the neurocognitive functioning and has been associated with the main clinical features observed in individuals with 17q21.31 microdeletion [Koolen et al., 2006; Shaw-Smith et al., 2006; Wray, 2013].
The corticotropin-releasing hormone receptor 1 gene (CRHR1) encodes a G protein-coupled receptor that binds neuropeptides of the corticotropin-releasing hormone family, which are major regulators of the hypothalamic-pituitary-adrenal pathway and mediate stress-induced endocrine, behavioral, autonomic, and immune responses [Yang et al., 2015]. According to the authors, peculiar genes within the 17q21.31 microdeletion conferring some seizure susceptibility and the haploinsufficiency of the CRH1 gene may predispose to having seizures, especially infantile spasms [Wray, 2013; Bernardo et al., 2016]. Furthermore, CRHR1 signaling is involved in memory and learning, and a dosage deficit of this gene may contribute to the presence of global developmental delay [Sharkey et al., 2009].
CMA analysis is a powerful diagnostic tool and it is an effective method to identify the 17q21.31 microdeletion associated with KDVS in our probands with idiopathic intellectual disability. To our knowledge, the cases reported here are similar to the first case of 17q21.31 microdeletion identified in Central Brazil [Dornelles-Wawruk et al., 2013], since CMA technology has been employed in the region, confirming its usefulness to increase the diagnostic yield of undiagnosed intellectual disability. The authors believe that further investigation is needed to allow adequate phenotypic classification of probands, especially in the event of undiagnosed rare genomic diseases. Although the phenotype found in our probands shows similar features of KDVS, only additional gene expression studies will confirm the association between these genes and KDVS. Thus, international collaborative studies will need to be designed and carried out to increase the likelihood of finding a large cohort of patients to validate the findings of individual studies. Notwithstanding, CMA is an efficient method to delineate phenotypic variations, allowing adequate clinical management and better follow-up of the probands and their families.
Statement of Ethics
This study was approved by the Institutional Ethics Committee. The family provided written informed consent.
Disclosure Statement
The authors declare that they have no competing interests.
Acknowledgments
The authors wish to thank Dr. Rinaldo W. Pereira and the Rede ExeGenes for supporting the improvement of genetic diagnosis in Central Brazil. This work was sponsored by grants from CNPq (Edital 031/564465/2010-10) and FAPEG (2011.6002.19.1847.1134-03).
References
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Barone C, Novelli A, Capalbo A, Del Grano AC, Giuffrida MG, et al. An additional clinical sign of 17q21.31 microdeletion syndrome: preaxial polydactyly of hands with broad thumbs. Am J Med Genet A. 2015;167:1671–1673. doi: 10.1002/ajmg.a.37054. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013;17:589–599. doi: 10.1016/j.ejpn.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Bernardini L, Alesi V, Loddo S, Novelli A, Bottillo I, et al. High-resolution SNP arrays in mental retardation diagnostics: how much do we gain? Eur J Hum Genet. 2010;18:178–185. doi: 10.1038/ejhg.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo P, Madia F, Santulli L, Del Gaudio L, Caccavale C, et al. 17q21.31 microdeletion syndrome: description of a case further contributing to the delineation of Koolen-de Vries syndrome. Brain Dev. 2016;38:663–668. doi: 10.1016/j.braindev.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Dornelles-Wawruk H, Pic-Taylor A, Rosenberg C, Krepischi AC, Safatle HP, et al. Complex phenotype associated with 17q21.31 microdeletion. Mol Syndromol. 2013;4:297–301. doi: 10.1159/000354120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger JI, Wingbermühle E, Verhoeven WM, Dijkman M, Radke S, et al. Hypersociability in the behavioral phenotype of 17q21.31 microdeletion syndrome. Am J Med Genet A. 2013;161A:21–26. doi: 10.1002/ajmg.a.35652. [DOI] [PubMed] [Google Scholar]
- Egloff M, Encha-Razavi F, Garel C, Bonnière-Darcy M, Millischer AE, et al. 17q21.31 microdeletion: brain anomalies leading to prenatal diagnosis. Cytogenet Genome Res. 2014;144:178–182. doi: 10.1159/000369117. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Sharp AJ, Hurst JA, Firth HV, Knight SJ, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Kramer JM, Neveling K, Nillesen WM, Moore-Barton HL, et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- Moreno-Igoa M, Hernández-Charro B, Bengoa-Alonso A, Pérez-Juana-del-Casal A, Romero-Ibarra C, et al. KANSL1 gene disruption associated with the full clinical spectrum of 17q21.31 microdeletion syndrome. BMC Med Genet. 2015;16:68. doi: 10.1186/s12881-015-0211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Mercier E, Dastan J, Hurlburt J, McGillivray B, et al. Copy number variants (CNVs) analysis in a deeply phenotyped cohort of individuals with intellectual disability (ID). BMC Med Genet. 2014;15:82. doi: 10.1186/1471-2350-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey FH, Morrison N, Murray R, Iremonger J, Stephen J, et al. 17q21.31 microdeletion syndrome: further expanding the clinical phenotype. Cytogenet Genome Res. 2009;127:61–66. doi: 10.1159/000279260. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Violet M, Delattre L, Tardivel M, Sultan A, Chauderlier A, et al. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front Cell Neurosci. 2014;8:84. doi: 10.3389/fncel.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlchova M, Hancarova M, Drabova J, Slamova Z, Koudova M, et al. Monozygotic twins with 17q21.31 microdeletion syndrome. Twin Res Hum Genet. 2014;17:405–410. doi: 10.1017/thg.2014.29. [DOI] [PubMed] [Google Scholar]
- Wray C. 17q21.31 microdeletion associated with infantile spasms. Eur J Med Genet. 2013;56:59–61. doi: 10.1016/j.ejmg.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Yang G, Zou LP, Wang J, Shi XY, Yang XF, et al. Association analysis of polymorphisms of the CRHR1 gene with infantile spasms. Mol Med Rep. 2015;12:2539–2546. doi: 10.3892/mmr.2015.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollino M, Orteschi D, Murdolo M, Lattante S, Battaglia D, et al. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat Genet. 2012;44:636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]