Abstract
Prostate cancer incidence is 1.6-fold higher in African Americans than in other populations. The risk factors that drive this disparity are unknown and potentially consist of social, environmental, and genetic influences. To investigate the genetic basis of prostate cancer in men of African ancestry, we performed a genome-wide association meta-analysis using two-sided statistical tests in 10 202 case subjects and 10 810 control subjects. We identified novel signals on chromosomes 13q34 and 22q12, with the risk-associated alleles found only in men of African ancestry (13q34: rs75823044, risk allele frequency = 2.2%, odds ratio [OR] = 1.55, 95% confidence interval [CI] = 1.37 to 1.76, P = 6.10 × 10−12; 22q12.1: rs78554043, risk allele frequency = 1.5%, OR = 1.62, 95% CI = 1.39 to 1.89, P = 7.50 × 10−10). At 13q34, the signal is located 5’ of the gene IRS2 and 3’ of a long noncoding RNA, while at 22q12 the candidate functional allele is a missense variant in the CHEK2 gene. These findings provide further support for the role of ancestry-specific germline variation in contributing to population differences in prostate cancer risk.
The incidence of prostate cancer (PCa) in African American men is 1.6-fold higher than in other racial/ethnic populations (1), remaining one of the most important health disparities globally. Reasons for this disparity likely involve a multitude of factors, including social and environmental factors and inherited susceptibility. Genome-wide association studies (GWAS) have identified more than 100 common risk alleles for PCa (2–7), including the susceptibility region on chromosome 8q24, which harbors multiple variants that have been suggested to contribute to racial/ethnic differences in PCa risk (8,9).
To search for additional PCa risk variants in men of African ancestry that may contribute to their greater disease incidence, we combined genetic association results from the African Ancestry Prostate Cancer GWAS Consortium (AAPC; 4853 case subjects and 4678 control subjects) (9), the Ghana Prostate Study (474 case subjects and 458 control subjects) (10), the Kaiser/ProHealth Prostate Cancer Study (601 case subjects and 1650 control subjects) (11), and the ELLIPSE/PRACTICAL OncoArray Consortium (4274 case subjects and 4024 control subjects) (Supplementary Table 1, available online). Subjects provided written informed consent to participate in the study. The protocol and consent documents were approved by the institutional review boards at each of the participating institutions. A total of 17.8 million genotyped and imputed single nucleotide polymorphisms (SNPs) and insertion/deletion variants with frequencies of 1% or more were tested for an association with PCa risk. For each SNP, per-allele odds ratios (ORs) and 95% confidence intervals (CIs) were estimated using unconditional logistic regression, and we tested for allele dosage effects through a 1-degree of freedom Wald trend test. All statistical tests were two-sided. Results from each study were combined through a meta-analysis of 10 202 case subjects and 10 810 control subjects (Supplementary Methods, available online). The cut-point for genome-wide statistical significance was a P value of less than 5.00×10-8.
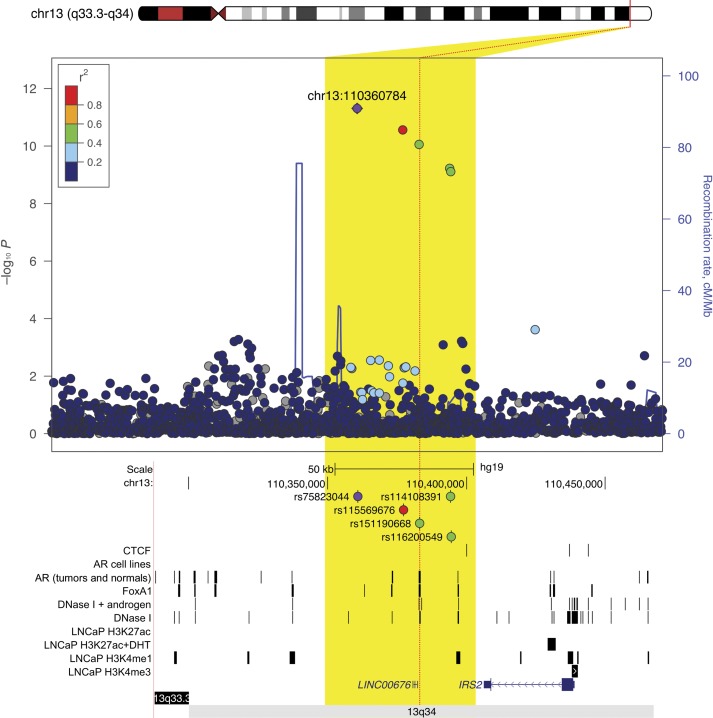
Only minor evidence of inflation in the test statistic was observed following adjustment for global genetic ancestry (λ = 1.04). In the meta-analysis, 775 alleles achieved genome-wide statistical significance (P < 5.00 × 10-8). These alleles were located at the 8q24 risk region (543 alleles) and other known susceptibility regions on chromosomes 2p15(EHBP1), 2q37(MLPH), 6q22(RFX6), 8p21(NKX3-1), 10q11(MSMB), 11q13(MYEOV), 12q13(KRT8), 17q21(ZNF652), and Xp11(NUDT11/LINC01496) (Supplementary Figure 1A, available online). Outside of these regions, genome-wide statistically significant associations were also observed on chromosomes 13q34 and 22q12.1 (Table 1; Supplementary Figure 1B, available online), with the risk-associated alleles found almost exclusively in men of African ancestry. At 13q34, marker rs75823044 (2.2% frequency) was associated with an odds ratio of 1.55 (95% CI = 1.37 to 1.76, P = 6.10×10-12). This marker is located within a cluster of five moderately correlated alleles (r2 > 0.30) approximately 45 kb 3’ of the gene insulin receptor substrate 2 (IRS-2), a signaling protein that mediates the effect of insulin and insulin-like growth factor 1 (12), and 20 kb 5’ of a long noncoding RNA (LINC00676). Of these five variants, rs151190668 (OR = 1.67, 95% CI = 1.43 to 1.96, P = 1.70×10-10) appears to be the best functional candidate because it is located in a region containing epigenetic chromatin modifications and androgen receptor and FOXA1 binding consistent with regulatory sequences (Figure 1; Supplementary Methods, available online).
Table 1.
Association results for prostate cancer risk variants at 13q34 and 22q12.1 in men of African ancestry
| SNP ID chr. position* | Nearby genes | AAPC |
Ghana |
Kaiser/ProHealth |
ELLIPSE OncoArray |
Meta-analysis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alleles† | OR (95% CI) | P† | RAF§ | OR (95% CI) | P‡ | RAF§ | OR (95% CI) | P‡ | RAF§ | OR (95% CI) | P‡ | RAF§ | OR (95% CI) | P‡ | ||
| rs75823044 | IRS2 | T/C | 1.47 (1.22 to 1.76) | 3.73 × 10‐5 | 0.022 | 2.66 (1.59 to 4.47) | 2.04 × 10-4 | 0.029 | 1.27 (0.80 to 2.00) | .31 | 0.021 | 1.60 (1.31 to 1.96) | 4.84 × 10-6 | 0.020 | 1.55 (1.37 to 1.76) | 6.10 × 10-12 |
| 13q34 | ||||||||||||||||
| 110,360,784 | ||||||||||||||||
| rs78554043 | CHEK2 | C/G | 1.60 (1.27 to 2.00) | 5.02 × 10‐5 | 0.015 | 2.45 (1.33 to 4.52) | .004 | 0.019 | 1.17 (0.69 to 1.99) | .55 | 0.017 | 1.66 (1.30 to 2.13) | 5.98 × 10-5 | 0.013 | 1.62 (1.39 to 1.89) | 7.50 × 10-10 |
| 22q12.1 | ||||||||||||||||
| 28,374,943 | ||||||||||||||||
Genome build 37/HG19. chr. = chromosome; CI = confidence interval; OR = odds ratio; RAF = risk allele frequency.
Risk allele/reference allele.
Allele dosage effects were tested through a 1-degree of freedom Wald trend test. All P values are two-sided.
Risk allele frequency in controls.
Figure 1.
Regional plot of a novel genome-wide statistically significant prostate cancer risk region at chromosome 13q34. Single nucleotide polymorphisms (SNPs) are plotted by their position 110 kb on either side of the index SNP (purple diamond) on the chromosome against their association (−log10P) with prostate cancer risk in men of African ancestry. SNPs surrounding the index SNP are colored to indicate the local linkage disequilibrium (LD) structure using pairwise r2 data from the African ancestry samples panel of the 1000 Genomes Project (November 2014 phase III). Below are peaks from transcription factor (TF) and histone modification ChIP-seq experiments in the same genomic window (see the Supplementary Methods, available online). All ChIP-seq in LNCaP unless otherwise indicated. AR = androgen receptor; CTCF = CCCTC-binding factor; LNCaP = Lymph Node Carcinoma of the Prostate cell line.
At 22q12.1, the association signal was also defined by multiple low-frequency African ancestry–specific variants spanning approximately 944 kb, with rs78554043 being the most statistically significant variant (1.5% frequency, OR = 1.62, 95% CI = 1.39 to 1.89, P = 7.50×10-10) (Table 1). The variant rs78554043 is correlated (r2 = 1) with a missense polymorphism (rs17886163, Ile448Ser, P = 1.38×10-9) in the CHEK2 gene (Supplementary Table 4, available online), which is a likely candidate for the underlying biologically functional allele. Although the Ile448Ser missense is characterized as “benign” by Polyphen2 and ClinVar and “tolerated” by Sifting Intolerant from Tolerant (SIFT) (Supplementary Methods, available online), it involves a nonconservative nonpolar to polar change in the amino acid. While the possibility of rare regulatory variation cannot be excluded, this nonconservative change provides support for previous studies suggesting that rare/less common missense variants in CHEK2 may be important in PCa development (13).
The risk alleles rs75823044 and rs78554043 are found almost exclusively in African ancestry populations. In the 1000 Genomes Project populations (n = 2504 subjects), the risk allele for rs75823044 was found in 48 of 661 African ancestry samples (AFR), one of 85 Peruvians, and one of 96 Punjabi. For rs78554043, the risk allele was found in 30 of 661 AFR samples, one of 104 Puerto Ricans, and one of 94 Colombians (data not shown).
At 13q34 and 22q12.1, no nominally statistically significant (P < .05) evidence of effect heterogeneity was noted by age (above vs below the median age in case subjects plus control subjects of 64, P ≥ .27) or disease aggressiveness (high-risk vs low-risk PCa, P ≥ .20) (Supplementary Methods, available online). GWAS of high-risk disease (vs controls) and high- (n = 2984) vs low-risk (n = 3012) disease (Supplementary Methods, available online) did not reveal any novel PCa loci of genome-wide statistical significance that could differentiate risk by disease aggressiveness (Supplementary Figure 1, C and D, available online). In addition, aside from 8q24 (14), admixture mapping using 220 474 genotyped SNPs in case-case and case-control comparisons of local ancestry (Supplementary Methods, available online) failed to identify any novel risk regions harboring risk alleles that are highly differentiated in frequency between men of African and European ancestry (data not shown) (Supplementary Figure 2A, available online).
The most statistically significant PCa risk association genome wide was observed with a novel triallelic (A/T/G) variant at 8q24, with the T allele found in approximately 12% of case subjects and approximately 6% of control subjects (rs72725854 at position 128,074,815 located in “region 2”) (8). The risk allele (T) is only found in populations of African ancestry with a per-allele odds ratio of 2.33 (95% CI = 2.16 to 2.50, P = 1.08×10-109) (Supplementary Figure 2B, available online) and is in linkage disequilibrium with African ancestry–specific risk alleles rs114798100 (4%, OR = 2.43, 95% CI = 2.21 to 2.66, P = 4.07×10-81) and rs111906932 (2%, OR = 1.92, 95% CI = 1.70 to 2.16, P = 1.44×10-26) (8,9). These SNPs are not correlated (r2 = 0 for rs114798100 and rs111906932) but define all observed haplotypes with the risk allele T of rs72725854, and thus describe the same association signal. In stepwise models, four additional variants were found to capture risk (P < 10-5) across the 8q24 locus (127.8–128.8 Mb) in men of African ancestry (Supplementary Table 2, available online).
Of the 100 reported PCa risk loci, 94 variants are polymorphic with an MAF of 0.05 or greater, 81 are directionally consistent with previous results in other populations (OR > 1), and 47 are nominally statistically significant associations (P < .05) in men of African ancestry (data not shown). Based on a polygenic risk score (Supplementary Methods, available online) comprising these risk variants as well as novel variants at 13q34 and 22q12.1 and variants shown to capture risk at 8q24 in men of African ancestry (n = 5), the 10% of men with the highest polygenic risk scores have a 3-fold (95% CI = 2.52 to 3.63) of PCa compared with men with “average risk” (polygenic risk scores in the 25th to 75th percentiles) (Supplementary Table 3, available online), which is comparable with that observed for the top 10% of the risk score distribution in men of European ancestry (OR = 2.93, 95% CI = 2.75 to 3.12) (2). Estimates for the top 1% of the polygenic risk scores in each population are 4.23 and 5.65, respectively.
A main limitation of this study is suboptimal statistical power (<80%) to detect modest effects (OR < 1.22) at genome-wide levels of statistical significance for common alleles with minor allele frequencies of less than 10%, particularly in analyses stratified by disease aggressiveness. Another limitation is the lack of understanding regarding the biological mechanisms through which genetic variation in these susceptibility regions influences risk.
Our findings substantiate the importance of conducting large-scale genetic studies in diverse populations for the discovery of novel risk loci that are ancestry specific (15). Further discovery efforts and fine-mapping of known loci will be needed to better understand the contribution of germline variation to PCa in men of African ancestry.
Funding
This work was supported by the National Cancer Institute at the National Institutes of Health, ELLIPSE GAME-ON U19 initiative for prostate cancer (U19 CA148537).
Notes
Authors: David V. Conti, Kan Wang, Xin Sheng, Jeannette T. Bensen, Dennis J. Hazelett, Michael B. Cook, Sue A. Ingles, Rick A. Kittles, Sara S. Strom, Benjamin A. Rybicki, Barbara Nemesure, William B. Isaacs, Janet L. Stanford, Wei Zheng, Maureen Sanderson, Esther M. John, Jong Y. Park, Jianfeng Xu, Victoria L. Stevens, Sonja I. Berndt, Chad D. Huff, Zhaoming Wang, Edward D. Yeboah, Yao Tettey, Richard B. Biritwum, Andrew A. Adjei, Evelyn Tay, Ann Truelove, Shelley Niwa, Thomas A. Sellers, Kosj Yamoah, Adam B. Murphy, Dana C. Crawford, Susan M. Gapstur, William S. Bush, Melinda C. Aldrich, Olivier Cussenot, Gyorgy Petrovics, Jennifer Cullen, Christine Neslund-Dudas, Mariana C. Stern, Zsofia-Kote Jarai, Koveela Govindasami, Anand P. Chokkalingam, Ann W. Hsing, Phyllis J. Goodman, Thomas Hoffmann, Bettina F. Drake, Jennifer J. Hu, Peter E. Clark, Stephen K. Van Den Eeden, Pascal Blanchet, Jay H. Fowke, Graham Casey, Anselm J. M. Hennis, Ying Han, Alexander Lubwama, Ian M. Thompson, Jr, Robin Leach, Douglas F. Easton, Fredrick Schumacher, David J. Van den Berg, Susan M. Gundell, Alex Stram, Peggy Wan, Lucy Xia, Loreall C. Pooler, James L. Mohler, Elizabeth T. H. Fontham, Gary J. Smith, Jack A. Taylor, Shiv Srivastava, Rosalind A. Eeles, John Carpten, Adam S. Kibel, Luc Multigner, Marie-Elise Parent, Florence Menegaux, Geraldine Cancel-Tassin, Eric A. Klein, Laurent Brureau, Daniel O. Stram, Stephen Watya, Stephen J. Chanock, John S. Witte, William J. Blot, Brian E. Henderson†, Christopher A. Haiman; for the PRACTICAL/ELLIPSE Consortium
†Deceased.
Affiliations of authors: Department of Preventive Medicine (DVC, KW, XS, SAI, MCS, YH, DJVdB, SMG, AS, PW, LX, LCP, DOS, BEH, CAH) and Department of Translational Genomics (JC), Keck School of Medicine, and Norris Comprehensive Cancer Center (SAI, MCS, DOS, BEH, CAH), University of Southern California, Los Angeles, CA; Department of Epidemiology (JTB) and Lineberger Comprehensive Cancer Center (JTB, JLM), University of North Carolina at Chapel Hill, Chapel Hill, NC; Bioinformatics and Computational Biology Research Center, Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA (DJH); Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD (MBC, SIB, SJC); University of Arizona College of Medicine and University of Arizona Cancer Center, Tucson, AZ (RAK); Department of Epidemiology, University of Texas MD Anderson Cancer Center, Houston, TX (SSS, CDH); Department of Public Health Sciences, Henry Ford Hospital, Detroit, MI (BAR, CND); Department of Preventive Medicine, Stony Brook University, Stony Brook, NY (BN, AJMH); James Buchanan Brady Urological Institute, Johns Hopkins Hospital and Medical Institution, Baltimore, MD (WBI); Division of Public Health Sciences (JLS) and SWOG Statistical Center (PJG), Fred Hutchinson Cancer Research Center, Seattle, WA; Department of Epidemiology, School of Public Health, University of Washington, Seattle, WA (JLS); Division of Epidemiology, Department of Medicine, Vanderbilt Epidemiology Center, Vanderbilt University School of Medicine, Nashville, TN (WZ, WJB); Department of Family and Community Medicine, Meharry Medical College, Nashville, TN (MS); California Prevention Institute of California, Fremont, CA (EMJ); Department of Health Research and Policy (Epidemiology) (EMJ) and Department of Medicine (AWH), Stanford Cancer Institute, Stanford University School of Medicine, Stanford, CA; Department of Cancer Epidemiology (JYP, TAS) and Department of Radiation Oncology and Cancer Epidemiology (KY), Moffitt Cancer Center and Research Institute, Tampa, FL; Program for Personalized Cancer Care and Department of Surgery, NorthShore University HealthSystem, Evanston, IL (JX); Epidemiology Research Program, American Cancer Society, Atlanta, GA (VLS, SMG); Department of Computational Biology, St. Jude Children's Research Hospital, Memphis, TN (ZW); University of Ghana Medical School, Accra, Ghana (EDY, YT, RBB, AAA, ET); Korle Bu Teaching Hospital, Accra, Ghana (EDY, YT, RBB, AAA, ET); Westat, Rockville, MD (AT, SN); Department of Urology, Northwestern University, Chicago, IL (ABM); Institute for Computational Biology, Department of Epidemiology and Biostatistics (DCC, WSB) and Department of Epidemiology and Biostatistics (FS), Case Western Reserve University, Cleveland, OH (DCC, WSB); Division of Epidemiology, Department of Thoracic Surgery (MCA) and Department of Medicine and Urologic Surgery (JHF), Vanderbilt University Medical Center (PEC), Nashville, TN; CeRePP, GRC No. 5 ONCOTYPE-URO, Institut Universitaire de Cancérologie, UPMC Univ Paris 6, Paris, France (OC, GCT); Department of Surgery, Center for Prostate Disease Research, Uniformed Services University of the Health Sciences, Bethesda, MD (GP, JC, SS); The Institute of Cancer Research, Sutton, London, UK (ZKJ, RAE); Oncogenetics Team, The Institute of Cancer Research and Royal Marsden NHS Foundation Trust, Sutton, UK (KG); School of Public Health, University of California, Berkeley, Berkeley, CA (APC); Department of Epidemiology and Biostatistics (TH, JSW) and Institute for Human Genetics (JSW), University of California, San Francisco, CA (TH, JSW); Department of Surgery, Division of Public Health Sciences, Washington University School of Medicine, St. Louis, MO (BFD); Sylvester Comprehensive Cancer Center and Department of Public Health Sciences, University of Miami Miller School of Medicine, Miami, FL (JJH); Division of Research, Kaiser Permanente Northern California, Oakland, CA (SKVDE); University Hospital of Pointe-à-Pitre, Guadeloupe (PB, LB); FWI and Inserm U1085-IRSET, Rennes, France (PB, LB); French West Indies University, Pointe-à-Pitre, Guadeloupe, FWI (PB, LB); Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, Charlottesville, VA (GC); Chronic Disease Research Centre and Faculty of Medical Sciences, University of the West Indies, Bridgetown, Barbados (AJMH); School of Public Health, Makerere University College of Health Sciences, Kampala, Uganda (AL, SW); Department of Urology, University of Texas Health Science Center at San Antonio, San Antonio, TX (IMTJr, RL); Centre for Cancer Genetic Epidemiology, Department of Oncology, University of Cambridge, Cambridge, UK (DFE); Department of Urology, Roswell Park Cancer Institute, Buffalo, NY (JLM, GJS); School of Public Health, Louisiana State University Health Sciences Center, New Orleans, LA (ETHF); Epigenetic and Stem Cell Biology Laboratory and Epidemiology Branch, National Institute of Environmental Health Sciences, Research Triangle Park, NC (JAT); Royal Marsden NHS Foundation Trust, London, UK (RAE); Division of Urology, Brigham and Women's Hospital/Dana-Farber Cancer Institute, Boston, MA (ASK); Washington University, St. Louis, MO (ASK); Inserm U1085 – IRSET, Rennes, France (LM); INRS-Institut Armand-Frappier, Institut National de la Recherche Scientifique, University of Quebec, Laval, Quebec, Canada (MEP); Inserm, Team Cancer-Environment, Center for Research in Epidemiology and Population Health, Université Paris-Saclay, Université Paris-Sud, Villejuif, France (FM); Glickman Urological and Kidney Institute, Cleveland Clinic, Cleveland, OH (EAK); Uro Care, Kampala, Uganda (SW).
Additional members and affiliations appear in the Supplementary Note (available online).
This study is dedicated to the memory of Brian E. Henderson.
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
A full listing of acknowledgements is detailed in the Supplementary Methods (available online).
Supplementary Material
References
- 1. Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: Baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al Olama AA, Kote-Jarai Z, Berndt SI, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46(10):1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eeles RA, Olama AA, Benlloch S, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45(4):385–391, 391e1–391e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44(12):1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takata R, Akamatsu S, Kubo M, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42(9):751–754. [DOI] [PubMed] [Google Scholar]
- 7. Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. [DOI] [PubMed] [Google Scholar]
- 8. Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39(5):638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han Y, Rand KA, Hazelett DJ, et al. Prostate cancer susceptibility in men of African Ancestry at 8q24. J Natl Cancer Inst. 2016;108(7):djv431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cook MB, Wang Z, Yeboah ED, et al. A genome-wide association study of prostate cancer in West African men. Hum Genet. 2014;133(5):509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann TJ, Van Den Eeden SK, Sakoda LC, et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015;5(8):878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patti ME, Sun XJ, Bruening JC, et al. 4PS/insulin receptor substrate (IRS)-2 is the alternative substrate of the insulin receptor in IRS-1-deficient mice. J Biol Chem. 1995;270(42):24670–24673. [DOI] [PubMed] [Google Scholar]
- 13. Southey MC, Goldgar DE, Winqvist R, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J Med Genet. 2016;53(12):800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103(38):14068–14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popejoy AB, Fullerton SM.. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.